Abstract
The innate immune system recognizes viral dsRNA through two distinct pathways; the Toll-like receptor 3 (TLR3) pathway detects dsRNA phagocytosed in endosomes; the helicases retinoic acid-induced protein I (RIG-I) and melanoma differentiation-associated gene-5 (mda-5) detect cytoplasmic dsRNA generated during viral replication. Both RIG-I and mda-5 can bind polyriboinosinic:polyribocytidylic acid (polyI:C), the synthetic analog of viral dsRNA, and mediate type I IFN responses to polyI:C and multiple RNA viruses in vitro. We generated mda-5-deficient mice and showed that mda-5 is the dominant receptor mediating type I IFN secretion in response to polyI:C in vitro and in vivo. Moreover, mda-5−/− mice exhibited a selectively impaired antiviral response to encephalomyocarditis picornavirus, indicating functional specialization of mda-5 in vivo.
Keywords: innate immunity, virus
To detect RNA viruses, the innate immune system must be able to sense conserved viral components (1). During infection of host cells, these viruses generate RNA–RNA strand pairs in the process of RNA-dependent RNA synthesis. Some DNA viruses also produce dsRNA during their life cycle. Thus, dsRNA can function as a pathogen-associated molecular pattern (PAMP) signaling viral infection (1). Indeed, this PAMP is recognized by the innate immune system, eliciting a prompt antiviral response. The synthetic analog of viral dsRNA, polyriboinosinic:polyribocytidylic acid (polyI:C), triggers the innate immune system to secrete the antiviral cytokines IFN-β and IFN-α as well as cytokines that induce an inflammatory response.
The innate immune system has developed two pathways for the recognition of dsRNA (2). One pathway is mediated by Toll-like receptor 3 (TLR3) (3). Because of its endosomal location (4), TLR3 allows cells to detect dsRNA that is phagocytosed from the extracellular space where it is released by virally infected cells that undergo lysis or necrosis (5). TLR3 may also allow detection of dsRNA viruses that are internalized from the extracellular space through receptor-mediated endocytosis. TLR3 signals through the TIR domain-containing adaptor TRIF (6, 7), which activates TANK-binding kinase-1 through TRAF3 (8, 9) and the inducible I-κB kinase IKK-ε. These kinases phosphorylate and activate IFN regulatory factors 3 and 7 (10, 11), which mediate transcriptional activation of IFN-β and IFN-α genes as well as IFN-inducible genes (12, 13). TRIF also triggers a signaling cascade that activates NF-κB and the transcription of proinflammatory cytokine genes (2). The TLR3 pathway has been implicated in the host responses to respiratory syncytial virus and influenza A virus infections in vitro (14, 15) and West Nile virus and murine cytomegalovirus (MCMV) infections in vivo (7, 16). However, additional in vivo studies with lymphocytic choriomeningitis virus, reovirus, and MCMV have indicated that antiviral responses are also mediated by pathways independent of TLR3 (17, 18).
A second pathway for detection of dsRNA is mediated by cytosolic sensors of dsRNA, which allow all cells to directly detect intracellular viral infection (2, 19). The prototypic cytosolic sensor is the dsRNA-dependent protein kinase (PKR). PKR is a serine–threonine kinase that binds dsRNA in its N-terminal regulatory region and induces phosphorylation of the α subunit of the eukaryotic protein synthesis initiation factor 2 (eIF2α), blocking cellular protein synthesis (20). PKR-deficient mouse embryonic fibroblasts have defective type I IFN responses to polyI:C and some RNA viruses, such as the encephalomyocarditis virus (EMCV) (21). However, this defect is completely corrected by pretreatment of cells with type I IFN, suggesting the existence of type I IFN-inducible mechanisms for the recognition of dsRNA. Moreover, PKR is not essential for in vivo responses to RNA viruses (21, 22). Recently, the IFN-inducible helicase retinoic acid-induced protein I (RIG-I) has been shown to bind polyI:C and mediate type I IFN responses to polyI:C in transfected cells (23, 24). RIG-I contains a DExD/H box RNA helicase domain, which unwinds dsRNA through its ATPase activity, and a caspase recruitment domain (CARD). Overexpression of RIG-I in transfected cells enhanced cellular secretion of type I IFN in response to Newcastle disease virus (NDV), vesicular stomatitis virus (VSV), and EMCV. RIG-I-deficient primary cells revealed reduced type I IFN responses to NDV, VSV, Sendai virus, hepatitis C virus, and West Nile virus infections (25–28). Because RIG-I−/− mice are embryonic lethal or die within a few weeks of birth (25), the function of RIG-I in antiviral responses in vivo is unclear.
The melanoma differentiation-associated gene-5 (mda-5), also known as Helicard, is another cytoplasmic sensor of dsRNA that contains a helicase domain and a CARD (29–32). mda-5 is ubiquitously expressed in low abundance and is induced by IFN-β and TNF-α. In transfected cells, mda-5 triggers type I IFN responses to NDV; inhibits EMCV, vesicular stomatitis virus, and NDV replication; and induces type I IFN-mediated inhibition of tumor cell growth (24, 30, 31). An essential role for mda-5 in antiviral responses is suggested by the existence of paramyxovirus proteins that antagonize mda-5 function, most likely to neutralize host responses (24, 30). Additionally, it has been shown that mda-5 is cleaved by apoptotic cells, and the processed protein significantly sensitizes cells to DNA degradation (32). Nonetheless, the function of mda-5 in antiviral responses in primary cells and in vivo remains uncertain.
