Abstract
Homo(purine•pyrimidine) sequences (R•Y tracts) with mirror repeat symmetries form stable triplexes that block replication and transcription and promote genetic rearrangements. A systematic search was conducted to map the location of the longest R•Y tracts in the human genome in order to assess their potential function(s). The 814 R•Y tracts with ≥250 uninterrupted base pairs were preferentially clustered in the pseudoautosomal region of the sex chromosomes and located in the introns of 228 annotated genes whose protein products were associated with functions at the cell membrane. These genes were highly expressed in the brain and particularly in genes associated with susceptibility to mental disorders, such as schizophrenia. The set of 1957 genes harboring the 2886 R•Y tracts with ≥100 uninterrupted base pairs was additionally enriched in proteins associated with phosphorylation, signal transduction, development and morphogenesis. Comparisons of the ≥250 bp R•Y tracts in the mouse and chimpanzee genomes indicated that these sequences have mutated faster than the surrounding regions and are longer in humans than in chimpanzees. These results support a role for long R•Y tracts in promoting recombination and genome diversity during evolution through destabilization of chromosomal DNA, thereby inducing repair and mutation.
INTRODUCTION
Chromosomal DNA exists principally as a right-handed double helix (B-DNA). However, other conformations, such as triplexes, tetraplexes, slipped structures with hairpin loops, left-handed Z-DNA and cruciforms are also known [reviewed in (1–7)]. These alternative (non-B DNA) conformations are formed at specific sequence motifs and are therefore possible only at discrete chromosomal locations. More than 15 genomic disorders, including neurofibromatosis type I, chronic myeloid leukemia, spermatogenetic failure and recurrent constitutional translocations, have recently been associated with rearrangements mediated by recombination between blocks of repetitive DNA (from a few hundred base pairs to several hundred kilobase pairs in length) almost exclusively composed of direct repeats (DR) and inverted repeats (IR) [reviewed in (8)]. Since double-strand breaks (DSBs) are localized at hotspots within these blocks in most cases, factors other than the primary DNA sequence must be involved, and indeed the locations of the rearrangement fusions are generally found at sequences known to adopt non-B DNA conformations (8–16). This conclusion is further supported by the finding that translocation frequencies correlate with genetic variations in the general population affecting the stability of the putative non-B DNA conformations at breakpoints (16).
Triplex DNA requires homo(purine•pyrimidine) sequences (R•Y tracts) and is stabilized by Hoogsteen hydrogen bonds between the purines in the Watson–Crick duplex and a third strand in the major groove, which may be composed of either pyrimidines bound in the parallel orientation (YRY triplexes) or purines (RRY triplexes) bound in the antiparallel orientation (1,3,6,17–19). Specific interactions consist of T-A•T and C+-G•C triads for YRY triplexes, and G-G•C and A-A•T triads for RRY triplexes; hence, mirror repeat symmetries within R•Y tracts yield fully paired and stable triplexes. The YRY triplexes are additionally stabilized by cytosine protonation at N3, a process that requires low pH in nucleotides, but which occurs cooperatively and at neutral pH in clustered cytosines in a long polynucleotide chain (20–22). Finally, the third strand may be provided by the folding back of a single R•Y tract with mirror repeat symmetry (intramolecular triplex), by the interaction between two R•Y tracts separated by some distance on the same or on two different DNA molecules (intermolecular triplex), or by a single-stranded oligonucleotide composed of either DNA or RNA (6,19,23).
R•Y tracts are genetically unstable in experimental model systems. Whereas numerous biological functions have been attributed to triplexes, including the blockage of DNA replication (24–26) and the interference with transcription (26,27), recent studies have provided revealing insights. First, studies conducted in live mice, mammalian cell cultures and Escherichia coli indicate that R•Y tracts are highly mutagenic (28–30), induce DSBs (30,31) and are frequently found at the breakpoint sites of gross deletions and other rearrangements (9,31). Indeed, three types of biochemical and genetic studies have shown that these genomic instabilities were due to the non-B conformations adopted by the R•Y tracts and not to the tracts in their orthodox right-handed B-form (32). Second, long GAA•TTC repeats in the first intron of the frataxin (FXN) gene adopt several structures including triplex and sticky DNA, which inhibit transcription of the gene thus reducing the expression of frataxin (26,33,34). Hence, triplexes may be involved in the etiology of Friedreich's ataxia. Third, triplexes may play critical roles in the bcl-2 major breakpoint region with respect to the RAG-dependent t(14;18) translocation associated with follicular lymphomas (35–37). Fourth, R•Y tracts elicit a biological response in the context of mammalian chromatin also in the absence of perfect mirror repeat symmetry and at moderate-to-short lengths (∼20 bp or less) (35–38), indicating that the energetic barrier associated with the duplex-to-triplex structural transitions may be easily overcome.
Long (several hundred base pairs) R•Y tracts in the human genome have been known for >20 years (6,17,39,40), and previous limited studies suggested their abundance in mammalian genomes (41–43). To date, no genome-wide queries, which might be informative with respect to the potential function(s) of these sequences, have been reported. Knowledge about the size and location of non-B DNA conformations in vertebrate genomes would be expected to give critical clues as to their biological functions. The human genome has so far only been surveyed for IR sequences (44), which can form cruciforms. Warburton et al. (44) reported that the large IR may have a role in testis gene expression and genome integrity.
