Abstract
Hereditary tyrosinemia type 1 (HT1) is a severe autosomal recessive metabolic disease associated with point mutations in the human fumarylacetoacetate hydrolase (FAH) gene that disrupt tyrosine catabolism. An acute form of HT1 results in death during the first months of life because of hepatic failure, whereas a chronic form leads to gradual development of liver disease often accompanied by renal dysfunction, childhood rickets, neurological crisis, and hepatocellular carcinoma. Mice homozygous for certain chromosome 7 deletions of the albino Tyr; c locus that also include Fah die perinatally as a result of liver dysfunction and exhibit a complex syndrome characterized by structural abnormalities and alterations in gene expression in the liver and kidney. Here we report that two independent, postnatally lethal mutations induced by N-ethyl-N-nitrosourea and mapped near Tyr are alleles of Fah. The Fah6287SB allele is a missense mutation in exon 6, and Fah5961SB is a splice mutation causing loss of exon 7, a subsequent frameshift in the resulting mRNA, and a severe reduction of Fah mRNA levels. Increased levels of the diagnostic metabolite succinylacetone in the urine of the Fah6287SB and Fah5961SB mutants indicate that these mutations cause a decrease in Fah enzymatic activity. Thus, the neonatal phenotype present in both mutants is due to a deficiency in Fah caused by a point mutation, and we propose Fah5961SB and Fah6287SB as mouse models for acute and chronic forms of human HT1, respectively.
Hereditary tyrosinemia type 1 (HT1) is an autosomal recessive disease caused by a deficiency of fumarylacetoacetate hydrolase (FAH), the last enzyme in the catabolic pathway of tyrosine (1). FAH deficiency results in the accumulation of fumarylacetoacetate, maleylacetoacetate, and succinylacetone (SA) in the liver and kidney, which is believed to be responsible for certain pathological aspects of the disorder (2, 3). The two major clinical forms of the disease, acute and chronic, can be distinguished on the basis of the severity and the progression of the disease in patients based on the residual amount of liver FAH activity (4, 5). In the acute form, symptoms appear during the first months of life, and death caused by hepatic failure usually occurs during the first year. The chronic form results in a gradual liver disease from which individuals develop childhood rickets, suffer from renal tubule defects, and often develop hepatocarcinoma in their teens. Biochemically, the disease is characterized by the accumulation of tyrosine, tyrosine metabolites, methionine, and SA in serum and/or urine. SA accumulation is unique to FAH deficiency (6, 7), and its presence is diagnostic for the disease (2, 8, 9). The disease has a worldwide distribution, with a high incidence in the French-Canadian and Scandinavian populations (1, 6).
The human FAH gene is located in chromosome 15q23-q25 (10). At least 26 different mutations in FAH have been identified in HT1 patients, including missense, nonsense, and splice consensus site mutations (5, 11–17). Likewise, it has been shown that targeted disruption of the Fah gene in mouse chromosome 7 (18) causes death in homozygotes within 12 hours after birth from liver dysfunction and hypoglycemia; a phenotype quite similar to the neonatal death observed for mice homozygous for certain deletions of the Tyr locus (19–22) also proved to include Fah (23, 24). The null phenotype, then, whether from chromosomal deletion or Fah-gene targeting, is neonatal lethality in mice. Phenotype correction experiments using Fah transgenes (25) expressed in mice homozygous for an Fah deletion, c14CoS, showed that Fah complements all aspects of the null phenotype except in lines manifesting very low liver expression of the transgene.
Whereas the Fah null allele in mice causes early lethality (within hours of birth), the known cases of human HT1 result from point mutations within FAH. Human HT1 may be modeled more suitably by similar types of point mutations in mice, such as those easily induced by the presumed point-mutation inducer N-ethyl-N-nitrosourea (ENU; ref. 26). We report here two ENU-induced point mutations of the Fah gene, designated Fah5961SB and Fah6287SB, that were recovered as postnatally lethal mutations mapping near Tyr; c in a screen for recessive mutations mapping to a 6- to 11-centimorgan region of mouse chromosome 7 (27, 28). The molecular and phenotypic characteristics of one of these mutants suggest its utility as a mouse model for the chronic human forms of HT1.
Materials and Methods
Mice.
All mice were bred at the Life Sciences Division of the Oak Ridge National Laboratory. Generation of ENU-induced mutations mapping to the large 6- to 11-centimorgan c26DVT deletion has been described (27, 28). Briefly, BALB/cRl (albino, c/c) males treated with ENU were mated to (C57BL/10Rl × C3Hf/Rl) G0 females (+/+). Then G1 females carrying the mutagenized paternal chromosome 7 were mated to males carrying the long deletion (cch/c26DVT; cch is the chinchilla allele at c, and cch/c gives a light chinchilla coat color). Albino G2 progeny were examined for expression of recessive mutations (m), because this phenotypic class carries the mutagenized chromosome 7 opposite the deletion chromosome (c m/c26DVT). All mutations can be maintained by crossing heterozygous carriers (cch +/c m) if the hemizygotes and homozygotes cannot be bred. Newborn mutants lack eye pigment and can be distinguished from wild-type pigmented littermates.
