Abstract
Defensins are members of a large diverse family of cationic antimicrobial peptides that share a signature pattern consisting of six conserved cysteine residues. Defensins have a wide variety of functions and their disruption has been implicated in various human diseases. Here we report the characterization of DEFB119–DEFB123, five genes in the human β-defensin cluster locus on chromosome 20q11.1. The genomic structures of DEFB121 and DEFB122 were determined in silico. Sequences of the five macaque orthologs were obtained and expression patterns of the genes were analyzed in humans and macaque by semiquantitative reverse transcription polymerase chain reaction. Expression was restricted to the male reproductive tract. The genes in this cluster are differentially regulated by androgens. Evolutionary analyses suggest that this cluster originated by a series of duplication events and by positive selection. The evolutionary forces driving the proliferation and diversification of these defensins may be related to reproductive specialization and/or the host–parasite coevolutionary process.
Keywords: β-defensins, epididymis, reproduction, immunity, molecular evolution
Introduction
The production of natural antibiotic peptides such as defensins has emerged as an important mechanism of innate immunity in animals and plants. Most defensins are two exon genes and this family of cationic peptides contains a pattern of six conserved cysteine residues. The α- and β-forms are distinguishable by the spacing and connectivity of the conserved cysteine residues within the mature peptide.1 In humans more than 30 β-defensins are organized in five syntenic chromosomal regions.2 Two of these functional human defensin gene clusters were previously investigated.3,4
Human β-defensins (DEFB) are expressed by epithelial cells lining tissues facing the external environment. They display antibacterial activity in vitro and are likely to be involved in innate immunity.5 DEFB1 is suggested to play an important role in innate immunity of the lung and is inactivated in cystic fibrosis.6 In contrast, over-expression of DEFB2 is associated with inflammatory skin lesions such as psoriasis.7 Further, it is suggested that defensins may contribute to HIV-1 resistance in the male and female genitourinary tracts.8 Not only are human β-defensins involved in innate immunity; they also display chemo-attractant activity for immature dendritic cells and memory T cells linking the innate and adaptive immune systems.9,10
β-Defensin involvement in reproduction is suggested by mounting evidence that the majority of β-defensins are most highly expressed in the functionally unique tissues of the male reproductive tract.3,4,11 In addition, the β-defensin-like epididymal protein (EP2)/sperm-associated antigen 11 (SPAG11) isoforms are also predominantly expressed in the male reproductive tract.12 Recent studies specifically implicate β-defensins in pivotal roles in reproductive biology including ESP13.2/DEFB126 in sperm capacitation13 and Bin1b/EP2E in sperm maturation,14 thereby redefining our concepts concerning the multifunctional roles of β-defensins.
In the epididymis, where numerous defensins are expressed, sperm maturation is promoted by a gradient of luminal environments created by region-specific and cell-specific patterns of gene expression.15 The underlying mechanisms regulating epididymal gene expression patterns are not well understood, but they are largely dependent on testicular androgens.16,17 Prior to exposure to epididymal gene products, spermatozoa are immotile and unable to bind or penetrate the ovum.18
Gene duplication and positive Darwinian selection are hallmarks of vertebrate host defense and in genes involved in reproduction.19,20 Evolution of immune system gene families is driven by gene duplication, interlocus recombination, and positive Darwinian selection favoring diversity at the amino-acid level.21 Evidence of such duplication and positive selection has been detected in mammalian β-defensins.4,22,23 Positive selection is pervasive among mammalian reproductive proteins, especially several sperm proteins that have been implicated in binding the egg promoting fertilization.24 The rapid evolution of reproductive proteins may be driven by the selective forces of sperm competition, sexual selection and sexual conflict, individually, or in combination.20
Previously, we identified the novel epididymal sperm-binding protein DEFB11825 and recently showed that DEFB118 has potent antibacterial activity.26 The potential importance of the male reproductive tract defensins in host defense as well as male reproductive functions prompted us to investigate the 20q β-defensin gene cluster. Herein we present the characterization and evolutionary analysis of human and macaque β-defensin orthologs, which give new insights into adaptive functional roles.
Results
To better characterize the 20q β-defensin gene cluster, we amplified their expressed sequences by polymerase chain reaction with reverse transcription (RT-PCR). Previously unknown splicing variants of DEFB119 (accession number AY501001), DEFB121 (AY501000), DEFB122p (AY500999), and DEFB123 (AY501002) were identified. The human DEFB122p appeared to be a pseudogene that resulted from a shift in the mRNA splice site. The signal peptide and defensin-like motif of DEFB122p are encoded in different reading frames and a stop codon close to the signal peptide truncates the protein.
Our results confirm the Unigene-defined two-exon structures of DEFB119, DEFB120, and DEFB123 and reveal previously unknown exon 1 sequences for DEFB121 and DEFB122p that are predicted to encode signal peptides closely related to the others in this cluster (Figure 1). The cDNAs of the functional macaque orthologs of the human DEFB119, DEFB120, DEFB121, DEFB122, and DEFB123 genes were amplified by RT-PCR and the sequences deposited in GenBank under sequence accession numbers AY499406, AY499407, AY499408, AY499409, and AY500998, respectively. Alignment of the human and macaque deduced amino-acid sequences reveals that all contain the conserved six-cysteine defensin signature motif (Figure 2). DEFB119 and DEFB120 represent alternatively spliced products of the same gene and thus share the common first exon and have different second exons spliced at the same splice junction.
Figure 1.