Herein, we have applied a data-mining approach to conduct the first systematic search for the longest uninterrupted R•Y tracts in the human genome and tested specific hypotheses by employing experimental methodologies in silico. We show that these sequences cluster specifically in the pseudoautosomal region of the sex chromosomes and are found predominantly in genes that are highly expressed in the brain. These determinations, along with comparative analyses of the mouse and chimpanzee genomes, indicate that long R•Y tracts constitute mutational hotspots and are likely to have played a key role in genome plasticity and evolution.
MATERIALS AND METHODS
Computer searches
All R•Y tracts were found using the program PTRfinder (45). The algorithm first maps the DNA sequence to R or Y for purine and pyrimidine, respectively. Then the program locates tandem repeats with this reduced alphabet and given the minimum lengths input by the user. The results were then imported into the Genomic Resource Information Database (GRID, http://grid.abcc.ncifcrf.gov/) (J. R. Collins, R. M. Stephens and J. Shan, manuscript in preparation) for web access and for generating queries to correlate R•Y tracts with gene location and overlap. Genomic build parameters describing tracts of pure R or Y were extracted from the GRID database using the dataset from the Human Genome Browser at the University of California, Santa Cruz (UCSC), which was based on the hg17, NCBI Build 35, May 2004 assembly. The following SQL query was used to retrieve all tracts of type R or Y with length ≥250 bp: ‘SELECT * from ‘pupy’ WHERE ‘len’>=250 AND length(‘type’)=1 ORDER BY ‘chrom’, ‘len’’. This query retrieved 818 records, 814 with fully determined coordinates, which were imported into MSExcel spreadsheets. An additional table column with 814 links to the UCSC Human Genome Browser was created using a URL pattern to link each record to the exact genomic position. By opening the links, each record was visually checked for being within or between gene sequences. Thus, 241 of the 814 tracts were found to occur within annotated genes. Of the 228 non-redundant R•Y tract-containing genes, information was gathered for 156 genes, either through OMIM or PubMed (http://www.ncbi.nlm.nih.gov).
In order to compare the 814 tracts and their surrounding sequences in human with their orthologous counterparts in chimpanzee (Pan troglodytes), five PERL (Practical Extraction and Reporting Language) scripts were written. A standalone BLAT (Blast-like Alignment Tool) program was downloaded from http://www.soe.ucsc.edu/~kent/src/ and installed on a computer with Linux FC3. The human and chimpanzee (NCBI Build 1 version 1, November 2003) genomes were downloaded from the UCSC website. In addition, the free package CLUSTAL W 1.83 and the EMBOSS application DOTMATCHER (Ian Longden, Sanger Institute, Cambridge, UK), which is under GNU license (http://www.gnu.org), were downloaded and installed.
The first PERL script ‘1_queryTOfasta.pl’ processed the 814-record table containing the repeat data. Coordinates were transformed so as to include 2500 bp of flanking sequences from both sides of each tract. For each record, the respective DNA sequence was extracted from the local human genome files and added to a file in FASTA format containing all records. A second PERL script ‘2_saBLATfastaVSchromosomes.pl’ automated the BLAST search of every record against the local chimpanzee genome. To speed up this process, a previously made chimpanzee ‘11.ooc’ R or Y tracts file was used (for details, see BLAT FAQs and tutorials at the website previously cited). The output was a directory with BLAT-hits results in chimpanzee in PLSX table format, one file for each record. A third PERL script ‘selectFROMfile.pl’ processed the PLSX-files directory picking up the best score from each file and merging all the best-scores into a single SELECT file, which also contained 814 records. A fourth PERL script ‘4_plsxTOfastaCHR.pl’ processed the 814-record SELECT table in the same way that the first script processed the GRID-database table. It extracted from the local chimpanzee genome each corresponding segment of DNA sequence and added them to a FASTA file. The product of running the first through fourth scripts are two multiple entry DNA FASTA files; one with human queries and the other with chimpanzee matches. A fifth script ‘5_html_u.pl’ compared all query and match sequences from the two files above. It also ran CLUSTAL W and DOTMATCHER for each of the pairs of records. Subsequently, it created an HTML document presenting the results. For ease in troubleshooting, the job was performed in several steps using separated scripts, even though it could readily be integrated into one. The results for the R•Y tracts present within genes have been posted at the following website: http://home.ncifcrf.gov/ccr/lgd/dean_lab/pure_R_or_Y_Comparison.
Similarly, 5 kb human DNA fragments containing the R•Y tracts centrally were used as queries in BLAT (http://genome.ucsc.edu/cgi-bin/hgBlat) searches on the mouse (Mus musculus, Mm5, NCBI build 33, May 2004), macaque (Macaca mulatta, NCBI, Mmul_0.1, January 2005) and dog (Canis familiaris, NCBI, v1.0, July 2004) genomes. Genomic interspersed repeats were identified by RepeatMasker (http://www.repeatmasker.org).