Through the CARD, both RIG-I and mda-5 recruit an adaptor protein containing an N-terminal CARD, designated IPS-1, MAVS, VISA, or Cardif (24, 33–36). This molecule is present in the outer mitochondrial membrane and mediates sequential recruitment and activation of TANK-binding kinase-1, inducible I-κB kinase (IKK-ε), and IFN regulatory factor 3, ultimately leading to type I IFN secretion. The RIG-I/mda-5/IPS-1 pathway is targeted by viral and endogenous inhibitors. As a means of immune evasion, hepatitis C virus targets IPS-1 through the protease NS3-4a and attenuates type I IFN responses (24, 26, 36–38). The endogenous protein LGP2, which contains a helicase domain but lacks a CARD, has been proposed as a negative regulator of RIG-I and mda-5 (24, 39).
Because both mda-5 and RIG-I can detect dsRNA in the cytosol, induce type I IFN responses through the same signaling pathway, and are targeted by common inhibitors (24), it is unclear whether RIG-I and mda-5 serve redundant functions or specialize in the recognition of different viruses. To address the role of mda-5 in vivo, particularly in mediating type I IFN responses to dsRNA and viruses, we generated mda-5−/− mice. Challenge of mda-5−/− dendritic cells (DC) and macrophages in vitro and mda-5−/− mice in vivo with polyI:C demonstrated that mda-5 is the main cytosolic receptor for polyI:C. mda-5−/− DC and macrophages exhibited a selective impairment of type I IFN and proinflammatory cytokine secretion in response to the picornavirus EMCV, whereas responses to other RNA viruses were slightly impaired, if at all. Moreover, mda-5−/− mice succumbed earlier than WT mice after EMCV infection in vivo. These results reveal a unique role of mda-5 in the recognition of polyI:C and an unexpected viral specificity within the cytosolic sensors of dsRNA.
Results
Normal Development of mda-5−/− Mice.
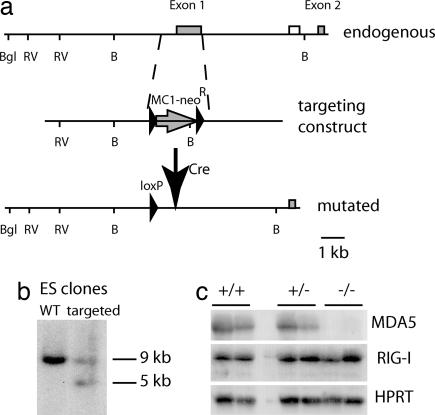
mda-5−/− mice were born at the expected Mendelian frequency and survived normally until at least 6–8 weeks of age. The viability of mda-5−/− mice is in striking contrast with that of RIG-I−/− mice, most of which were embryonic lethal or died within a few weeks of birth (25). RNA blot (Fig. 1) and RT-PCR (data not shown) analyses revealed that abrogation of mda-5 transcript in mda-5−/− mice did not affect expression of RIG-I transcript. Flow cytometric analysis of mda-5−/− mice did not reveal major differences in the lymphoid cell populations (B, CD4+ T, CD8+ T, γδ T, natural killer cells, DC, and plasmacytoid DC) in the spleen, thymus, peripheral blood, and lymph nodes or in bone marrow precursor populations (data not shown). Therefore, in contrast to RIG-I, mda-5 appears to be dispensable for development, including that of the immune system.
Fig. 1.
Generation of mda-5−/−mice. (a) Map of the endogenous mda-5 allele, targeting construct, and the mutated mda-5 allele after the removal of MC1-neoR by Cre recombination. mda-5 exons are shown as gray boxes, the external probe as an open box, and the MC1-neoR construct as an arrow. LoxP sites are designated by filled arrowheads. Restriction enzyme sites: B, BamHI; Bgl, BglII; and RV, EcoRV. (b) DNA blot analysis of ES cell DNA. Genomic DNA was cut with BamHI, and the blot was hybridized with the probe indicated in a. (c) RNA blot analysis of total liver and kidney RNA of WT, heterozygous, and mda-5−/− littermates. Mice were injected with polyI:C 3 days before collection of organs. For each mouse, the left lane represents liver, and the right lane represents kidney. mda-5 (Top), RIG-I (Middle), and hypoxanthine-guanine phosphoribosyl transferase (HPRT) (Bottom) blots are shown.
PolyI:C-Induced Secretion of Type I IFN by Bone Marrow-Derived DC and Macrophages Is Controlled by mda-5.