Deletion Mapping.
To map ENU-induced mutations relative to deletion endpoints, proved cch +/c m males were crossed to cch/Del(c) females, where Del(c) is one of several c locus deletions. The progeny of these mapping crosses were observed at 3 weeks after birth for the number of albinos present. Any particular Del(c) was noncomplementing for m if there were either abnormal albinos or no albinos in the progeny of the cross. A cch +/c m male was considered proved if he produced either abnormal albinos or no albinos in 30 progeny from a testcross to a cch/Del(c)26DVT female (27, 28).
Complementation Analysis.
Trans complementation testing between the two ENU-induced mutations was done by reciprocally intercrossing mutation carriers (cch +/c m5961SB × cch+/c m6287SB and cch +/c m6287SB × cch +/c m5961SB) and examining the phenotype of albinos (c m5961SB/c m6287SB).
RNA Isolation and Northern Blot Analysis.
Cellular RNAs were isolated from the fetal livers at embryonic day 17.5 of albino mice hemizygous for the large deletion Del(c)26DVT and carrying a chromosome from BALB/cRl [c/Del(c)26DVT] or from an Fah5961SB mutant [c Fah5961SB/Del(c)26DVT] or an Fah6287SB mutant [c Fah6287SB/Del(c)26DVT]. The genotype of embryos was determined by visual examination for the absence of eye pigment. Total and poly(A)+ RNAs were isolated by using kits (5 Prime→3 Prime) according to the manufacturer's instructions and were analyzed by standard procedures (29, 30). Poly(A)+ RNA (3 μg) was loaded into each lane of a formaldehyde-containing gel (1.0% agarose), transferred onto a nylon membrane (Duralon-UV membrane, Stratagene), and hybridized with a full-length murine Fah cDNA (23) labeled with [α-32P]dCTP by using the random-primer method. Hybridization was carried out at 42°C overnight in 1× SSCP (120 mM NaCl/5 mM sodium citrate/20 mM sodium phosphate, pH 6.8), 2% (vol/vol) SDS, 10% (vol/vol) dextran sulfate, and 50% (vol/vol) formamide. Washing stringency was 0.1× SSCP with 0.1% SDS at 65°C. A loading control was done by reprobing the blot with a β-actin cDNA.
cDNA Production, PCR, and Sequencing.
Double-stranded Fah cDNAs (≈1.4 kb) were produced by reverse transcription (RT) of total murine fetal liver RNA and then PCR amplified by using gene-specific oligonucleotides (5′-GCCCGGTGCTCGTCAGCATGTCC-3′ and 5′-AGCATGATCCTCATAAGGCAAGGG-3′). PCR-amplified products were cloned into the plasmid vector pCRII, provided in a One-Shot TA Cloning Kit (Invitrogen), and sequenced. When sequencing of the RT-PCR product from Fah5961SB revealed the deletion of exon 7 in the cDNA, primer sets were designed to amplify the genomic sequences surrounding exon 7 for analysis of the consensus splice sites. One primer pair (5′-GAAGGATCAGAAAGGCCTGG-3′ and 5′-GGAGATTGTGGTTCCAAAGC-3′) amplified a 1,108-bp fragment from exon 6 through exon 8, and a second pair (5′-GAA GGA TCA GAA AGG CCT GG-3′ and 5′-GTA CAG CTC TTC ATC AGC AG-3′) amplified a 579-bp fragment containing all of exon 7 with its flanking intronic sequences. Sequencing of these genomic fragments detected a G-to-A mutation at the last base in exon 7. Amplifications were performed over 35 cycles, each cycle consisting of 30 sec of denaturation at 94°C, 2 min of annealing at 60°C, and 4 min of extension at 72°C. PCR-amplified products were cloned into the plasmid vector pCRII, provided in a One-Shot TA Cloning Kit (Invitrogen) and sequenced. Sequencing of control (c/c) and mutant [c m/Del(c)26DVT] cDNAs was performed manually on both cDNA strands by the dideoxy chain-termination method (31) with synthetic oligodeoxynucleotides as primers and a Sequenase Version 2.0 DNA Sequencing kit (United States Biochemical) or later on an ABI 377 DNA sequencer with the ABI PRISM Big Dye terminator kit by following the manufacturer's specifications. Sequences were analyzed by using SEQUENCER 3.1 software (Gene Codes, Ann Arbor, MI).