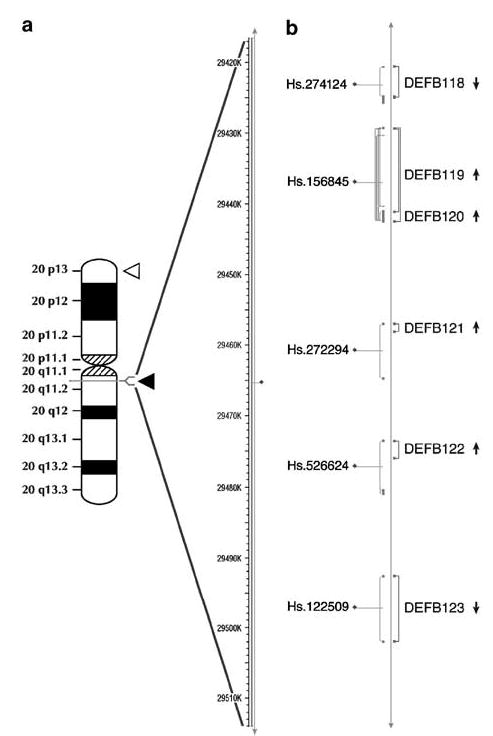
Human β-defensin genes located on chromosome 20. (a) An ideogram of chromosome 20 is given in the left. Filled triangle indicates the location of the β-defensin gene cluster at 20q11.1. The empty triangle indicates the location of another β-defensin cluster at 20p13. Arrangement of the β-defensin genes near the centromeric region and distributed over 80 kilobases. (b) The Unigene accession numbers and Unigene-defined exons are indicated on the left. The positions of exons that we determined are given on the right with the respective defensin gene names. Arrows indicate the direction of transcription of each gene. This figure is based on human genome Build 35.1 available at MapView (http://www.ncbi.nlm.nih.gov/mapview).
Figure 2.

Multiple sequence alignment of human and macaque β-defensin protein sequences. The amino acids shaded in gray represent highly conserved sequences and the six-cysteine motifs are shaded in black. The sequences shown are DEFB118 (AF347073), DEFB119 (AF479698), DEFB120 (AF479699), DEFB121 (AY501000), and DEFB123 (AY501002), and macaque DEFB118 (AF207834), DEFB119 (AY499406), DEFB120 (AY499407), DEFB121 (AY499408), DEFB122 (AY499409), and DEFB123 (AY500998). The amino-acid sequence of E. coioides (orange-spotted grouper) reproductive regulator 1(AY129305) is also aligned with the β-defensin cluster. The arrow indicates the signal peptide region deduced using SignalP.40
The defensins in the 20q cluster lack a long propiece, except for DEFB118 that contains the propiece in an unusual location, after the six-cysteine motif (defined in Swiss-Prot number Q96PH6). This propiece has a calculated isoelectric point (pI) of 4.73. The overall pI of DEFB118 mature protein is 6.46, though its six-cysteine array is cationic (pI 9.13). It is the only anionic β-defensin in this cluster, while the others are cationic.
Further, we conducted a systematic evaluation of relative steady-state mRNA levels in 16 human tissues. Interestingly, only tissues of the male reproductive tract showed expression of these functional β-defensin genes (Figure 3). DEFB118 was expressed only in the testis and epididymis. Expression was abundant in the proximal epididymis (caput and corpus), while expression was low in the distal region of the epididymis and testis. Similar trends in the expression were observed for DEFB120 and DEFB121. However, DEFB119 and DEFB123 expression was abundant in testis and the caput region of epididymis, but low in the corpus region. In addition to the fully spliced mature DEFB119 transcript, two alternatively spliced longer transcripts were detected as higher molecular weight bands in human epididymis and testis (Figure 3). We deposited the sequence of the middle transcript in GenBank as DEFB119 isoform b, accession number AY501001. The higher molecular weight band for DEFB123 in human testis also represents an alternatively spliced transcript and is deposited as DEFB123 isoform b under accession number AY501002. Surprisingly, the expression of DEFB122p transcript was found only in the testis and not in any region of the epididymis. In order to investigate the expression of DEFB118 to DEFB123 in the female reproductive tract, mRNA from human uterine endometrial samples from proliferative and mid-secretory stages and from the endometrial epithelial adenocarcinoma cell lines, Ishikawa and RL95-2, were analyzed (data not shown). The expression of DEFB118–DEFB123 was not detected in any of these endometrial tissues or cell lines.
Figure 3.
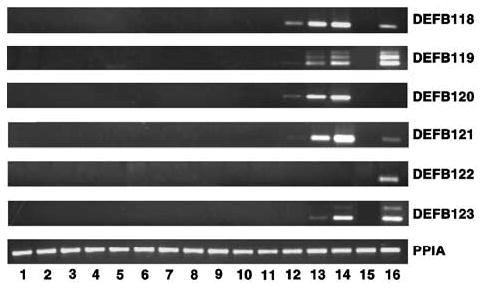
Expression of novel human β-defensin genes in different tissues. Total RNAs isolated from human (1) parotid glands, (2) thyroid, (3) breast, (4) lungs, (5) liver, (6) spleen, (7) kidney, (8) colon, (9) adrenal, (10) ovary, (11) seminal vesicle, (12) cauda, (13) corpus, (14) caput, (15) prostate, and (16) testis were used for RT-PCR analysis. PPIA was amplified as a control to check the quality and quantity of the template.