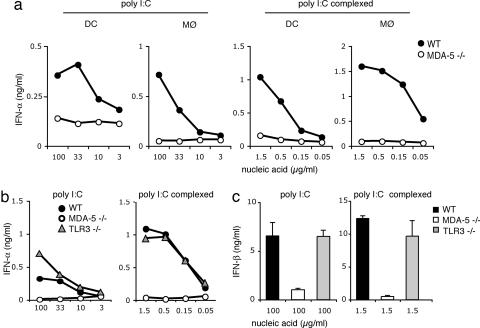
Previous studies have shown that mda-5 mediates type I IFN responses to polyI:C in transfected cells (24, 30). However, because primary cells express additional sensors of polyI:C, such as TLR3 and RIG-I, it remains unknown whether mda-5 is essential for polyI:C recognition or is redundant. To address the contribution of mda-5 to polyI:C responses in primary immune cells, DC and macrophages were generated from bone marrow cells of mda-5−/− and WT mice and were stimulated with polyI:C to induce type I IFN secretion. We used high concentrations of naked polyI:C (100–3 μg/ml) as well as low concentrations of polyI:C (1.5–0.05 μg/ml) complexed with a transfection reagent that optimizes polyI:C effects by facilitating intracellular delivery. Strikingly, secretion of IFN-α in response to polyI:C was completely abrogated in mda-5−/− cells as compared with WT cells (Fig. 2a). mda-5 deficiency also abrogated type I IFN responses to dsRNA generated by in vitro transcription (Fig. 7, which is published as supporting information on the PNAS web site). Because a prior study demonstrated that dsRNA induces up-regulation of costimulatory molecules through TLR3-dependent and independent mechanisms (40), we investigated the contribution of mda-5 to this up-regulation. Interestingly, mda-5−/− and WT bone marrow-derived DC exhibited similar cell surface expression profiles of B7.1 (CD80), B7.2 (CD86), and CD40 (data not shown).
Fig. 2.
mda-5 is critically required for the type I IFN response of bone marrow-derived DC and macrophages to polyI:C. DC and macrophages derived from WT (filled symbols), mda-5-deficient (open symbols, a–c), and TLR3-deficient mice (gray symbols, b and c) were stimulated in vitro, as indicated, with synthetic polyI:C or with polyI:C in complex with Fugene. Cell culture supernatants were assessed after 24 h of stimulation by ELISA for IFN-α (a and b) and IFN-β (c). Data shown are representative of four (a) and two (b and c) independent experiments.
To compare the relative contributions of mda-5 and TLR3/TRIF to polyI:C-induced secretion of type I IFN, we stimulated mda-5−/− and TLR3−/− bone marrow-derived DC with polyI:C and measured the levels of IFN-α and IFN-β by ELISA. mda-5 deficiency abrogated secretion of IFN-α and IFN-β, whereas TLR3 deficiency had little or no effect (Fig. 2 b and c). Similar results were obtained with bone marrow-derived macrophages (data not shown). We also compared the contributions of mda-5 and TLR3 to polyI:C-induced secretion of proinflammatory cytokines and chemokines in DC. We observed only a limited reduction of IL-6 secretion in mda-5−/− DC (Fig. 8, which is published as supporting information on the PNAS web site). Our data suggest that mda-5 is absolutely required for type I IFN responses of bone marrow-derived DC and macrophages to polyI:C in vitro, whereas it has a modest effect on proinflammatory cytokines and no effect on cell surface expression of costimulatory molecules. We conclude that, at least in bone marrow-derived DC and macrophages, mda-5 is functionally dominant over TLR3 for type I IFN responses to polyI:C.
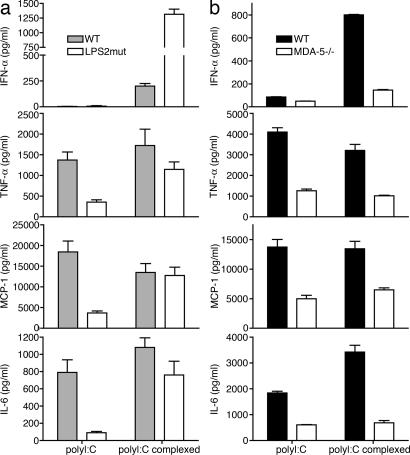
We also analyzed the response of thioglycollate-induced peritoneal macrophages to polyI:C administered either in solution or transfected as a complex with cationic lipid. To address the contributions of TLR3-TRIF and mda-5 to polyI:C-induced cytokine responses, we compared peritoneal macrophages from WT and TrifLPS2 mice (7), which carry a single base-pair deletion in the TRIF gene that causes instability or inactivation of the mutant protein. In contrast to bone marrow-derived DC and macrophages, thioglycollate-induced macrophages produced low amounts of IFN-α when stimulated with naked polyI:C. However, we observed considerable induction of inflammatory cytokines that was severely impaired in TrifLPS2 cells (5- to 10-fold reduction), indicating a significant contribution of the TLR3-TRIF pathway to polyI:C recognition in these cells (Fig. 3a). PolyI:C delivered with a transfection reagent induced higher amounts of IFN-α, which were not affected by the TrifLPS2 mutation; in fact, TRIF-deficient macrophages secreted higher amounts of IFN-α than WT controls in several of these assays. Other inflammatory cytokines were also independent of TRIF signaling when polyI:C was delivered in this manner.
Fig. 3.
Distinct and complementary contributions of mda-5 and TLR3-TRIF to polyI:C-induced responses in thioglycollate-induced peritoneal macrophages. Macrophages (2 × 105 per well) from WT and TRIFLPS2 mice (a) or WT and mda-5−/− mice (b) were collected 48 h after i.p. thioglycollate injection and stimulated with polyI:C either naked (100 μg/ml) or in complex with Fugene (1.5 μg/ml). After 24 h, cell culture supernatants were assessed for IFN-α by ELISA and for TNF-α, MCP-1, and IL-6 by cytokine bead array. Data shown are representative of two independent experiments.