Biochemical Analysis.
Urine from Fah5961SB and Fah6287SB heterozygotes or hemizygotes, heterozygous carriers, and wild-type siblings was collected by free catch and stored at −20°C.
Extraction and derivatization of SA.
First, the SA was oximated by using a modification of the procedure described by Tuchman (32). Then HPLC-grade water (0.3 ml), hydroxylamine hydrochloride (0.2 ml, 200 mg/ml in water), HCl (40 μl, 6 M), and mouse urine (5–100 μl) were added to 4-ml ReactiVials (Pierce). The samples were spiked with 5 μl of oximated 5,7-dioxooctanic acid (Ox-DOOA, 40 μg/ml in methanol) as an internal reference. (DOOA is a homolog of SA and contains an additional CH2 group.) The samples were heated at 80°C for 30 min to oximate the SA present and convert it to 5(3)-methyl-3(5)-isoxazole propionic acid. After cooling, the samples were extracted twice with 3 ml of ether each time. (In earlier extractions, NaCl was added to saturate the samples with salt (33), but this method was found to be unnecessary for good recoveries of Ox-SA.) The ether extracts were blown gently to dryness at 40°C with nitrogen. To form the tert-butyl-dimethylsilyl derivative of the Ox-SA, 20 μl of N-methyl-N-(tert-butyl-dimethylsilyl)trifluoracetamide (MTBSTFA) and 20 μl of pyridine were added to each vial. The samples were vortexed in 2-ml ReactiVials and heated at 80°C for 30 min.
Gas chromatography/mass spectrometry.
Analyses were carried out by using a Hewlett Packard 5890 Series II GC interfaced to a 5989A mass spectrometer. An HP 5-MS (crosslinked 5% diphenyl/95% dimethylsiloxane) capillary column (30-m × 0.25-mm × 0.25-μm film) was used for the chromatographic separations. The injector and detector were set at 250°C and 280°C, respectively. The source and analyzer were set at 200°C and 100°C, respectively. The initial column temperature was 140°C; this temperature was held for 5 min, then ramped at 15°C/min to 260°C, and held for 3 min. The injections (1 μl) were splitless, with the purge valve turned on at 1 min. The oximated and derivatized analytes were detected by selected ion monitoring (SIM) with 212 and 138 m/z ions for SA and 226 and 152 m/z ions for DOOA.
Results
Postnatal Death of 5961SB and 6287SB Homo- and Hemizygotes.
Two independent, postnatally lethal mutations (5961SB and 6287SB) were among 31 mutations detected in a long-term effort to recover recessive ENU-induced mutations mapping within the limits of the 6- to 11-centimorgan c26DVT deletion (27, 28). Males heterozygous for either the 5961SB or 6287SB mutation (cch +/c m) were bred to female heterozygotes (cch +/c m) or to females carrying the 26DVT deletion [cch +/Del(c)26DVT], and litters containing albino offspring were observed daily. No obvious differences in morphology or size were noted immediately after birth, and all pups appeared healthy. In 5961SB litters, however, no live albino pups (identified by lack of eye pigment) were found past 24 h after birth. In 6287SB litters, albino pups died 10–20 days after birth, with the majority dying at days 12–14. No 6287SB albino mutants survived past 1 month.
Deletion Mapping and Complementation Analysis.
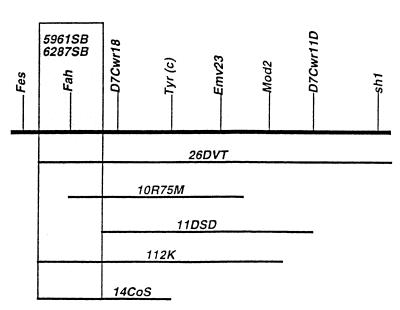
Matings of male carriers (cch +/c m) of either 5961SB or 6287SB to females heterozygous for the albino deletions Del(c) (14CoS), Del(c) (10R75 M), or Del(c)112K produced either abnormal or no albino offspring (Table 1). Normal albinos were produced, however, when Del(c)11DSD deletion heterozygotes were mated to either mutation. These data suggest that both of these mutations map to the 315-kb region of nonoverlap between the proximal ends of the c14CoS and c11DSD deletions, the region known to include Fah (ref. 23; Fig. 1).
Table 1.