Tissue expression of R•Y tract-containing genes
Tissue gene expression data were downloaded from the Genomic Institute of the Novartis Research Foundation (GNF, http://web.gnf.org/index.shtml), and comprised the Affymetrix U133A chip (∼22 000 probe sets) plus a custom GNF Affymetrix chip with ∼11 000 probe sets analyzed on 79 human tissues and cell lines. Approximately 17 000 known human genes were mapped to these probe sets and their relative expression levels were analyzed in the 79 tissue types to identify genes preferentially expressed in each tissue. Preferential expressed genes are defined as follows: the expression levels of a given gene across all tissues were transformed to a Z-score and genes with a Z-score > 1 (i.e. >1 SD above the mean) in a given tissue were classified as highly expressed in that tissue. The Z-score was averaged when more than one probe set hybridized to the same gene and for duplicate tissues. A total of ∼1 390 000 Z-score values for all genes and all tissues were obtained, ∼10% of which qualified as highly expressed. P-values were obtained by comparing the fraction of the ∼17 000 genes highly expressed in a given tissue with the fraction observed for the set of genes containing either the pure R•Y tracts ≥250 bp or the pure R•Y tracts ≥100 bp in length, assuming a binomial distribution. P-values were corrected (Bonferroni) for multiple comparison of 79 tissues.
Functional category enrichment analysis and creation of gene-term association networks (GTANs)
Functional category enrichment analyses were performed using a software tool, WholePathwayScope (WPS) (46), developed at the ABCC (NCI-Frederick, MD). Briefly, this analysis is based on Fisher Exact Test for 2 × 2 contingency tables (gene list versus functional category) to estimate and rank the statistical significance of the enrichment of functional categories within a given system (GO terms, BioCarta pathways, KEGG pathways, gene-disease associations, protein interaction partners and protein families) for genes harboring ≥250 pure R•Y tracts (RY250) or ≥100 pure R•Y tracts (RY100). The databases used, which were collected in the WPS database, were as follows: the GO terms and gene-GO term association tables were downloaded from the Gene Ontology Consortium (http://www.geneontology.org); the BioCarta pathways were kindly provided by the CGAP group (Cancer Genome Anatomy Project: http://cgap.nci.nih.gov/), which originated from the Bio Carta pathway collections (http://www.biocarta.com/genes/allPathways.asp); the KEGG pathways were downloaded from the KEGG pathway collections (Kyoto Encyclopedia of Genes and Genomes, http://www.genome.ad.jp/kegg); the gene disease associations were downloaded from the Genetic Association Database (http://geneticassociationdb.nih.gov/); information on Protein Interaction Partners and Protein families (Pfam) information was kindly provided by the DAVID (Database for Annotation, Visualization and Integrated Discovery) group (http://david.niaid.nih.gov). Gene-Term Association Networks (GTANs) were created within WPS, such that for any given gene in a gene list (RY100 or RY250), its associated term(s) was sought in the WPS database. The pairwise gene-term relationships were represented as a graphical layout of a Gene-Term Association Network within WPS, in which any gene and its associated term were linked with an edge to indicate the gene-term association relationship.
Gene size
The sizes for annotated genes were obtained from the lengths of pre-mRNA transcripts, which were downloaded from the National Center for Biotechnology Information (NCBI) website (ftp://ftp.ncbi.nih.gov/genomes/H_sapiens/RNA/). The set of 1377 genes used to determine the size distribution of genes expressed in brain was obtained from the tissue distribution analysis, with genes with a Z-score > 1 in brain tissues being selected. In most cases, the SigmaPlot 2002 program, version 8.02 (SPSS, Chicago, IL) was used to represent the results graphically and to determine the best fit to the data.
Clustering of R•Y tracts
The order statistics, r-scans, as described in Karlin and Macken (47), were used to detect significant clustering of the R•Y tracts observed along each chromosome, by comparing their distribution with that of a uniform Poisson distribution. We assume that {Xi} are the distances between adjacent R•Y tracts (n tracts in total) along an individual chromosome that are not necessarily independently and identically distributed (iid). Note that all points are mapped into a [0,1] interval. Denote by
the distance from the j-th R•Y tract to its r-th nearest neighbor. There is an order statistic associated with the sequence of partial sums
and a minimum defined as
If we assume that distances {Xi} are iid, then according to Karlin and Macken (47),
We declare significant clustering when the observed minimum for the limit distribution has <0.01 (or <0.05) probability of occurring by chance and therefore
Several R•Y tract clusters were detected (after correcting for adjacent tracts separated by single nucleotide interruptions) by testing the first minimum of r-scans (where r = 1, … , 16).
Complexity analysis
Complexity analysis, as devised by Gusev et al. (48), was employed in a search for DR, IR and mirror repeats (MR) in the pseudoautosomal region (PAR1) and the downstream 4.4 Mb region (After_PAR), in the 5 kb sequences flanking the R•Y tracts in the PRKCB1 and ADAM18 genes, in the 3′-untranslated region (3′-UTR) of the DISC1 gene and in the comparative analyses of the R•Y tracts in the mammalian TIAM1 gene.
RESULTS
R•Y tracts within genes
The human genome was screened for the largest R•Y tracts with the expectation that these could shed light on their potential biological role(s). The total number of tracts equaling or exceeding an arbitrary length cutoff of 250 pure R (adenine, A and guanine, G) or Y (cytosine, C and thymine, T) bases was 814. Approximately 30% (241/814) were located within annotated genes (228, some genes contained more than one tract, Supplementary Table 1), all within introns. The longest tract (1303 bp) was present in the CENTA1 gene on chromosome 7, which was the only pure intragenic sequence to exceed 1 kb. Tracts were then analyzed to determine the R•Y tract length if occasional interruptions (mostly single nt changes) were ignored. The 2.5 kb PKD1 R•Y tract (49), which has been instrumental in the characterization of the mutational properties of R•Y sequences (9,25,32), was the third longest, preceded only by a 3.1 kb tract in the DKFZp434G0625 gene and a tract of nearly 4 kb in the LOC348094 gene (both encoding products of unknown function). Hence, the PKD1 gene possesses the longest gene-associated R•Y tract in the human genome for genes of known function. A double logarithmic plot of these tracts arranged in order of size exhibited a linear relationship (y = 3.499 – 0.458x, r2 = 0.99, data not shown), indicating an exponential decay distribution. This distribution suggests that selection against or in favor of any R•Y tract length is not exercised to a detectable level within the existing envelope.