In contrast, mda-5−/− peritoneal macrophages responded poorly to both naked and transfected polyI:C; in comparison with WT controls, cytokine secretion was reduced 2- to 3-fold in both cases, and IFN-α secretion strikingly diminished in response to transfected polyI:C (Fig. 3b). This result suggests that the endosomal and cytoplasmic pathways may directly or indirectly, through type I IFN secretion, synergize in this particular case. Together, these data imply that, depending on the specific cell type analyzed, mda-5 may either exercise a dominant role or cooperate with the TLR system for antiviral responses to dsRNA.
mda-5 Deficiency Attenuates PolyI:C-Induced Type I IFN Secretion in Vivo.
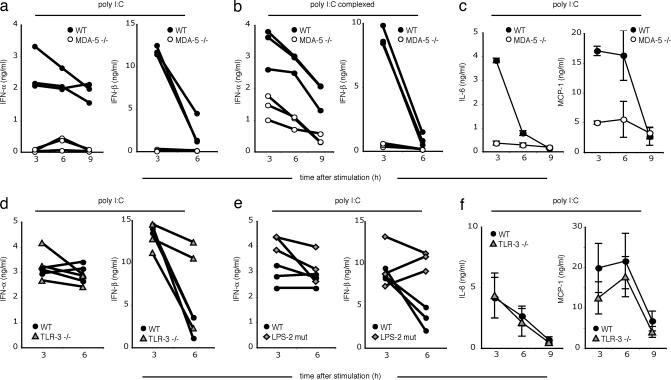
To determine whether mda-5 plays also a major role in responses to polyI:C in vivo, mda-5−/− and WT mice were i.v.-injected with polyI:C, either naked or in complex with a transfection reagent. Serum concentrations of IFN-α, IFN-β, and proinflammatory cytokines and chemokines were measured at different time points after polyI:C injection. IFN-α and IFN-β responses to naked polyI:C were abrogated in mda-5−/− mice (Fig. 4a). When polyI:C was complexed with transfection reagent, the IFN-β response was abrogated, but IFN-α was partially mda-5-independent (Fig. 4b). Additionally, a deficiency of mda-5 significantly decreased serum levels of the proinflammatory cytokines IL-6 and monocyte chemoattractant protein-1 (MCP-1) (Fig. 4c).
Fig. 4.
mda-5 deficiency strongly impairs the type I IFN response to polyI:C in vivo. WT, mda-5-deficient, TLR3-deficient, or TRIFLPS-2 (LPS-2 mutant) mice were injected i.v., as indicated, with either 100 μg of polyI:C or 10 μg of polyI:C complexed with Fugene. Serum samples were taken 3, 6, and 9 h after stimulation and were analyzed by ELISA for IFN-α and IFN-β (a, b, d, and e). Lines show cytokine kinetics in individual mice. IL-6 and MCP-1 were assessed by cytokine bead array (c and f). Error bars indicate SEM.
To address the relative contribution of TLR3-TRIF and mda-5 to polyI:C-induced cytokine responses in vivo, we i.v. injected polyI:C in TLR3-deficent and TrifLPS2 mice. In both cases, we observed no significant reduction of serum levels of IFN-α, IFN-β, IL-6, or MCP-1 in response to polyI:C, either naked (Fig. 4 d–f) or in association with a transfection reagent (data not shown). These results demonstrate that mda-5 is functionally dominant over TLR3 for type I IFN responses to polyI:C in vivo.
mda-5 Is Required for Recognition of EMCV in Vitro and Host Responses in Vivo.
The essential role of mda-5 in response to polyI:C in primary cells and in vivo suggested that mda-5 may be essential for responses to specific RNA viruses that produce dsRNA intermediates during their life cycles. However, dsRNA produced by RNA viruses could be alternatively and dominantly sensed by RIG-I. To address the question of whether mda-5 and RIG-I are redundant or specialize in the recognition of different viruses, we assessed type I IFN secretion of mda-5−/− and WT bone marrow-derived DC and macrophages after in vitro stimulation with an array of different RNA and DNA viruses, including West Nile virus, reovirus, Sindbis virus, influenza A, MCMV, herpes simplex virus-1, and EMCV. The viral panel also included a recombinant influenza A virus expressing an RNA-binding-defective NS1 protein (R38A NS1) (22, 41). NS1 binds dsRNA and sequesters it from intracellular sensors, thereby promoting viral evasion from host responses (42). Thus, the recombinant influenza A virus expressing R38A NS1 protein is attenuated and induces high levels of type I IFN (41).