A similar analysis of expression of DEFB118–DEFB123 in macaque (Macaca mulatta) tissues revealed that DEFB118 was restricted to the caput and corpus regions of epididymis (Figure 4). DEFB119 and DEFB123 were expressed abundantly in testis, while expression in epididymis decreased gradually from caput to cauda. In contrast, the expression of DEFB120 and DEFB121 was lower in testis, while the caput and corpus showed relatively abundant expression. Interestingly, macaque DEFB122 message was abundant in the testis and corpus, while caput expressed relatively lower levels. Altogether, the functional β-defensin genes examined are predominantly expressed in the testis, caput, and corpus regions of the epididymis, both in humans and macaques. However, faint expression of DEFB119 was also observed in the adrenal and urinary bladder of macaques.
Figure 4.
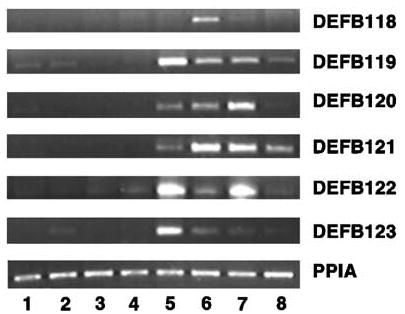
Expression patterns of novel macaque β-defensin genes in male tissues. Total RNAs isolated from macaque (1) adrenal, (2) bladder, (3) seminal vesicle, (4) prostate, (5) testis, (6) caput, (7) corpus, and (8) cauda were used for RT-PCR analysis. PPIA was amplified as a control to check the quality and quantity of the template.
Age-dependent regulation of the functional defensin genes was analyzed in the epididymis from macaques of different ages starting from day 1 to 3.9 years (Figure 5). Each defensin showed a unique pattern of developmental expression. DEFB118 and DEFB123 were specifically expressed in the pubertal and adult epididymis. Expression of DEFB119 was observed in the total epididymis of the new-born (1-day-old) macaque and pre-pubertal and adult stages, but not in 5.6-months-old and 2-years-old epididymis. DEFB120, DEFB121, and DEFB122 were expressed at all ages, but were most abundant in the adult. The higher molecular weight bands for DEFB122 appear not to represent DEFB122 transcripts.
Figure 5.
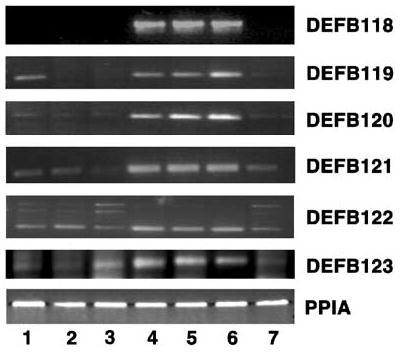
Developmental regulation of macaque β-defensin genes. Total RNA isolated from the macaque (1) pooled epididymides of 1-day-old, (2) pooled epididymides of 5.6-months-old, (3) pooled epididymides from 2-years-old, (4) pooled epididymides of 3.9-years-old, (5) caput, (6) corpus, and (7) cauda from 4.2-years-old macaque were used for RT-PCR analysis. PPIA was amplified as a control to check the quality and quantity of the template.
Regional expression of the functional defensin genes was androgen regulated in the epididymis. Levels of expression of all the defensin mRNAs in the caput region were downregulated by androgen withdrawal except DEFB121, while testosterone replacement maintained the mRNA levels comparable to the sham-operated caput (Figure 6). On the contrary, in the corpus region the expression of DEFB118–DEFB123 was little affected by castration. DEFB119 and DEFB123 were weakly expressed in the cauda region, and castration further decreased expression.
Figure 6.
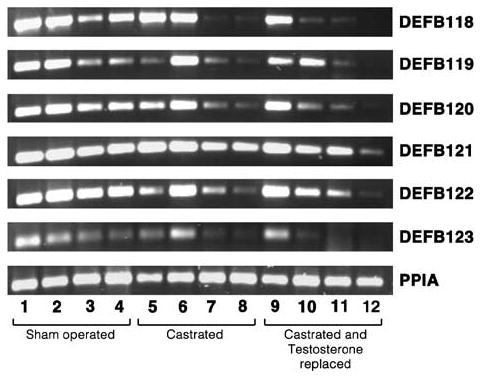
Androgen regulation of β-defensin genes. M. mulatta were sham operated, castrated, or castrated and androgen replaced with immediate injection of 400 mg of testosterone enanthate. Epididymides were removed 6 days post-castration. Total RNA isolated from each tissue was used for RT-PCR analysis of β-defensin genes. PPIA was amplified as a control to check the quality and quantity of the template. Lanes 1, 5, and 9: caput; lanes 2, 6, and 10: corpus; lanes 3, 7, and 11: proximal cauda; and lanes 4, 8, and 12: distal cauda.
The evolution of this multigene family was analyzed using the neighbor-joining (NJ) method27 with the evolutionary divergence based on the transversional differences in the third codon position (Figure 7). Most of these six paralogous genes in the 20q cluster appear to have diverged in rapid succession by tandem duplication of individual genes, rather than by duplication of gene clusters (Figure 7). The latter is excluded because closely located genes are also found to have diverged more recently. The same inference can be made whether we use the putative defensin, the Epinephelus coioides reproduction regulator 1 (rr1) sequence, to root this tree or use the mid-point rooting method. However, there is only modest bootstrap support for the interrelationship of the paralogous genes. The phylogeny inferred using the protein sequences (Figure 8) shows a topology similar to that obtained using the third codon positions (Figure 7), except for the location of the cluster containing h119 and m119. Again, the bootstrap test does not provide conclusive support for this phylogeny, but its congruence with the third codon position data provides additional support. The deepest splits in the 20q cluster can be inferred to be at least as old as the fish-primate divergence about 450 million years ago28 because fish-primate sequence divergence is smaller than many other divergences in this defensin phylogeny. Many of the duplication events that led to the generation of the 20q cluster may have predated even the origin of vertebrates. Separate analysis of the two exons shows that the exon 1 protein sequence has undergone a smaller amount of change (thus evolved slowly) compared to exon 2 (data not shown).