Complexity analysis was employed to identify MR separated by any distance within these R•Y tracts. The average fractions of non-redundant MR elements (with no interruptions) per R•Y tract were 16, 4 and 2 for MR lengths of 10, 20 and 30 bp, respectively. In addition, all R•Y tracts had at least one 10 bp-long MR element. Hence, based on the work conducted on the PKD1 and other R•Y tracts in vivo (9,28,30,32,35,37,50,51), we conclude that most, if not all, R•Y tracts with ≥250 uninterrupted base pairs have the potential to form intramolecular and/or intermolecular triplexes in vivo. However, future work will be required to experimentally evaluate triplex formation by these R•Y tracts.
To determine whether there was preferential association within certain categories of genes, we compared the set of R•Y tract-containing genes with that of the human (reference) dataset in proteomic databases (Supplementary Table 2). For the Gene Ontology (GO) Molecular Function, eight terms exceeded a P-value of 10−4 and included seven terms for channel activities and one term for glutamate receptor activity, consistent with strong enrichment for genes encoding transmembrane proteins in the brain (Supplementary Table 2A). The GO Biological Process analysis revealed 13 terms which exceeded P-values of 10−4, and included three terms for cell adhesion and cell communication, four for neuronal function and three for ion transport (Supplementary Table 2B), indicative of a preferential distribution within genes involved in specialized functions at the cell membrane. The GO Cellular Component analysis (Supplementary Table 2C) yielded four terms that exceeded P-values of 10−4 and which were associated with localization to the cell membrane.
The fibronectin type III, C-cadherin and α-catenin domains, present in many cell surface receptors and cell adhesion molecules, represented the most highly enriched categories (P-values ∼10−4) in the Protein Families (Pfam) and Protein Information Resource (PIR) databases (Supplementary Tables 2D and 2E), whereas eight terms including the large neuroactive ligand interaction pathway, which comprises an array of neuronal receptors implicated in neuropeptide and small molecule signaling pathways, were found to be enriched in the more limited BioCarta and Kyoto Encyclopedia of Genes and Genomes databases (Supplementary Tables 2F and 2G). Analysis of the Protein Interaction Partners database (Supplementary Table 2H) indicated 34/91 enrichments led by DLG4 (psd-95, P ∼ 8 × 10−4), a component of the post-synaptic density structure involved in receptor clustering. Finally, analysis of the Disease Association database indicated that the most significant enrichment (P ∼ 5 × 10−3) was for candidate genes for schizophrenia susceptibility (Supplementary Table 2I). In summary, the longest R•Y tracts were non-randomly distributed, and were disproportionately associated with genes whose products are localized to the plasma membrane and which perform cell communication and transport functions.
Distinct gene categories
The ≥250 bp R•Y tract-containing genes represented ∼1% of the annotated human gene dataset. We therefore sought to determine whether lowering the R•Y length threshold would yield a larger set of genes with the same non-random distribution profile. A search for genes with known functions containing pure R•Y tracts ≥100 bp in length yielded a total of 2886 hits and a non-redundant gene set of 1957. Comparison of the distribution of these R•Y tract-containing genes with that of the reference dataset (Supplementary Table 3) revealed that the terms most highly enriched were common to those identified for the genes containing the ≥250 R•Y tracts (Table 1). This correlation was particularly striking for gene products involved in signal transduction pathways at synapses, which also showed strong associations with susceptibility to schizophrenia (Supplementary Figures 1 and 2, Supplementary Table 4 and Supplementary Text).
Table 1.