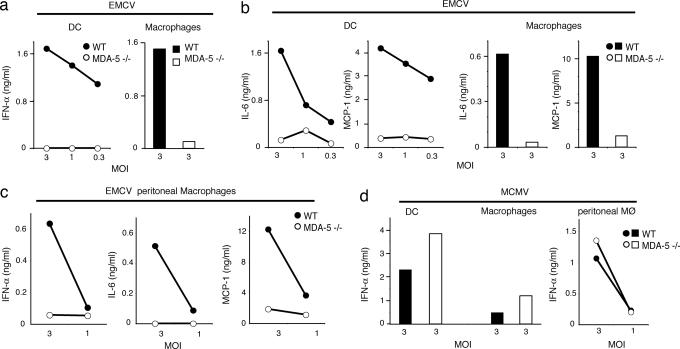
Remarkably, mda-5 deficiency completely abrogated DC and macrophage secretion of IFN-α only in response to EMCV (Fig. 5a). mda-5 deficiency also compromised IL-6 and MCP-1 secretion in response to EMCV (Fig. 5b). These results were confirmed in thioglycollate-induced peritoneal macrophages (Fig. 5c). In contrast, we observed only limited (≈2-fold) reduction of IFN-α secretion in response to recombinant influenza A with the R38A NS1 mutation, West Nile virus, Sindbis virus, and herpes simplex virus-1 (data not shown) and no reduction at all with MCMV (Fig. 5d) and reovirus (data not shown). These results demonstrate a remarkable specificity of mda-5 for the detection of EMCV, although we cannot exclude a contribution of mda-5 to the recognition of dsRNA from other viruses.
Fig. 5.
mda-5 is critically required for type I IFN and cytokine response to EMCV. Bone marrow-derived DC (a, b, and d), bone marrow-derived macrophages (a, b, and d), and thioglycollate-induced peritoneal macrophages (c and d) from WT (filled symbols) and mda-5-deficient (open symbols) mice were stimulated in vitro with EMCV or MCMV at indicated multiplicities of infection. Production of IFN-α (a and c) and IL-6 and MCP-1 (b and c) in response to EMCV. (d) IFN-α response to MCMV. Cell culture supernatants were examined for IFN-α by ELISA and for IL-6 and MCP-1 by cytokine bead array. Data shown are representative of two independent experiments.
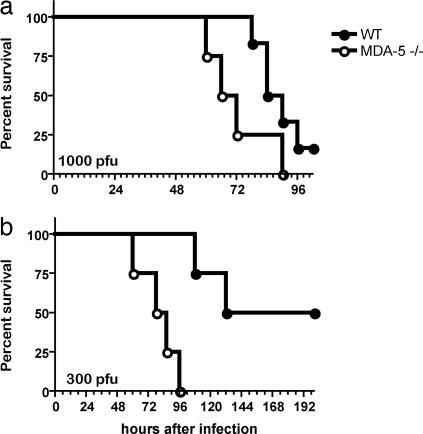
To assess the involvement of mda-5 in the control of EMCV infection in vivo, mda-5−/− and WT littermate control mice were injected i.v. with a lethal dose of EMCV and monitored for survival. After i.v. injection, EMCV infects the central nervous system, causing lethal encephalitis within 72–96 h (43). mda-5−/− developed symptoms of hind-limb paralysis ≈24–72 h earlier than WT mice and succumbed sooner to the infection (Fig. 6), consistent with an essential role of mda-5 in host resistance to EMCV infection in vivo.
Fig. 6.
mda-5-deficient mice show increased susceptibility to lethal infections with EMCV. (a) mda-5−/− mice (n = 4, open symbols) and WT littermate controls (n = 6, filled symbols) on a pure 129 SvJ background were injected i.v. with 1,000 plaque-forming units (pfu) of EMCV and then monitored for survival. mda-5−/− mice died after a mean survival time of 69 h, whereas WT mice survived for a mean of 87 h (P = 0.07). (b) mda-5−/− mice (n = 5, open symbols) and WT littermate controls (n = 5, filled symbols) on a B6x129 SvJ background were injected i.v. with 300 pfu of EMCV and monitored for survival. mda-5−/− mice died after a mean survival time of 82 h, whereas WT mice survived for a mean of 166 h (P = 0.008).
Discussion
Our study demonstrates that mda-5 is remarkably important for type I IFN responses to polyI:C by DC and macrophages in vitro and in mice in vivo. This result is surprising, given previous demonstrations that both the TLR3/TRIF pathway and RIG-I can mediate type IFN responses to polyI:C. Why does mda-5 predominate over TLR3/TRIF in the type I IFN response to polyI:C? One important difference between mda-5 and TLR3 is that mda-5 is located in the cytosol, whereas TLR3 is found in endosomal compartments (4). Thus, naked polyI:C or polyI:C in complex with a transfection reagent may predominantly reach the cytosol rather than endosomes, resulting in preferential activation of mda-5. Cytosolic entry of polyI:C may be facilitated by a transmembrane transporter or channel that may be mimicked by the transfection reagent used in our experiments.
TLR3/TRIF may play a relevant role in vivo when polyI:C or viral dsRNA reach appropriate endosomal compartments. This event could occur when cells phagocytose dsRNA released into the extracellular space by virally infected cells undergoing lysis or necrosis or when cells internalize viruses through receptor-mediated endocytosis. Consistent with this, phagocytosis of virus-infected cells or cells containing synthetic dsRNA enhances antigen presentation via stimulation of TLR3 in antigen-presenting cells (5). In our study, mda-5 was essential for bone marrow-derived DC and macrophage type I IFN responses to polyI:C. In contrast, TLR3-TRIF induced responses to polyI:C in peritoneal macrophages, consistent with previous studies (3, 6, 7, 44). In vivo, in the spleen, TLR3 is found primarily in CD8+ DC but not in marginal-zone DC (45). Because the marginal zone of the spleen is the first to encounter the contents of the blood, it may be a major target of polyI:C sensing after i.v. administration. Thus, the dominant role of mda-5 over TLR3 in systemic responses to polyI:C could reflect the preferential involvement of tissues that express relatively low levels of TLR3.
mda-5 appeared to predominate over RIG-I in type I IFN responses to polyI:C. One possible explanation is that responses to polyI:C require the concerted activation of RIG-I and mda-5. In this case, mda-5 deficiency would inhibit this signaling cascade, preempting RIG-I function. However, it is also possible that mda-5 has a higher affinity for polyI:C than RIG-I or recognizes a specific feature of this particular dsRNA mimic. Therefore, even if RIG-I can bind polyI:C and trigger IFN-α secretion in transfected cells in vitro (23), it may not be required, and mda-5 in fact may be the dominant cytosolic receptor for polyI:C in vitro and in vivo.