Figure 7.
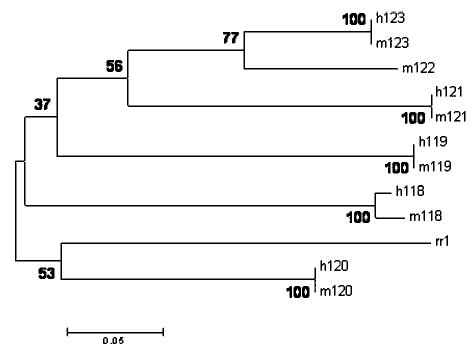
Phylogenetic tree of functional human and macaque β-defensin cDNA sequences. The NJ tree was rooted with a putative defensin of E. coioides reproductive regulator 1 sequence using transversional differences from the third codon position with bootstrap confidence values of 500. The confidence values (expressed as %) obtained using bootstrap are indicated on nodes. The scale bar indicates the number of substitutions per site.
Figure 8.
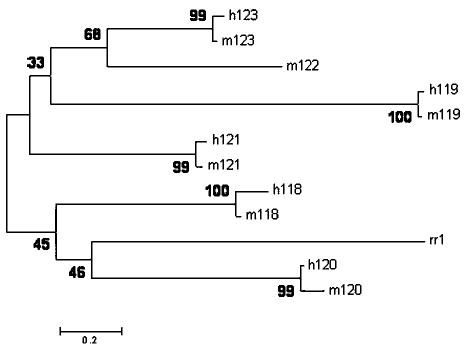
Phylogenetic tree of functional human and macaque β-defensin amino-acid sequences. The NJ tree was rooted with E. coioides reproductive regulator 1 sequence and the reliability of each branch was assessed using bootstrap replications (500). The numbers at the nodes are bootstrap percentage values and the scale bar represents the number of amino-acid substitutions per site.
Estimations of synonymous (dS) and nonsynonymous (dN) substitutions show that synonymous differences are generally saturated in paralog comparisons, because the gene duplications occurred a long time ago. In this case, one could compare the cDNA sequences of closest paralogs (m123 and m122) to compare dN and dS in the mature peptide encoded by exon 2. The modified Nei-Gojobori method produces a dN–dS value of 0.22, which is only significant at a 10% level. The lack of power in rejecting the null hypothesis of neutral evolution in exon 2 is probably caused by the fact that it is rather short (104 codons), but still dN is larger than dS in this exon, which could be taken as a sign of positive selection. The null hypothesis of neutrality can be rejected for the comparison of m123 and h123, as the dN–dS difference is 0.03 and significant at a 5% level, indicating positive selection.
Discussion
In this analysis of the human β-defensin gene cluster at 20q, we present five functional human genes DEFB119, DEFB120, DEFB121, and DEFB123 and their macaque orthologs. We demonstrate that, similar to other β-defensins,3,4,11 the expression of these novel 20q defensins is restricted to the testis and epididymis. DEFB118 location on sperm,25 DEFB126 involvement in the capacitation process,13 the Bin1b role in sperm motility and maturation,14 taken together with the male tract specific expression and differential regional expression of the 20q defensins provide evidence that these β-defensins have evolved to perform reproduction-specific tasks, combining immunity and fertility in their functional repertoires. In addition, the male organs may require more elaborate protection against pathogens than other organ systems. Both explanations are consistent with recent investigations that have shown expression of a wide variety of α- and β-defensins in the male urogenital tract.4
Although the 20q defensins are structurally distinct from DEFB1 and DEFB2, they maintain the cationic property of the defensin signature six-cysteine array that is essential for antimicrobial action.29 The antibacterial property is likely to be conserved throughout the cluster since the lead member, DEFB118, displays this activity26 and the human 20q genes and macaque orthologs show greater sequence conservation than the members of other defensin clusters. Defensins of the 20p cluster exhibit long C-terminal domains in contrast to the 8p cluster short C-terminal domains. On the other hand, 20q defensins have a high degree of variation in this region. At present, functions of the C-terminal elongations remain to be analyzed. The antibacterial peptide, DEFB118, alone is anionic in this cluster, while the 8p cluster has two anionic defensins. The initial interaction of the cationic β-defensin with the anionic membrane lipids is thought to be central to defensin-mediated bacterial membrane destruction; hence the relative increase in anionic residues would be expected to affect the antimicrobial function of the protein.30
Developmentally regulated expression of DEFB118–DEFB123 in the macaque epididymis also supports the concept of acquired functions in addition to immunity. DEFB118 and DEFB123, expressed only in the pubertal and adult macaque epididymis, may have roles in sperm maturation similar to Bin1b.14,31 Defensins such as DEFB119 and DEFB122 expressed at early stages of development may have roles in the growth and maintenance of the epididymis epithelium as demonstrated in an earlier study of mitogenic stimulation of the epithelial cells by defensins.32 Thus, the precise spatial and temporal regulation of β-defensins may be related to different functions in different regions during the development of the epididymis.