Major enriched categories for genes with ≥ 250 bp and ≥ 100 bp R•Y
| Combined Rank | Category (term—pathway) | P-value | Fold enrichment | ||
|---|---|---|---|---|---|
| ≥250 R•Y | ≥100 R•Y | ≥250 R•Y | ≥100 R•Y | ||
| GO molecuar function | |||||
| 10 | Ion channel activity | 1.95E−05 | 5.92E−09 | 4.4 | 2.2 |
| 12 | Protein binding | 3.14E−03 | 6.25E−15 | 1.7 | 1.6 |
| 20 | Glutamate receptor activity | 6.11E−04 | 1.92E−07 | 10.3 | 4.3 |
| GO biological process | |||||
| 5 | Cell adhesion | 1.11E−04 | 3.36E−12 | 3.4 | 2.2 |
| 7 | Cell communication | 2.19E−04 | 5.24E−15 | 1.6 | 1.4 |
| 15 | Transmission of nerve impulse | 1.83E−04 | 5.24E−08 | 5.1 | 2.5 |
| GO cellular component | |||||
| 2 | Membrane | 2.78E−06 | 2.46E−14 | 1.6 | 1.3 |
| 15 | Synapse | 2.18E−02 | 7.69E−05 | 5.0 | 2.8 |
| 19 | Extracellular matrix | 4.96E−02 | 1.33E−08 | 2.3 | 2.2 |
| Pfam family | |||||
| 2 | Fibronectin type III | 2.84E−05 | 1.43E−12 | 5.1 | 2.8 |
| 15 | C-cadherin | 7.76E−05 | 3.14E−05 | 17.0 | 4.3 |
| 17 | C2 | 1.17E−04 | 3.95E−05 | 5.4 | 2.1 |
| PIR family | |||||
| 3 | α-catenin | 3.55E−04 | 1.20E−03 | 60.4 | 9.4 |
| 11 | Cadherin | 1.81E−02 | 4.04E−04 | 9.5 | 4.0 |
| KEGG pathway | |||||
| 5 | Neuroactive ligand–receptor interaction | 7.06E−03 | 2.52E−03 | 2.9 | 1.5 |
| 6 | Phosphatidylinositol signaling system | 2.30E−02 | 1.87E−05 | 4.8 | 2.6 |
| 7 | Cholera—infection | 1.26E−02 | 3.39E−03 | 6.0 | 2.2 |
| Protein interaction partners | |||||
| 22 | DLG4 | 7.98E−04 | 2.06E−03 | 45.1 | 6.1 |
| 23 | RAC1 | 8.34E−03 | 9.78E−04 | 7.1 | 2.4 |
| Disease association | |||||
| 4 | Schizophrenia | 5.23E−03 | 2.33E−03 | 7.5 | 2.6 |
Combined Rank, sum of the two categories ranks from Supplementary Tables 2 and 3; categories with one entry were excluded; when more than one category with largely redundant gene entries was present, only one was chosen.
Since P-values are sensitive to sample size, we next determined whether the terms were more enriched in ≥250 or ≥100 bp R•Y tract-containing genes based on ‘fold-enrichment’. Consistently greater enrichments were noted for the ≥250 bp R•Y tract-containing genes (Table 1). Hence, as the R•Y tract length increases, so does the probability of their association with genes involved in specific functions at the plasma membrane.
A second set of terms was found to be highly enriched in the ≥100 bp R•Y tract-containing genes but not in the ≥250 bp R•Y tract-containing genes. These included terms containing transferase and kinase activities (GO_MF) and protein/receptor phosphorylation of genes implicated in development and morphogenesis (GO_BP), implying an enrichment in genes involved in signal transduction pathways (Supplementary Figure 3 and Supplementary Text).
Hence, we conclude that the longest R•Y tracts in the human genome are distributed between two main pools of genes in a length-dependent fashion. The first group, comprising shorter length tracts (100 bp ≤ R•Y ≤ 250 bp), co-localized preferentially with genes involved in signal transduction pathways associated with development, whereas a second group, comprising longer tracts (R•Y ≥ 250 bp), co-localized with genes mostly involved in ion transport, cell adhesion and neurogenesis. Since several terms exceeded P-values of 10−10 and the different databases (particularly the large GO_MF, GO_BP, GO_CC, and Pfam Family) were in agreement in identifying terms that contained the same genes, the conclusions are strongly supported by the data.
Tissue gene expression
To determine whether the ≥250 bp R•Y tract-containing genes (250-set) and the ≥100 bp R•Y tract-containing genes (100-set) expressed a greater proportion of transcripts in specific tissues as compared with the reference dataset (All-set), we examined the Affymetrix U133A tissue expression dataset, derived from 79 tissues, from the Genomic Institute of the Novartis Research Foundation (GNF). For a given tissue, the fraction of highly expressed genes in the 250-set (and 100-set) was then compared with the fraction of highly expressed genes in the reference dataset (All-set).
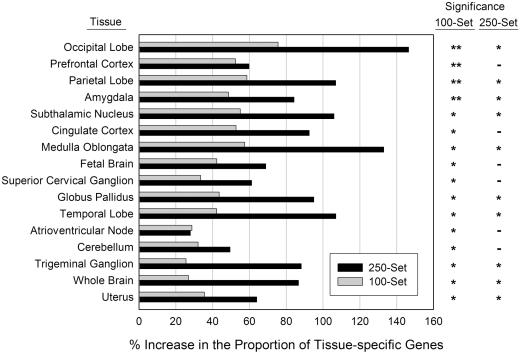
The fractions of highly expressed genes containing R•Y tracts (250-set and 100-set) were significantly greater than the corresponding fractions of the All-set in all brain tissues examined, in the atrioventricular node of the fetus and the uterus (Figure 1; P-values ranged from 4 × 10−2 to <1 × 10−13 for the 100-set and from 2 × 10−2 to 6 × 10−5 for the 250-set). These fractions were also consistently more enriched in genes with longer R•Y tracts (250-set) than with shorter ones (100-set), regardless of the P-values. In summary, these data are compatible either with the view that R•Y tracts with lengths ≥100 bp (including ≥250) co-localize preferentially with genes highly expressed in the brain, or that these tracts may perform a role in mediating gene transcription in brain tissues.
Figure 1.
Tissue distribution of R•Y tract-containing genes. The percentage increase in the fraction of R•Y-containing genes (100-set and 250-set) highly expressed (Z-score >1) in a given tissue relative to the fraction of all genes highly expressed in the same tissue is displayed on the x-axis. Asterisks, significantly enriched tissues as determined by binomial probability after a Bonferroni multiple comparison correction. Significance: **, P < 10−7; *, 10−7 ≤ P < 0.05, -, not significant.