Deletion mapping of the 5961SB and 6287SB mutations recovered in the c region hemizygosity screen
| Deletion [Del(c)] |
m
|
|
|---|---|---|
| 5961SB | 6287SB | |
| Del(c)14CoS | 0/40* (5)† | 0/422 (8) |
| Del(c)10R75M | 0/30 (7) | 3s/42 (11) |
| Del(c)112K | 0/31 (1) | 0/23 (10) |
| Del(c)11DSD | 5n/16 (5) | 8n/35 (4)‡ |
The ratios represent the number of albinos present at weaning (25% expected) over the total number of progeny born from the cross cch +/Del(c) × cch +/c m. “n” designates albinos that were externally normal, and “s” designates albinos that were runted.
Number in parentheses gives the number of albinos present at birth. Albinos are distinguished at birth by the absence of eye pigment.
Six nonalbinos, in addition to the four albinos indicated, were also lost between birth and weaning.
Figure 1.
Deletion mapping of two ENU-induced postnatally lethal mutations to the Fah region of mouse chromosome 7. Horizontal lines beneath the chromosome (heaviest line) represent a subset of Tyr-locus deletions used for the mapping of the ENU-induced mutations 5961SB and 6287SB. Genetic markers are shown above the chromosome in italics. Detailed definitions and descriptions of DNA markers and loci have been published (28). The proximal breakpoints of the Del(c)14CoS and Del(c)11DSD deletions flank the Fah region. The proximal breakpoint of the Del(c)10R75 M deletion maps <1 kb 5′ to the start of Fah transcription and is believed to remove sequences essential for expression of Fah (23).
Because both mutations mapped to the same deletion interval, they were tested for allelism. Among 35 total progeny from the cross cch +/c m5961SB × cch +/c m6287SB, 16 albinos were present as neonates but were missing by weaning. Among 34 total progeny of the reciprocal cross (cch +/c m6287SB × cch +/c m5961SB +), six albinos were present as neonates but also were missing by weaning. The absence of normal albinos demonstrates noncomplementation between the two mutations, suggesting that they are alleles of a single locus mapping at or very near Fah.
Expression of the Normal Fah mRNA in the Fetal Livers of 5961SB and 6287SB Mice.
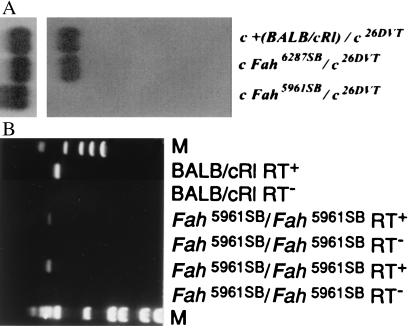
Northern blot hybridization to the full-length Fah cDNA probe was performed to determine whether there was any significant alteration in the amount and/or size of the Fah mRNA in fetal livers of the mutant mice. In control [c + (BALB/cRl)/c26DVT] fetal livers from embryonic day 17.5 embryos, the mRNA migrates at ≈1.4 kb. (Fig. 2A, lane 1; ref. 23). In the 6287SB mutant [c Fah6287SB/Del(c)26DVT], the transcript appears to be of wild-type size and expression level (Fig. 2A, lane 2). In the 5961 mutant [cFah5961SB/Del(c)26DVT], there is no apparent Fah transcript (Fig. 2A, lane 3).
Figure 2.
Northern blot analysis of the Fah6287SB and Fah 5961 alleles and RT-PCR analysis of the Fah 5961 allele. (A) Expression of Fah mRNA (1.4-kb transcript) in 3 μg of poly(A)+ fetal liver RNA from control [BALB/cRl/Del(c)26DVT] mice (lane 1), Fah6287SB/Del(c)26DVT (lane 2), and Fah5961SB/Del(c)26DVT (lane 3) mutants is shown. The genotype of the animals is given above; the probe for Upper is the full-length Fah cDNA and for Lower is β-actin (hybridized sequentially). (B) PCR-amplification of BALB/cRl and homozygous 5961SB mutant Fah cDNA with primers that amplify exon 7. Genotypes are given above. Fragments in lanes 2, 4, and 6 were amplified in the presence of reverse transcriptase, whereas reactions in lanes 3, 5, and 7 contained no reverse transcriptase. Lanes 1 (øX174/HaeIII) and 8 (1-kb DNA ladder) contain molecular-mass standards (M).
5961SB and 6287SB Have Altered Fah cDNA Sequence.