Our observation that mda-5 is selectively required for cytokine responses to EMCV but not other viruses supports a model in which mda-5 and RIG-I have different specificities for dsRNA molecules. mda-5 deficiency completely abolished type I IFN and cytokine responses to EMCV in DC and macrophages in vitro, and mda-5−/− mice were highly susceptible to EMCV infection. In contrast, cytokine responses to other viruses were only marginally affected, if at all. Because the amino acid sequences of the helicase domains of mda-5 and RIG-I are only ≈35% identical, it is possible that this genetic diversity results in different specificity for distinct dsRNA conformations, resulting in preferential recognition of different viruses. EMCV and EMC-like viruses share structural characteristics in their dsRNA, such as the presence of a homopolymeric polyC acid tract within the 5′ untranslated sequence (46). These polyC tracts are retained during viral replication in vitro and in vivo and are associated with virulence. Thus, it will be important to determine whether mda-5 is required for the recognition of other ECM-like viruses and polyC-less EMCV. In addition, it will be crucial to assess the mda-5 contribution to innate responses to other members of the picornavirus family, which includes important human and agricultural pathogens (43).
The lack of a dramatic effect of mda-5 deficiency on type I IFN secretion by other viruses we tested could be due to the presence of RIG-I or additional yet-uncharacterized sensors that contribute to type I IFN secretion. Additionally, some of these viruses could encode novel immune evasion molecules that specifically antagonize mda-5 function. Indeed, recent studies have shown that human and murine paramyxoviruses encode such a protein, which inhibits mda-5 function and interferes with type I IFN responses (24, 30). A more detailed study of the RNA viruses that are not affected by mda-5 deficiency may identify additional virally encoded mda-5 inhibitors. Such investigation will help elucidate the pathogenesis of these viruses and facilitate the development of specific antagonists that enhance innate immunity against these viruses and possibly improve clinical outcome.
Materials and Methods
Generation of mda-5−/− Mice.
The targeting construct was designed to replace the first exon and nucleotides ≈400 bp upstream of mda-5 with an MC1-neor expression cassette (Fig. 1). Correctly targeted ES cells (129x1/SvJ) were injected into C57BL/6 (B6) blastocysts. Chimeras were bred to B6 transgenic mice expressing Cre recombinase under the cytomegalovirus promoter (47) to delete the MC1-neor cassette. The resulting mda-5−/+ heterozygotes were intercrossed to obtain mda-5−/− mice. Chimeras were also bred to 129x1/SvJ mice and mda-5−/+ heterozygotes to obtain mda-5−/− mice on a pure 129x1/SvJ background.
Mice.
TLR3−/− (3) and TRIFLPS (7) mice were on a C57BL/6 background. Age-matched WT control C57BL/6 mice were obtained from Taconic Farms.
Cell Cultures and Stimulation in Vitro.
Mouse bone marrow-derived DC and macrophages were generated as described (22). To elicit primary macrophages in mice, 1.5 ml of 2% thioglycollate media was injected i.p., and cells were isolated by peritoneal lavage. All cells were stimulated for 18–24 h, as indicated in the figure legends. Synthetic polyI:C (Amersham Pharmacia) was used as such or complexed with Fugene (3 μl/μg nucleic acid) (Roche Applied Science, Indianapolis). dsRNA was generated by in vitro transcription of a 500-bp template within the firefly luciferase ORF by using the MEGAscript RNA transcription kit (Ambion, Austin, TX). EMCV (EMCV-k) was obtained from R. Silverman (Cleveland Clinic, Cleveland) and passaged in L929 cells.
Stimulations and Infections in Vivo.
Mice were injected with 100 μg of polyI:C or 10 μg of polyI:C complexed with 30 μl of Fugene. For in vivo infections with EMCV, 1,000 or 300 plaque-forming units were injected i.v. in mda-5−/− mice on either a mixed or 129x1/SvJ background with appropriate controls.
Cytokine Analysis and Flow Cytometry.
IFN-α and IFN-β were detected in cell-free supernatants and mice sera by ELISA (PBL Biomedical Laboratories, New Brunswick, NJ). TNF-α, MCP-1, and IL-6 were measured by cytometric bead array (BD Biosciences).
Supplementary Material
Acknowledgments
We thank Julia Klesney-Tait, Isaiah Turnbull, and Rachel Presti for helpful critiques; and Andrew Pekosz, Skip Virgin, David Leib, and Wayne Yokoyama (Washington University School of Medicine, St. Louis); Robert Silverman (Cleveland State University, Cleveland); and Terence Dermody (Vanderbilt University School of Medicine, Nashville, TN) for viruses. L.G. is a postdoctoral fellow of the Cancer Research Institute, and B.B. is supported by National Institutes of Health Grant GM060031.