Gene size distributions
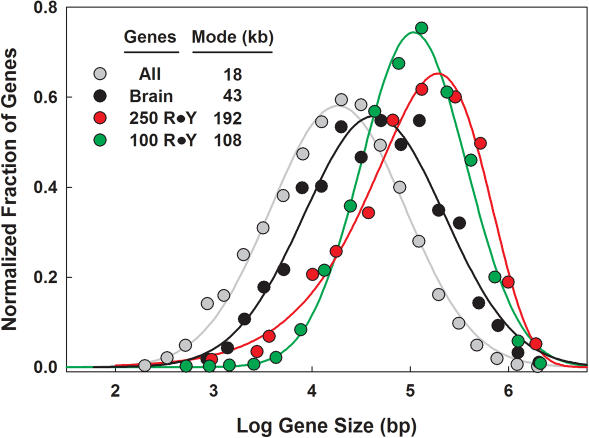
Some of the genes expressed in human brain tissues, such as contactin-associated protein-like 2 (CNTNAP2), dystrophin (DMD), and ataxin-2 binding protein (A2BP1), are among the largest known, each spanning >1.5 Mb of genomic DNA. We therefore posed the question as to whether the preferential finding of R•Y tracts in brain-expressed genes might be associated with larger gene size. The size distribution for the annotated human gene population followed a Gaussian distribution when the logarithms of gene lengths were analyzed (Figure 2, gray). According to this distribution, the average gene length is ∼18 kb (Figure 2—mode and mean coincide for Gaussian distributions). In contrast, the set of 1377 genes highly expressed in the brain yielded a Gaussian distribution peaking at ∼43 kb (black). Thus, human genes predominantly expressed in brain tissues are on average more than twice as long as those from the total gene population when their log-normal distributions are compared. This notwithstanding, the size distributions of the R•Y tract-containing genes were of disproportionately greater size, with the ≥100 bp R•Y tract-containing gene set peaking at 108 kb (green) and the ≥250 bp R•Y tract-containing gene set peaking at 192 kb (red). In addition, the ≥250 bp R•Y tract-containing genes displayed a pronounced negative skewness, and hence a non-Gaussian behavior. We therefore conclude that larger human genes also tend to host longer R•Y tracts.
Figure 2.
Relationship between gene size and the presence of R•Y tracts. The fractions of genes with log gene size falling within 0.2 (0.25 for ≥100 bp R•Y tract-containing genes and 0.3 for ≥250 bp R•Y tract-containing genes) log-intervals plotted as a function of their mean values. Gray, 22,799 human genes; black, 1377 brain-specific human genes; green, 1891 ≥100 bp R•Y tract-containing human genes; red, 200 ≥250 bp R•Y tract-containing human genes.
Next, we investigated whether larger genes also contained a greater number of R•Y tracts. By analyzing the ≥100 bp R•Y tract-containing gene set for all pure R•Y tracts ≥50 bp in length, 33 genes were found to harbor between 20 and 57 R•Y tracts (Supplementary Table 5). The size of these genes ranged from 38 kb to 2.3 Mb, with a median of ∼800 kb and an average of ∼910 kb, indicating that larger genes also tend to host more R•Y tracts, as predicted.
The gene size distributions of Figure 2 also raise the question as to whether the percent increases in the proportions of highly expressed R•Y tract-containing genes in the brain (Figure 1) could be accounted for by the overall greater length of the genes expressed in this organ. Considering that the distributions PAll, PBrain, PRY > 100 and PRY > 250 (Figure 2, gray, black, green and red lines, respectively) are interdependent and log-normal (with the exception of PRY > 250), the predicted ratio increase may be calculated from the following relationship:
where PRY is either PRY>100 or PRY>250, using the means and standard deviations that defined the respective curves. The values obtained for the ≥100 and ≥250 bp R•Y tract-containing gene sets were 1.30 and 1.32, respectively, for the integrals bound by the log gene sizes of 0 and 8 (which extends beyond all gene sizes), yielding a predicted percent increase of ∼30%. For the 250-set, all percentage increases were greater than this value of 30% for the 13 brain tissues examined (Figure 1). About 2/3 of the 100-set percentage increases in brain tissues were >30% (up to ∼75%), whereas 1/3 were close to 30%. In summary, these data indicate that the number of R•Y tract-containing genes (but not the number of R•Y tracts per kilobase pair) expressed in the brain is greater than expected based on their longer lengths.
The pseudoautosomal region
The total number of pure R•Y tracts ≥250 bp in the human genome was 814. We evaluated whether these tracts were evenly dispersed following a Poisson-like distribution throughout chromosomes, or whether instead they were clustered in specific regions. By testing each chromosome separately, four small clusters were noted on chromosomes 5, 6 and 12, each involving 2–5 tracts (Supplementary Figure 4). In contrast, two large clusters were identified on the X and Y chromosomes, of 17 and 11 tracts respectively, starting from the telomeric p-arm and extending for 6.2 and 2.6 Mb, respectively. The sequence of the first 2.6 Mb (pseudoautosomal region, or PAR1) is identical on both sex chromosomes, and is essential for homologous recombination during male meiosis and chromosome pairing (52,53). This prompted speculation that the R•Y tracts might play a role in PAR1 function by virtue of their structural properties, perhaps in concert with other non-B DNA-forming sequences, such as IR, DR and MR.