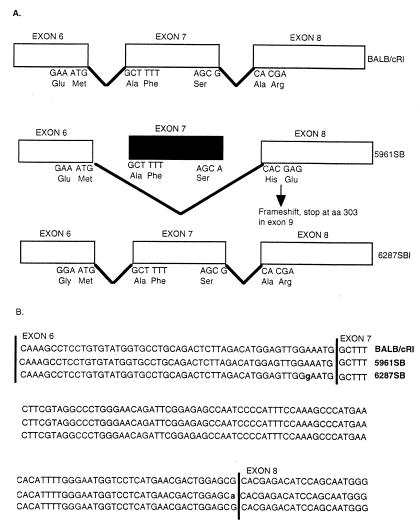
All Fah exons from mutant cDNAs amplified by PCR were screened for point mutations by sequencing of two independently amplified fragments for each mutation. As an additional control, we resequenced BALB/cRl cDNA and found it identical to that previously reported for a full-length BALB/cRl Fah mouse cDNA clone (23), including an ORF of 1,257-bp coding for 419 amino acids starting from the first ATG at position 1 and ending at the TGA at position 1,258. Sequencing of the 6287SB cDNA revealed an A→G transition causing a missense mutation in nucleotide position 602 (Fig. 3), introducing glycine in place of glutamic acid in amino acid position 201 (E201G). Sequencing of the complete 5961SB cDNA product revealed that nucleotides 607–706, which constitute the seventh exon of murine Fah, are deleted (607d100, Fig. 3). Then, sequencing of the genomic DNA surrounding exon 7 showed that the last base in exon 7, a G in BALB/cRl, is mutated to an A in 5961SB, affecting the consensus splicing sequence, leading to the splicing of exon 6 to exon 8, and resulting in a transcript that lacks exon 7 (Fig. 3). The deletion of exon 7 results in a frameshift and subsequently the introduction of a premature stop codon at amino acid position 303, such that the amino acids beginning with position 235 are mistranslated, and amino acids 304–419, in addition to those in exon 7, would be absent were the message stable. To ensure that the smaller product sequenced from 5961SB cDNA did not represent a minor splicing variant, we performed RT-PCR on liver RNAs with primers that flank exon 7. RT-PCR fragments amplified from BALB/cRl or 6287SB (data not shown) RNA were of the expected size (497 bp), whereas the 5961SB RT-PCR fragments were ≈100 bp shorter in length (Fig. 2B, lanes 4 and 6). It is noteworthy that this shorter RT-PCR product was the only fragment amplified from 5961SB RNA in multiple independent experiments, thus making it unlikely that the cDNA lacking exon 7 was derived from a minor, alternatively spliced RNA.
Figure 3.
Schematic representation of the murine Fah6287SB and Fah5961SB point mutations and their effect on the processing of the Fah mRNA. (A) Top illustrates the normal splicing pattern for Fah followed by the altered pattern in Fah5961SB and a similar schematic for Fah6287SB. (Middle) As indicated, the Fah5961SB mutation results in an altered pattern of mRNA splicing, causing the loss of exon 7 from the Fah mRNA and a subsequent shift in the reading frame to introduce a premature stop codon in exon 9. Bottom shows the relative position of the missense mutation in Fah6287SB. (B) Nucleotide sequences of Fah exons 6, 7, and 8 of control BALB/cRl (line 1), mutant 6287SB (line 2), and mutant 5961SB (line 3). Mutant Fah6287SB has a point mutation at nucleotide 602, as indicated by the “g” in exon 6 in line 2. Mutant Fah5961SB has a point mutation in the last position of exon 7, as indicated by the “a” in line 3, that destroys the consensus splice donor sequence.
Biochemical Abnormality in Fah5961SB and Fah6287SB Mutant Mice.
The presence of high concentrations of SA in urine is a specific abnormality caused by FAH deficiency (6, 7). SA levels in Fah5961SB and Fah6287SB hemizygous mutants were compared with those of control mice of the same age. Table 2 shows mean SA values pooled by age and mutation. In all cases, mutants clearly had greater SA concentrations than like-aged controls. Wild-type (cch +/cch +) and heterozygous (cch +/c m) adults for either mutation have comparable urine-SA levels (76 ng/ml and 64 ng/ml, respectively; data not shown).
Table 2.
Levels of SA detected in urine of control and mutant mice
| Mutation | Genotype* | Age at sampling | SA, ng/ml | Range |
|---|---|---|---|---|
| 6287SB (control) | cch +/c + or cch +/Del | 7–9 days (n = 1)† | 44 | — |
| 6287SB (mutant) | c m/Del | 7–9 days (n = 4) | 2,329 | 1,197–3,138 |
| 6287SB (control) | cch +/c + or cch +/Del | 11 days (n = 4) | 80 | 0–170 |
| 6287SB (mutant) | c m/Del | 11 days (n = 4) | 1,715 | 605–2,867 |
| 5961SB (control) | cch +/c + or cch +/Del | 1 day (n = 3) | 43 | 0–71 |
| 5961SB (mutant) | c m/Del | 1 day (n = 6) | 552 | 347–837 |
cch, chinchilla allele at c; m, mutation; Del, c26DT.
n, number of animals in sample group.