Abbreviations
- polyI:C
polyriboinosinic:polyribocytidylic acid
- TLR3
Toll-like receptor 3
- MCMV
murine cytomegalovirus
- EMCV
encephalomyocarditis virus
- CARD
caspase recruitment domain
- DC
dendritic cell.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Janeway C. A., Jr, Medzhitov R. Annu. Rev. Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 2.Akira S., Uematsu S., Takeuchi O. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 3.Alexopoulou L., Holt A. C., Medzhitov R., Flavell R. A. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 4.Matsumoto M., Funami K., Tanabe M., Oshiumi H., Shingai M., Seto Y., Yamamoto A., Seya T. J. Immunol. 2003;171:3154–3162. doi: 10.4049/jimmunol.171.6.3154. [DOI] [PubMed] [Google Scholar]
- 5.Schulz O., Diebold S. S., Chen M., Naslund T. I., Nolte M. A., Alexopoulou L., Azuma Y. T., Flavell R. A., Liljestrom P., Reis e Sousa C. Nature. 2005;433:887–892. doi: 10.1038/nature03326. [DOI] [PubMed] [Google Scholar]
- 6.Yamamoto M., Sato S., Hemmi H., Hoshino K., Kaisho T., Sanjo H., Takeuchi O., Sugiyama M., Okabe M., Takeda K., Akira S. Science. 2003;301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 7.Hoebe K., Du X., Georgel P., Janssen E., Tabeta K., Kim S. O., Goode J., Lin P., Mann N., Mudd S., et al. Nature. 2003;424:743–748. doi: 10.1038/nature01889. [DOI] [PubMed] [Google Scholar]
- 8.Hacker H., Redecke V., Blagoev B., Kratchmarova I., Hsu L. C., Wang G. G., Kamps M. P., Raz E., Wagner H., Hacker G., et al. Nature. 2006;439:204–207. doi: 10.1038/nature04369. [DOI] [PubMed] [Google Scholar]
- 9.Oganesyan G., Saha S. K., Guo B., He J. Q., Shahangian A., Zarnegar B., Perry A., Cheng G. Nature. 2006;439:208–211. doi: 10.1038/nature04374. [DOI] [PubMed] [Google Scholar]
- 10.Fitzgerald K. A., McWhirter S. M., Faia K. L., Rowe D. C., Latz E., Golenbock D. T., Coyle A. J., Liao S. M., Maniatis T. Nat. Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 11.Sharma S., tenOever B. R., Grandvaux N., Zhou G. P., Lin R., Hiscott J. Science. 2003;300:1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- 12.Honda K., Yanai H., Takaoka A., Taniguchi T. Int. Immunol. 2005;17:1367–1378. doi: 10.1093/intimm/dxh318. [DOI] [PubMed] [Google Scholar]
- 13.Sato M., Suemori H., Hata N., Asagiri M., Ogasawara K., Nakao K., Nakaya T., Katsuki M., Noguchi S., Tanaka N., Taniguchi T. Immunity. 2000;13:539–548. doi: 10.1016/s1074-7613(00)00053-4. [DOI] [PubMed] [Google Scholar]
- 14.Rudd B. D., Burstein E., Duckett C. S., Li X., Lukacs N. W. J. Virol. 2005;79:3350–3357. doi: 10.1128/JVI.79.6.3350-3357.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guillot L., Le Goffic R., Bloch S., Escriou N., Akira S., Chignard M., Si-Tahar M. J. Biol. Chem. 2005;280:5571–5580. doi: 10.1074/jbc.M410592200. [DOI] [PubMed] [Google Scholar]
- 16.Wang T., Town T., Alexopoulou L., Anderson J. F., Fikrig E., Flavell R. A. Nat. Med. 2004;10:1366–1373. doi: 10.1038/nm1140. [DOI] [PubMed] [Google Scholar]
- 17.Edelmann K. H., Richardson-Burns S., Alexopoulou L., Tyler K. L., Flavell R. A., Oldstone M. B. Virology. 2004;322:231–238. doi: 10.1016/j.virol.2004.01.033. [DOI] [PubMed] [Google Scholar]
- 18.Schroder M., Bowie A. G. Trends Immunol. 2005;26:462–468. doi: 10.1016/j.it.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 19.Lopez C. B., Moltedo B., Alexopoulou L., Bonifaz L., Flavell R. A., Moran T. M. J. Immunol. 2004;173:6882–6889. doi: 10.4049/jimmunol.173.11.6882. [DOI] [PubMed] [Google Scholar]
- 20.Saunders L. R., Barber G. N. FASEB J. 2003;17:961–983. doi: 10.1096/fj.02-0958rev. [DOI] [PubMed] [Google Scholar]
- 21.Yang Y. L., Reis L. F., Pavlovic J., Aguzzi A., Schafer R., Kumar A., Williams B. R., Aguet M., Weissmann C. EMBO J. 1995;14:6095–6106. doi: 10.1002/j.1460-2075.1995.tb00300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barchet W., Krug A., Cella M., Newby C., Fischer J. A., Dzionek A., Pekosz A., Colonna M. Eur. J. Immunol. 2005;35:236–242. doi: 10.1002/eji.200425583. [DOI] [PubMed] [Google Scholar]