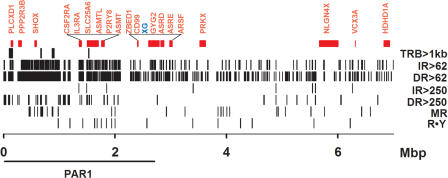
A search for pairs of DR, IR and MR with an arbitrary length cut-off of ≥62 bp in the first 7.0 Mb of the X chromosome revealed a significant (χ2 test) over-representation of DR of both ≥62 bp and ≥250 bp and IR of ≥62 bp in the PAR1 region by comparison with the 4.4 Mb region that follows (After_PAR, P < 0.0001). In contrast, MR of ≥62 bp and IR of ≥250 bp were not significantly over-represented (Figure 3 and Supplementary Text). In addition, the distribution of MR was characterized by a preponderance of (GAAA•TTTC)n motifs in PAR1 (10/11 in PAR1 and 8/15 in After-PAR). We surveyed the sequence composition of all DR and IR of ≥180 bp to determine whether they were uniquely represented in the PAR1 region or whether they corresponded to interspersed elements distributed genome-wide. No significant homology was found to any of the 201 repeats in PAR1. In contrast, 11/21 repeats in the After-PAR region exhibited extensive homologies with other chromosomal sites; indeed 9/11 tracts were identified as LINE1 (7/9), LINE1/LTR (1/9) or LTR (1/9) elements. Additional large segments of repetitive DNA were also identified in PAR1. Seven (>1 kb) were closely examined and found to comprise orderly arrays of tandem repeat blocks (TRB) containing multiple sequence motifs with few interruptions (Supplementary Figure 6). In summary, these data indicated that the clustered R•Y tracts ≥250 bp form part of a larger family of repetitive elements, mostly DR of unique composition and short IR that densely populate the PAR1 region.
Figure 3.
Repetitive elements in the pseudoautosomal region. The vertical lines represent the locations of repetitive elements in the first 7 Mbp of the human X chromosome containing the PAR1 region. R•Y, clustered R•Y repeats from Supplementary Figure 4; MR, mirror repeats ≥62 bp; DR>250, direct repeats ≥250 bp; IR>250, inverted repeats ≥250 bp; DR>62, direct repeats ≥62 bp but <250 bp; IR>62, inverted repeats ≥62 bp but <250 bp; TRB>1kb, sequences containing tandem repeat blocks >94.8% pure with a total length >1kb; red, annotated genes; XG (in blue) gene spanning the PAR1 boundary.
The (GAAA•TTTC)n repeats
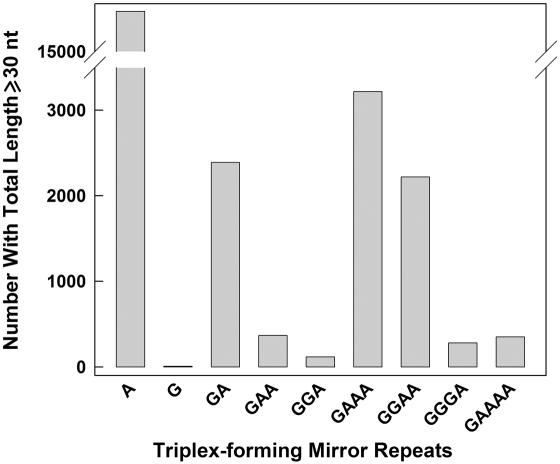
The recurrence of the (GAAA•TTTC)n repeat within PAR1 was intriguing since this sequence possesses the R•Y mirror symmetry that facilitates triplex-formation (1,3,6,17,18). In addition, its similarity to the (GAA•TTC)n motifs, which form extraordinarily stable non-B structures (26,34,54,55) raised the question as to whether these structural properties might have been functionally exploited throughout the genome. We addressed this issue by searching for R•Y tracts with mirror symmetry ≥30 bp in length (sufficient to yield stable triplexes) and comparing their frequencies with those of all microsatellites of comparable repeat units and total lengths. The search for microsatellites with unit lengths from 1 to 29 nt yielded a bimodal distribution with a first peak comprising the mono- to penta-nucleotide repeats (Supplementary Figure 5) and a second peak composed of ≥15 nt unit lengths, most of which could be decomposed into shorter repeat units displaying an even/odd length asymmetry, as previously noted for the short (1–6mer) repeats (56). Dinucleotides were the most abundant (55 196 copies), followed by tetra- (29 590 copies), mono- (16 686 copies), penta- (8699) and tri-nucleotides (6711). The distribution of R•Y tracts revealed an abundance of (A•T)n (16 679 copies) over (G•C)n (seven copies, Figure 4). At present, we do not understand this paucity of (G•C)n runs. The distributions were followed by (GAAA•TTTC)n (3217 copies), (GA•TC)n (2390 copies) and (GGAA•TTCC)n (2200 copies). All other combinations of G+A repeats contained <400 copies. Therefore, given that poly(A) sequences have the unique property of decreasing triplex stability with increasing length (57–59), these analyses (Figure 4 and Supplementary Figure 5) indicated that the (GAAA•TTTC)n repeats were over-represented with respect to the total microsatellite population, and were especially abundant among the triplex-forming motifs with mirror symmetry. However, the role, if any, of such tracts and the underlying ability to form intra- or intermolecular triplexes remains to be determined.
Figure 4.