Discussion
The ENU-induced mutations Fah5961SB and Fah6287SB cause Fah deficiency that leads to neonatal and juvenile death in mice, respectively. The acute form of human HT1 is characterized by severe symptoms caused by the lack of FAH enzymatic activity and is modeled by the phenotype of Fah5961SB; the phenotype of chronic patients who have milder features and reduced FAH levels (6) is modeled by the milder phenotype of Fah6287SB mice. Nonetheless, in either form, HT1 is a severe liver disease caused by a deficiency in FAH, and patients usually succumb to generalized liver failure, disease-related complications, and death (1, 6).
The more severe Fah5961SB allele has a mutation in a consensus sequence required for correct precursor-mRNA splicing, resulting in aberrant splicing with the loss of exon 7, an altered translational reading frame caused by the deletion of 100 bp of mRNA, and the introduction of a premature termination codon downstream of the skipped exon. Our presumption is that the change from a G to an A at the end of exon 7 causes skipping of exon 7, because it affects the consensus donor-site sequence at the exon 7–intron 7 boundary; this presumption is confirmed by the direct sequencing of the Fah5961SB cDNA, proving the absence of exon 7. A similar mutation, a G-to-C transversion in the splice donor site in exon 5 in the steroidogenic acute regulatory (StAR) gene in a patient with congenital lipoid adrenal hyperplasia, was proven to result in the skipping of exon 5, a premature stop codon, and an inactive protein product (34). In this case, as in the case of Fah5961SB, it appears that the wild-type G at the exon–intron boundary is essential to the integrity of the sequence context at the splice donor site, and that alteration to A in Fah5961SB or a C in the StAR gene results in incorrect splicing and a missing exon.
An altered reading frame and a premature stop codon usually result in the production of truncated protein and a reduction in mRNA abundance (35, 36), because misspliced RNAs are often unstable, and RNAs that harbor premature stop codons usually are degraded by cell-defense mechanisms developed to preclude the formation of deleterious proteins (36). The Fah5961SB mutation was analyzed for its effect on the level and processing of the Fah mRNA, and little or no Fah mRNA could be detected by Northern blot analysis. The low-template Fah-mRNA levels should have a direct effect on Fah protein abundance, and even if the remaining abnormal transcripts from the Fah5961SB mutation were translated, it likely would result in a structurally and/or functionally abnormal protein.
In the Fah6287SB allele, a missense transition mutation (A→G) in exon 6 was identified that changes a glutamic acid to a glycine at codon 201 (E201G). This glutamic acid 201 is conserved in human, rat, and mouse Fah, which may reflect an essential requirement for this amino acid at that position. Glutamic acid has a very polar acidic side chain; glutamic acid is found nearly always on the outside of protein molecules, whereas glycine is found commonly to the inside of proteins. This substitution could adversely affect protein stability and/or enzyme activity of the Fah protein. The role of residue 201 has not been investigated; however, the Fah6287SB allele indicates, by the loss of function consequent to this missense mutation, that this amino acid most likely plays a role in the normal function of the enzyme. Alternatively, this missense mutation could have a more global effect on the tertiary structure of Fah, thus compromising enzyme function. This E201G mutation is located only six amino acids downstream from the location of a missense mutation known to cause HT1 (G207D; ref. 17), which may suggest that the region represents a critical domain in the polypeptide. The Fah6287SB mutants can live into the 3rd week of life, suggesting the presence of at least some Fah activity and thus making Fah6287SB a murine model for the chronic form of HT1.
In humans, the primary deficiency of FAH leads to the accumulation of the alkylating metabolites maleylacetoacetate and fumarylacetoacetate, which are believed to be responsible for the liver and kidney damage found in HT1 (1, 6). Reduction and decarboxylation of these metabolites in vivo results in the production of SA. SA accumulation is thus the indirect result of the complete or incomplete block of the FAH enzyme, and SA accumulation has been linked to certain pathological aspects of the disorder (1, 2, 37, 38). Elevation of SA above normal levels (in serum, urine, and/or amniotic fluid) is the specific diagnostic indicator for HT1 (6, 7, 9, 37, 38).
The increased levels of SA in the urine of Fah5961SB and Fah6287SB mice confirm that the mutations are affecting Fah activity and causing an accumulation of the toxic metabolites fumarylacetoacetate and maleylacetoacetate. Variation in SA levels existed between animals, which is consistent with the finding that SA levels in HT1 patients can vary considerably (8, 39). Our SA data also suggest that lethality in Fah-deficient mice may not result directly from the amount of toxic metabolite accumulation (which is related to SA levels), because SA levels in Fah5961SB mice are, on average, considerably lower just hours before death than the levels found in Fah6287SB mice that live much longer. It may also be that lethality and SA phenotype are related in a more complex way not addressed by the experiments reported here. Mice heterozygous for either mutation do not exhibit an increase in SA levels, confirming that a rise in SA accompanies the disease state (40).