- 23.Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M., Taira K., Akira S., Fujita T. Nat. Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 24.Yoneyama M., Kikuchi M., Matsumoto K., Imaizumi T., Miyagishi M., Taira K., Foy E., Loo Y. M., Gale M., Jr, Akira S., et al. J. Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 25.Kato H., Sato S., Yoneyama M., Yamamoto M., Uematsu S., Matsui K., Tsujimura T., Takeda K., Fujita T., Takeuchi O., Akira S. Immunity. 2005;23:19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 26.Foy E., Li K., Sumpter R., Jr, Loo Y. M., Johnson C. L., Wang C., Fish P. M., Yoneyama M., Fujita T., Lemon S. M., Gale M., Jr Proc. Natl. Acad. Sci. USA. 2005;102:2986–2991. doi: 10.1073/pnas.0408707102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sumpter R., Jr, Loo Y. M., Foy E., Li K., Yoneyama M., Fujita T., Lemon S. M., Gale M., Jr J. Virol. 2005;79:2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fredericksen B. L., Gale M., Jr J. Virol. 2006;80:2913–2923. doi: 10.1128/JVI.80.6.2913-2923.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang D. C., Gopalkrishnan R. V., Lin L., Randolph A., Valerie K., Pestka S., Fisher P. B. Oncogene. 2004;23:1789–1800. doi: 10.1038/sj.onc.1207300. [DOI] [PubMed] [Google Scholar]
- 30.Andrejeva J., Childs K. S., Young D. F., Carlos T. S., Stock N., Goodbourn S., Randall R. E. Proc. Natl. Acad. Sci. USA. 2004;101:17264–17269. doi: 10.1073/pnas.0407639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kang D. C., Gopalkrishnan R. V., Wu Q., Jankowsky E., Pyle A. M., Fisher P. B. Proc. Natl. Acad. Sci. USA. 2002;99:637–642. doi: 10.1073/pnas.022637199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kovacsovics M., Martinon F., Micheau O., Bodmer J. L., Hofmann K., Tschopp J. Curr. Biol. 2002;12:838–843. doi: 10.1016/s0960-9822(02)00842-4. [DOI] [PubMed] [Google Scholar]
- 33.Kawai T., Takahashi K., Sato S., Coban C., Kumar H., Kato H., Ishii K. J., Takeuchi O., Akira S. Nat. Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 34.Seth R. B., Sun L., Ea C. K., Chen Z. J. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 35.Xu L. G., Wang Y. Y., Han K. J., Li L. Y., Zhai Z., Shu H. B. Mol. Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 36.Meylan E., Curran J., Hofmann K., Moradpour D., Binder M., Bartenschlager R., Tschopp J. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 37.Breiman A., Grandvaux N., Lin R., Ottone C., Akira S., Yoneyama M., Fujita T., Hiscott J., Meurs E. F. J. Virol. 2005;79:3969–3978. doi: 10.1128/JVI.79.7.3969-3978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li X. D., Sun L., Seth R. B., Pineda G., Chen Z. J. Proc. Natl. Acad. Sci. USA. 2005;102:17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rothenfusser S., Goutagny N., DiPerna G., Gong M., Monks B. G., Schoenemeyer A., Yamamoto M., Akira S., Fitzgerald K. A. J. Immunol. 2005;175:5260–5268. doi: 10.4049/jimmunol.175.8.5260. [DOI] [PubMed] [Google Scholar]
- 40.Hoebe K., Janssen E. M., Kim S. O., Alexopoulou L., Flavell R. A., Han J., Beutler B. Nat. Immunol. 2003;4:1223–1229. doi: 10.1038/ni1010. [DOI] [PubMed] [Google Scholar]
- 41.Donelan N. R., Basler C. F., Garcia-Sastre A. J. Virol. 2003;77:13257–13266. doi: 10.1128/JVI.77.24.13257-13266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang W., Riedel K., Lynch P., Chien C. Y., Montelione G. T., Krug R. M. RNA. 1999;5:195–205. doi: 10.1017/s1355838299981621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rueckert R. R. In: Fields Virology. Fields B. N., Knipe D. M., Howley P. M., editors. Vol. 1. Philadelphia: Lippincott–Raven; 1996. pp. 609–654. [Google Scholar]
- 44.Hemmi H., Takeuchi O., Sato S., Yamamoto M., Kaisho T., Sanjo H., Kawai T., Hoshino K., Takeda K., Akira S. J. Exp. Med. 2004;199:1641–1650. doi: 10.1084/jem.20040520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edwards A. D., Diebold S. S., Slack E. M., Tomizawa H., Hemmi H., Kaisho T., Akira S., Reis e Sousa C. Eur. J. Immunol. 2003;33:827–833. doi: 10.1002/eji.200323797. [DOI] [PubMed] [Google Scholar]
- 46.Osorio J. E., Martin L. R., Palmenberg A. C. Virology. 1996;223:344–350. doi: 10.1006/viro.1996.0485. [DOI] [PubMed] [Google Scholar]
- 47.Schwenk F., Baron U., Rajewsky K. Nucleic Acids Res. 1995;23:5080–5081. doi: 10.1093/nar/23.24.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.