The mRNA levels of all six defensin genes were downregulated in the caput region after androgen ablation, which is associated with altered principal cell morphology, protein secretion, and apoptotic cell death.33 The expression of defensins was not affected by castration in the corpus region, while caudal expression of defensins DEFB118–DEFB123 was not maintained at normal levels by exogenous testosterone, suggesting the requirement for additional testis factors as suggested previously.25 Principal cells are particularly sensitive to circulating androgens and the androgen regulation of beta defensins may be cell and tissue specific. Region-specific regulation of gene expression by androgens in the epididymis is complex and modulated by both epididymis and testis factors.16
We demonstrated that two (DEFB121 and DEFB122) of the five genes in the 20q cluster have novel structures, different from those predicted by computational methods. Similar deviations from the predicted gene structures were reported in the actual structures of three out of five defensins at the 8p cluster,4 underlining the importance of empirical validation of computational predications. DEFB120 has no detectable orthologs in the mouse and rat genomes (unpublished data). Similarly, rodents and other mammals lack the human DEFB104 and DEFB108 orthologs. This indicates that these genes arose by duplication after the rodent divergence from other mammals or the orthologs were lost in rodents.4 It is interesting to note that ‘gene sorting’,34 in which ‘birth’ implies gene duplication and ‘death’ refers to gene deactivation, accounts for certain murine β-defensins not being detected in some other species.19 Gene death appears to be in progress for the pseudogenes DEFB122p and DEFB109p4 following the ‘birth and death’ mechanism, which is well known in the Ig and MHC multigene families.35
Molecular evolutionary analysis suggests that the diverse cluster of defensin paralogs at 20q evolved by tandem duplication of individual genes rather than groups. All of these duplications occurred long prior to the human–macaque split, which is thought to have occurred more than 23 million years ago.28 Our data and other previous investigations of primate beta defensin evolution support this dating.4,36 Preliminary estimates suggest that the oldest duplication could be more than 450 MYA dating back to the appearance of primitive jawed fish. However, further studies are required including human–mouse comparisons in order to calibrate the molecular clock more precisely.
The functional divergence of the sequences in this locus has been caused by excess of nonsynonymous substitutions in the second exons, while the first exons show purifying selection. These findings are strikingly similar to the evolution of primate β-defensin locus on chromosome 8p.4 Purifying selection of Exon 1, which codes for the signal peptide, is expected since signal sequences have been conserved during evolution and have a well-characterized role in targeting and endoplasmic reticulum membrane insertion. Investigations showing mammalian β-defensin evolution by gene duplication and positive selection implicate selective pressures promoting structure–function variability.4,22,23,36 The divergence in the mature peptide of murine β-defensins is hypothesized to be a response to pathogen diversity, each gene performing a particular specialized function.19 The macaque β-defensin 2 more efficiently inactivates certain pathogens than the human ortholog, perhaps a response to environmental pressures to modulate the evolving activity.37 In this context, it is pertinent to note that the differences in gene structure, expression patterns, regulation and evolutionary divergence make each one of the defensins in this cluster unique. Each individual gene might be involved in different functions and could have evolved due to host–pathogen co-evolutionary processes. Further studies are required to understand the molecular evolutionary mechanisms acting on the three defensin clusters at 8p, 20p, and 20q, especially in the context of structure–function relationship. Recently, our group reported fertility-related and antimicrobial roles of the androgen-regulated sperm-binding protein, epididymal protease inhibitor (Eppin).38,39 Similar roles for the defensins are currently under investigation.
In conclusion, using both computational and molecular biology approaches, the 20q β-defensin locus was analyzed. Differential regional expression, developmental and hormonal regulations suggest the placement of the 20q defensins, where DEFB126 and Bin1b are established at the interface between innate immunity and fertility. Molecular evolutionary analysis of the clusters suggests their evolution by duplication and positive selection, which may either be related to functional specialization of the duplicated gene and/or to the necessity to respond to pathogen diversity.
Materials and methods
Our previously cloned DEFB118 (formerly ESC42) and the public domain search tools (BLAST) were used to identify novel human genomic sequences and expressed sequence tags (ESTs) substantially similar to DEFB118 (AAL27987) and derived from the same chromosomal region, 20q11.1. After determining by Northern Blot analysis that expression was highly male reproductive tract specific (data not shown), we deposited these sequences (ESC42 RELA-RELD) in Genbank (accession numbers AF479698–AF479701). To obtain the first exons of DEFB121 and DEFB122 which were not correctly annotated in the Unigene database, upstream sequences were translated in silico in three reading frames and analyzed for the hydrophobic amino-acid groups preceded by a methionine. These regions selected were then scanned for classical exon–intron boundaries. The predicted proteins were found to contain putative signal peptides using SignalP.40 Intron spanning primers were designed and using human epididymis mRNA as a template RT-PCR was performed. The specific products were sequenced and the corresponding exon/intron boundaries were assessed by alignment of cDNAs with the genomic sequences.