We propose the ENU-generated mutants Fah5961SB and Fah6287SB as mouse models for the acute and chronic forms of HT1, respectively; all phenotypic effects in these mice are attributable to FAH deficiency alone. The Fah5961SB and Fah6287SB mice represent model systems in which to assess the therapeutic effects of drugs and diets on disease progression and expression and to evaluate the efficacy of gene-therapy approaches to treating the disease.
Acknowledgments
Research was sponsored by the Office of Biological and Environmental Research, U.S. Department of Energy and managed by University of Tennessee-Batelle, LLC, under contract DE-AC05-00OR22725.
Abbreviations
- HT1
hereditary tyrosinemia type 1
- FAH
fumarylacetoacetate hydrolase
- SA
succinylacetone
- ENU
N-ethyl-N-nitrosourea
- RT
reverse transcription
References
- 1.Goldsmith L A, Laberge C. The Metabolic Basis of Inherited Disease. 6th Ed. Vol. 1. New York: McGraw–Hill; 1989. pp. 547–562. [Google Scholar]
- 2.Berger R, van Faassen H, Smith G P A. Clin Chim Acta. 1983;134:129–141. doi: 10.1016/0009-8981(83)90191-2. [DOI] [PubMed] [Google Scholar]
- 3.Overturf K, Al-Dhalimy M, Tanguay R, Brantly M, Ou C, Finegold M, Grompe M. Nat Genet. 1996;12:266–273. doi: 10.1038/ng0396-266. [DOI] [PubMed] [Google Scholar]
- 4.Tanguay R M, Valet J P, Lescault A, Duband J L, Laberge C, Lettre F, Plante M. Am J Hum Genet. 1990;47:308–316. [PMC free article] [PubMed] [Google Scholar]
- 5.Labelle Y, Phaneuf D, Leclerc B, Tanguay R M. Hum Mol Genet. 1993;2:941–946. doi: 10.1093/hmg/2.7.941. [DOI] [PubMed] [Google Scholar]
- 6.Kvittingen E A. Scand J Clin Lab Invest. 1986;46:27–34. [PubMed] [Google Scholar]
- 7.Pettit B R, MacKenzie F, King G S. J Inherited Metab Dis. 1984;7:135–136. doi: 10.1007/978-94-009-5612-4_42. [DOI] [PubMed] [Google Scholar]
- 8.Grenier A, Lescault A, Laberge C, Gagne R, Mamer O. Clin Chim Acta. 1982;123:93–99. doi: 10.1016/0009-8981(82)90117-6. [DOI] [PubMed] [Google Scholar]
- 9.Linstedt S, Holme E, Lock E A, Hjalmarson O, Strandvik B. Lancet. 1992;340:813–817. doi: 10.1016/0140-6736(92)92685-9. [DOI] [PubMed] [Google Scholar]
- 10.Phaneuf D, Labelle T, Berube D, Arden K, Cavenee W, Gagne R, Tanguay R M. Am J Hum Genet. 1991;4:525–535. [PMC free article] [PubMed] [Google Scholar]
- 11.Phaneuf D, Lambert M, Laframboise R, Mitchell G, Lettre F, Tanguay R M. J Clin Invest. 1992;90:1185–1192. doi: 10.1172/JCI115979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rootwelt H, Chou J, Gahl W A, Berger R, Coskun T, Brodtkorb E, Kvittingen E A. Hum Genet. 1994;93:615–619. doi: 10.1007/BF00201558. [DOI] [PubMed] [Google Scholar]
- 13.Rootwelt H, Kristensen T, Berger R, Hoie K, Kvittingen E A. Hum Genet. 1994;94:235–239. doi: 10.1007/BF00208276. [DOI] [PubMed] [Google Scholar]
- 14.St-Louis M, Leclerc B, Laine J, Salo M K, Holmberg C, Tanguay R M. Hum Mol Genet. 1994;3:69–72. doi: 10.1093/hmg/3.1.69. [DOI] [PubMed] [Google Scholar]
- 15.St-Louis M, Poudrier J, Phaneuf D, Leclerc B, Laframboise R, Tanguay R M. Hum Mol Genet. 1995;4:319–320. doi: 10.1093/hmg/4.2.319. [DOI] [PubMed] [Google Scholar]
- 16.Ploos van Amstel J K, Bergman A J I, van Beurden E A C M, Roijers J F M, Peelen T, van den Berg I E T, Poll-The B T, Kvittingen E A, Berger R. Hum Genet. 1996;97:51–59. doi: 10.1007/BF00218833. [DOI] [PubMed] [Google Scholar]