A panel of human and macaque (Macaca mulatta) tissues were obtained as described elsewhere.41 Total RNA was extracted from these tissues, including parotid glands, thyroid, breast, lungs, liver, spleen, kidney, colon, adrenal, ovary, seminal vesicle, cauda, corpus, caput, prostate and testis, using Trizol reagent (Invitrogen, Carlsbad, CA, USA). In order to understand the expression patterns of the 20q defensin genes in female reproductive tissues, endometrial samples were obtained under a protocol approved by the UNC School of Medicine IRB from Dr Bruce Lessey (Greenville Hospital, Greenville, SC, USA) as described.42 In all, 2 μg of total RNA was reverse transcribed using 50 U Stratascript™ (Stratagene, La Jolla, CA, USA) according to the supplier’s instructions with 0.5 μg of oligodT (Invitrogen). Gene-specific intron spanning primer sequences for β-defensin DEFB118–DEFB123 and peptidyl isomerase (PPIA) were as follows:
118 F 5′-CCATCTCCTGTTTCCCAAGCACC
R 5′-GACAGGGGTGAGTGACATTCGAG
119 F 5′-CTTCCCCTGCAGCTCAGCC
R 5′-GGGGTTGACAGCAGGTCTCTG
120 F 5′-CCTCTCAGGTCTGCCAGCC
R 5′-GTGGACCCTAAGCAACCTGAAAC
121 F 5′-GAAGCCTCCTATCCCTCCCTG
R 5′-GGTTGGAAGAGAGGTGTCAGAC
122 F5′-CGGACTTGCAGCTTCATTTTGGG
R 5′-CAAAGCAGTTAAAATGGCCACCAC
123 F5′-CGGACTTGCAGCTTCATTTTGGG
R 5′-CAAAGCAGTTAAAATGGCCACCAC
PPIA F 5′-GGGCCGCGTCTCCTTTGAGC
R 5′-CATTCCTGGACCCAAAGCGCTC
PCR reactions were carried out using 2 μl of the resultant cDNA according to the following procedures: 94°C for 1 min followed by 23–38 cycles at 94°C for 30 s, 58°C for 30 s and 72°C for 1 min, and a final round of extension for 5 min. Linear amplification range for each gene was tested on the epididymal cDNA. The less expressed transcript DEFB123 required 32 cycles of PCR for detection, while the rest of the genes required 28 cycles. Amplification of PPIA, which served as an internal control in this semiquantitative analysis, was carried out in parallel with similar conditions for 23 cycles. PCR amplified products were analyzed by electrophoresis on a 2% agarose gels. Gel-purified PCR products were sequenced at the UNC-CH Genome Analysis Facility using ABI PRISM model 377 DNA sequencer (PE Applied Biosystems, Foster City, CA, USA) as described previously.41 For analysis of the hormonal regulation of these defensins, castrated and hormone-replaced epididymides were used as previously reported.25 For studies of the developmental regulation of defensins, macaque epididymis was obtained from the Oregon National Primate Research Center, Portland, OR, USA.
All evolutionary analyses were carried out using MEGA 3 software.43 The amino-acid sequences were obtained by translating the cDNA sequences. The aminoacid alignment employing only the functional defensin genes for human and macaque shown in Figure 2 was constructed using the default options in Clustal W, embedded in MEGA3. Human DEFB122 was excluded from analysis because it is a pseudogene not subjected to the same evolutionary pressures as the defensin protein encoding paralogs. The corresponding cDNA alignments were generated using the protein sequence alignments as guides. These nucleotide-aligned sequences were used in most of the analyses. Phylogenetic relationships among sequences were inferred using the NJ method,27 where the Kimura two-parameter model was used to estimate evolutionary distances. To infer the evolutionary history, we used substitutions that are not likely to be under adaptive evolutionary forces. For these analyses we used only third codon positions. All sites containing alignment gaps and missing data were excluded from phylogenetic analyses. Exon 1 of DEFB119 was excluded from the analysis since it also appears in DEFB120. Using a concatenated sequence of β-defensins from 20q cluster, by tblastn translation we identified the sequence of E. coioides (orange-spotted grouper) reproduction regulator 1 (accession number: AY129305) as highly homologous to the human defensins. This putative defensin sequence aligned well with the 20q cluster; thus, we used it to root the trees. Confidence limits were calculated by the bootstrap re-sampling method (500 replicates).44 To detect the presence of positive selection, additional analyses were carried out using the modified-Nei-Gojobori method in MEGA3.
Additional data files
The following data files are available for this article: the aligned macaque and human DNA sequences (Data file 1), the aligned first exon macaque and human DNA sequences (Data file 2), the aligned second exon macaque and human DNA sequences (Data file 3), and the aligned macaque and human DNA sequences along with the E. coioides (rr1) sequence (Data file 4). The data files are in .meg format, and can be used with MEGA software, which can be downloaded from www.megasoftware.net. The data files are available from http://www.med.unc.edu/lrb/shall.htm.
Acknowledgments
We thank Ms Graziela Valente and Dr Sudhir Kumar from the Arizona State University for their help in evolutionary analysis and in the critical reading of the manuscript. We thank Dr Hemant Kelkar and Dr Jianping Jin, Center for Bioinformatics, University of North Carolina, Chapel Hill, for their suggestions. Support for this project (CIG-96-06-A) was provided by the CICCR Program of the Contraceptive Research and Development Program (CONRAD), Eastern Virginia Medical School. The views expressed by us do not necessarily reflect the views of CONRAD or CICCR. This work was also supported by grants from The Andrew W Mellon Foundation and by National Institutes of Health grants R37-HD04466 and U54-HD35041 as part of the Specialized Cooperative Centers Program in Reproduction Research, and by the Fogarty International Center Training and Research in Population and Health grant D43TW/HD00627.