- 17.Timmers C, Grompe M. Hum Mutat. 1996;7:367–369. doi: 10.1002/(SICI)1098-1004(1996)7:4<367::AID-HUMU14>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 18.Grompe M, Al-Dhalimy M, Finegold M, Ou C, Burlingame T, Kennaway N G, Soriano P. Genes Dev. 1993;7:2298–2307. doi: 10.1101/gad.7.12a.2298. [DOI] [PubMed] [Google Scholar]
- 19.Gluecksohn-Waelsch S. Cell. 1979;16:225–237. doi: 10.1016/0092-8674(79)90001-1. [DOI] [PubMed] [Google Scholar]
- 20.Russell L B, Montgomery C S, Raymer G D. Genetics. 1982;100:427–453. doi: 10.1093/genetics/100.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelsey G, Schedl A, Ruppert S, Niswander L, Magnuson T, Klebig M L, Rinchik E M, Schutz G. Genomics. 1992;14:275–287. doi: 10.1016/s0888-7543(05)80217-4. [DOI] [PubMed] [Google Scholar]
- 22.Schedl A, Ruppert S, Kelsey G, Thies E, Niswander L, Magnuson T, Klebig M L, Rinchik E M, Schutz G. Genomics. 1992;14:288–297. doi: 10.1016/s0888-7543(05)80218-6. [DOI] [PubMed] [Google Scholar]
- 23.Klebig M L, Russell L B, Rinchik E M. Proc Natl Acad Sci USA. 1992;89:1363–1367. doi: 10.1073/pnas.89.4.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruppert S, Kelsey G, Schedl A, Schmid E, Thies E, Schutz G. Genes Dev. 1992;6:1430–1443. doi: 10.1101/gad.6.8.1430. [DOI] [PubMed] [Google Scholar]
- 25.Kelsey G, Ruppert S, Beermann F, Grund C, Tanguay R M, Schutz G. Genes Dev. 1993;7:2285–2297. doi: 10.1101/gad.7.12a.2285. [DOI] [PubMed] [Google Scholar]
- 26.Russell W L, Kelley E M, Hunsicker P R, Bangham J W, Maddux S C, Phipps E L. Proc Natl Acad Sci USA. 1979;76:5818–5819. doi: 10.1073/pnas.76.11.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rinchik E M, Carpenter D A, Selby P B. Proc Natl Acad Sci USA. 1990;87:896–900. doi: 10.1073/pnas.87.3.896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rinchik E M, Carpenter D A. Genetics. 1999;152:373–383. doi: 10.1093/genetics/152.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maniatis T, Fritsch E F, Sambrook J, editors. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. [Google Scholar]
- 30.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current Protocols in Molecular Biology. New York: Wiley; 1989. [Google Scholar]
- 31.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tuchman M, Whitley C B, Ramnamraine M L, Bowers L D, Fregien K D, Krivit W. J Chromatogr Sci. 1984;22:211–215. doi: 10.1093/chromsci/22.5.211. [DOI] [PubMed] [Google Scholar]
- 33.Schierbeek H, Berger R. Clin Chim Acta. 1989;184:243–250. doi: 10.1016/0009-8981(89)90057-0. [DOI] [PubMed] [Google Scholar]
- 34.Katsumata N, Kawada Y, Yamamoto Y, Noda M, Nimura A, Horikawa R, Tanaka T. J Clin Endocrinol Metab. 1999;84:3983–3987. doi: 10.1210/jcem.84.11.6118. [DOI] [PubMed] [Google Scholar]
- 35.Dietz H C, Valle D, Francomano C A, Kendzior R J, Jr, Pyeritz R E, Cutting G R. Science. 1993;259:680–683. doi: 10.1126/science.8430317. [DOI] [PubMed] [Google Scholar]
- 36.Maquat L E. Am J Hum Genet. 1996;59:279–286. [PMC free article] [PubMed] [Google Scholar]
- 37.Sassa A, Kappas A. J Clin Invest. 1983;71:625–634. doi: 10.1172/JCI110809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitchell G. N Engl J Med. 1990;322:432–437. doi: 10.1056/NEJM199002153220704. [DOI] [PubMed] [Google Scholar]
- 39.Divry P, Vianey-Liaud C, Cotte J. Biomed Environ Mass Spectrom. 1987;14:663–668. doi: 10.1002/bms.1200141117. [DOI] [PubMed] [Google Scholar]
- 40.Tuchman M, Freese D K, Sharp H L, Whitley C B, Ramnaraine M L, Ulstrom R A, Najarian J S, Ascher N. J Inherited Metab Dis. 1985;8:21–24. doi: 10.1007/BF01805479. [DOI] [PubMed] [Google Scholar]