References
- 1.Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat Rev Immunol. 2003;3:710–720. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- 2.Schutte BC, Mitros JP, Bartlett JA, et al. Discovery of five conserved beta-defensin gene clusters using a computational search strategy. Proc Natl Acad Sci USA. 2002;99:2129–2133. doi: 10.1073/pnas.042692699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodriguez-Jimenez F-J, Krause A, Schulz S, et al. Distribution of new human [beta]-defensin genes clustered on chromosome 20 in functionally different segments of epididymis. Genomics. 2003;81:175–183. doi: 10.1016/s0888-7543(02)00034-4. [DOI] [PubMed] [Google Scholar]
- 4.Semple C, Rolfe M, Dorin J. Duplication and selection in the evolution of primate beta-defensin genes. Genome Biol. 2003;4:R31. doi: 10.1186/gb-2003-4-5-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang D, Biragyn A, Hoover DM, Lubkowski J, Oppenheim JJ. Multiple roles of antimicrobial defensins, cathelicidins, and eosinophil-derived neurotoxin in host defense. Annu Rev Immunol. 2004;22:181–215. doi: 10.1146/annurev.immunol.22.012703.104603. [DOI] [PubMed] [Google Scholar]
- 6.Goldman MJ, Anderson GM, Stolzenberg ED, Kari UP, Zasloff M, Wilson JM. Human beta-defensin-1 is a salt-sensitive antibiotic in lung that is inactivated in cystic fibrosis. Cell. 1997;88:553–560. doi: 10.1016/s0092-8674(00)81895-4. [DOI] [PubMed] [Google Scholar]
- 7.Schroder JM, Harder J. Human beta-defensin-2. Int J Biochem Cell Biol. 1999;31:645–651. doi: 10.1016/s1357-2725(99)00013-8. [DOI] [PubMed] [Google Scholar]
- 8.Lehrer RI. Primate defensins. Nat Rev Microbiol. 2004;2:727–738. doi: 10.1038/nrmicro976. [DOI] [PubMed] [Google Scholar]
- 9.Garcia JR, Jaumann F, Schulz S, et al. Identification of a novel, multifunctional beta-defensin (human beta-defensin 3) with specific antimicrobial activity. Its interaction with plasma membranes of Xenopus oocytes and the induction of macrophage chemoattraction. Cell Tissue Res. 2001;306:257–264. doi: 10.1007/s004410100433. [DOI] [PubMed] [Google Scholar]
- 10.Yang D, Chertov O, Bykovskaia SN, et al. Defensins: linking innate and adaptive immunity through dendritic and T cell CCR6. Science. 1999;286:525–528. doi: 10.1126/science.286.5439.525. [DOI] [PubMed] [Google Scholar]
- 11.Yamaguchi Y, Nagase T, Makita R, et al. Identification of multiple novel epididymis-specific {beta}-defensin isoforms in humans and mice. J Immunol. 2002;169:2516–2523. doi: 10.4049/jimmunol.169.5.2516. [DOI] [PubMed] [Google Scholar]
- 12.Avellar MCW, Honda L, Hamil KG, et al. Differential expression and antibacterial activity of epididymis protein 2 isoforms in the male reproductive tract of human and rhesus monkey (Macaca mulatta) Biol Reprod. 2004;71:1453–1460. doi: 10.1095/biolreprod.104.031740. [DOI] [PubMed] [Google Scholar]
- 13.Yudin AI, Tollner TL, Li MW, Treece CA, Overstreet JW, Cherr GN. ESP13.2, a member of the beta-defensin family, is a macaque sperm surface-coating protein involved in the capacitation process. Biol Reprod. 2003;69:1118–1128. doi: 10.1095/biolreprod.103.016105. [DOI] [PubMed] [Google Scholar]
- 14.Zhou CX, Zhang YL, Xiao L, et al. An epididymis-specific beta-defensin is important for the initiation of sperm maturation. Nat Cell Biol. 2004;6:458–464. doi: 10.1038/ncb1127. [DOI] [PubMed] [Google Scholar]
- 15.Rodriguez C, Kirby J, Hinton B. Regulation of gene transcription in the epididymis. Reproduction. 2001;122:41–48. doi: 10.1530/rep.0.1220041. [DOI] [PubMed] [Google Scholar]
- 16.Orgebin-Crist MC. Androgens and epididymal function. In: Bhasin SGH, Spieler JM, Swerdloff RE, Wang C, Kelly C (eds). Pharmacology, Biology, and Clinical Applications of Androgens Wiley-Liss: New York, 1996, pp 27–38.
- 17.Hinton BT, Lan ZJ, Rudolph DB, Labus JC, Lye RJ. Testicular regulation of epididymal gene expression. J Reprod Fertil Suppl. 1998;53:47–57. [PubMed] [Google Scholar]
- 18.Gould KG, Young LG. Acquisition of fertilizing capacity by chimpanzee sperm. Folia Primatol (Basel) 1990;54:105–108. doi: 10.1159/000156430. [DOI] [PubMed] [Google Scholar]
- 19.Morrison GM, Semple CA, Kilanowski FM, Hill RE, Dorin JR. Signal sequence conservation and mature peptide divergence within subgroups of the murine beta-defensin gene family. Mol Biol Evol. 2003;20:460–470. doi: 10.1093/molbev/msg060. [DOI] [PubMed] [Google Scholar]
- 20.Swanson WJ, Vacquier VD. The rapid evolution of reproductive proteins. Nat Rev Genet. 2002;3:137–144. doi: 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- 21.Hughes AL, Yeager M. Coordinated amino acid changes in the evolution of mammalian defensins. J Mol Evol. 1997;44:675–682. doi: 10.1007/pl00006191. [DOI] [PubMed] [Google Scholar]
- 22.Boniotto M, Tossi A, DelPero M, et al. Evolution of the beta defensin 2 gene in primates. Genes Immun. 2003;4:251–257. doi: 10.1038/sj.gene.6363958. [DOI] [PubMed] [Google Scholar]
- 23.Maxwell AI, Morrison GM, Dorin JR. Rapid sequence divergence in mammalian beta-defensins by adaptive evolution. Mol Immunol. 2003;40:413–421. doi: 10.1016/s0161-5890(03)00160-3. [DOI] [PubMed] [Google Scholar]
- 24.Swanson WJ, Nielsen R, Yang Q. Pervasive adaptive evolution in mammalian fertilization proteins. Mol Biol Evol. 2003;20:18–20. doi: 10.1093/oxfordjournals.molbev.a004233. [DOI] [PubMed] [Google Scholar]
- 25.Liu Q, Hamil KG, Sivashanmugam P, et al. Primate epididymis-specific proteins: characterization of ESC42, a novel protein containing a trefoil-like motif in monkey and human. Endocrinology. 2001;142:4529–4539. doi: 10.1210/endo.142.10.8422. [DOI] [PubMed] [Google Scholar]
- 26.Yenugu S, Hamil KG, Radhakrishnan Y, French FS, Hall SH. The androgen-regulated epididymal sperm-binding protein, human beta-defensin 118 (DEFB118) (formerly ESC42), is an antimicrobial beta-defensin. Endocrinology. 2004;145:3165–3173. doi: 10.1210/en.2003-1698. [DOI] [PubMed] [Google Scholar]
- 27.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 28.Kumar S, Hedges SB. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- 29.Hwang PM, Vogel HJ. Structure–function relationships of antimicrobial peptides. Biochem Cell Biol. 1998;76:235–246. doi: 10.1139/bcb-76-2-3-235. [DOI] [PubMed] [Google Scholar]
- 30.White SH, Wimley WC, Selsted ME. Structure, function, and membrane integration of defensins. Curr Opin Struct Biol. 1995;5:521–527. doi: 10.1016/0959-440x(95)80038-7. [DOI] [PubMed] [Google Scholar]
- 31.Li P, Chan HC, He B, et al. An antimicrobial peptide gene found in the male reproductive system of rats. Science. 2001;291:1783–1785. doi: 10.1126/science.1056545. [DOI] [PubMed] [Google Scholar]
- 32.Murphy CJ, Foster BA, Mannis MJ, Selsted ME, Reid TW. Defensins are mitogenic for epithelial cells and fibroblasts. J Cell Physiol. 1993;155:408–413. doi: 10.1002/jcp.1041550223. [DOI] [PubMed] [Google Scholar]
- 33.Ezer N, Robaire B. Gene expression is differentially regulated in the epididymis after orchidectomy. Endocrinology. 2003;144:975–988. doi: 10.1210/en.2002-220705. [DOI] [PubMed] [Google Scholar]
- 34.Zhang J, Dyer KD, Rosenberg HF. Evolution of the rodent eosinophil-associated RNase gene family by rapid gene sorting and positive selection. Proc Natl Acad Sci USA. 2000;97:4701–4706. doi: 10.1073/pnas.080071397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nei M, Gu X, Sitnikova T. Evolution by the birth-and-death process in multigene families of the vertebrate immune system. Proc Natl Acad Sci USA. 1997;94:7799–7806. doi: 10.1073/pnas.94.15.7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Del Pero M, Boniotto M, Zuccon D, et al. Beta-defensin 1 gene variability among non-human primates. Immunogenetics. 2002;53:907–913. doi: 10.1007/s00251-001-0412-x. [DOI] [PubMed] [Google Scholar]
- 37.Antcheva N, Boniotto M, Zelezetsky I, et al. Effects of positively selected sequence variations in human and Macaca fascicularis beta-defensins 2 on antimicrobial activity. Anti-microb Agents Chemother. 2004;48:685–688. doi: 10.1128/AAC.48.2.685-688.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yenugu S, Richardson RT, Sivashanmugam P, et al. Antimicrobial activity of human EPPIN, an androgen-regulated, sperm-bound protein with a whey acidic protein motif. Biol Reprod. 2004;71:1484–1490. doi: 10.1095/biolreprod.104.031567. [DOI] [PubMed] [Google Scholar]
- 39.O’Rand MG, Widgren EE, Sivashanmugam P, et al. Reversible immunocontraception in male monkeys immunized with eppin. Science. 2004;306:1189–1190. doi: 10.1126/science.1099743. [DOI] [PubMed] [Google Scholar]
- 40.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int J Neural Syst. 1997;8:581–599. doi: 10.1142/s0129065797000537. [DOI] [PubMed] [Google Scholar]
- 41.Hamil KG, Liu Q, Sivashanmugam P, et al. LCN6, a novel human epididymal lipocalin. Reprod Biol Endocrinol. 2003;1:112. doi: 10.1186/1477-7827-1-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Young SL, Lyddon TD, Jorgenson RL, Misfeldt ML. Expression of Toll-like receptors in human endometrial epithelial cells and cell lines. Am J Reprod Immunol. 2004;52:67–73. doi: 10.1111/j.1600-0897.2004.00189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 44.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]