Number of Triplex-forming sequences with mirror symmetry in the human genome. Gray bars, total numbers of uninterrupted R•Y tracts with mirror symmetry ≥30 bp in length.
Evolutionary comparisons
To determine whether long pure R•Y tracts (≥250 bp) are unique to human, we queried the chimpanzee, dog, mouse, rat and chicken genomes. Such R•Y tracts lengths were found to be common for all the mammalian/avian species examined (Supplementary Table 6). In addition, the number of pure sequences ≥50 bp varied from ∼8000 in chicken to >100 000 in murids, attesting to their abundance. A search of the mouse genome for genes with ≥250 bp R•Y tracts returned a set of 827 non-redundant entries and indicated that the main functional enrichment categories (Supplementary Table 7) largely overlapped those of the ≥100 bp R•Y tract-containing human gene set (Supplementary Table 3). This indicates that R•Y tracts have been retained within the same gene families since human–mouse divergence ∼80 million years ago (Mya), and also that human membrane-associated genes (Supplementary Table 2) highly expressed in brain have maintained longer R•Y tract lengths than other gene families.
This notwithstanding, little or no homology was found between human and mouse orthologous genes when 5 kb fragments flanking the ≥250 bp R•Y tracts of the 228 human genes (Supplementary Table 1) were compared. Similarly, analysis of the human and mouse orthologous genes which contained ≥250 bp R•Y tracts in both species (24 total) revealed little conservation in terms of either the number or location of the tracts (Supplementary Table 8). Thus, whereas R•Y tracts have been retained within gene families, their lengths and locations within individual genes have diverged substantially.
Human–chimpanzee orthologous R•Y tracts
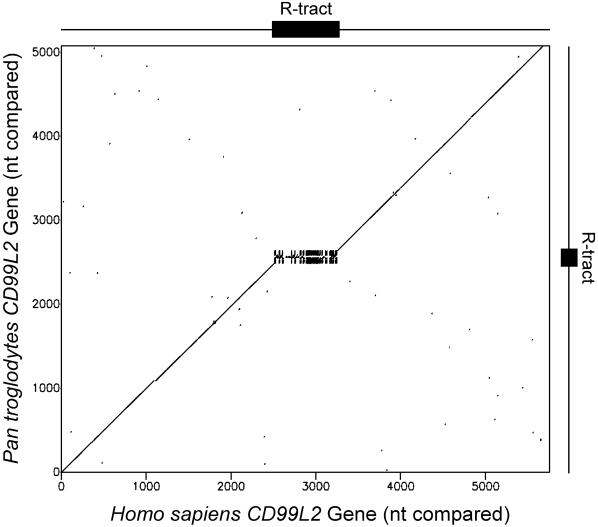
To determine more accurately the extent of sequence conservation, the human intragenic R•Y tracts ≥250 bp, together with ±2.5 kb of flanking sequence, were compared with their orthologous sites in the chimpanzee genome. These species diverged ∼5–8 Mya from their last common ancestor (LCA) and still share ∼98% sequence identity. Orthologous sequences were identified for most of the 239 tracts (Supplementary Text). A typical dot-plot depicting a comparison of 5 kb of the human CD99L2 gene with its chimpanzee counterpart is displayed in Figure 5. Sequence identity is evident throughout the region with the exception of the R tract which, although present in both species, is somewhat shorter in chimpanzee. Of all the 142 R•Y sequence pairs analyzed, most displayed the expected ∼98% homology in the R•Y tract-flanking sequences, but none showed sequence conservation within the tracts. We conclude that the R•Y tracts have mutated at a much faster rate than the surrounding sequences. Subsequent analyses (Supplementary Figure 7) provided evidence for a combination of slipped mispairing, recombination-mediated duplications, nucleotide substitutions and possibly also gene conversion, in mediating R•Y tract divergence.
Figure 5.
Comparative genomics of R•Y tracts. Dot-plot of the R-tract and 5 kb of flanking sequence in the human CD99L2 gene with the orthologous gene from chimpanzee.
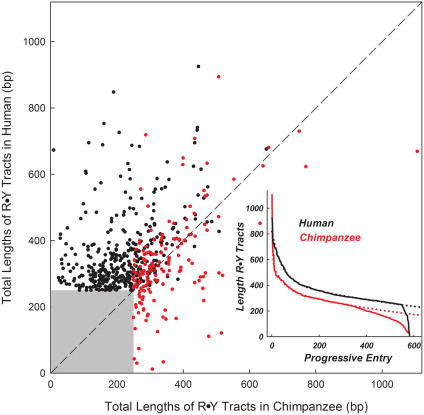
To determine whether R•Y tracts manifest a length bias in hominids, we compared the total length of all pure tracts ≥250 bp in both species with their orthologous counterparts (Supplementary Text); 80% of the 582 tracts analyzed were found to be longer in human than in the chimpanzee (Figure 6). This bias was unlikely to be due to the lower accuracy of assembly of the chimpanzee genome (∼3.6× coverage versus 6–10× for the human assembly). Not only did many long chimpanzee R•Y tracts with sequencing gaps match long tracts in the human assembly, but a pairwise plot of all tracts also displayed exponential decay when arranged by size. However, these sizes diverged from the curve fit for ∼150/582 tracts in the chimpanzee, but only for ∼30/582 tracts in the human (inset in Figure 6). These results suggest that long R•Y tracts may have been either more readily acquired or maintained in the human lineage than in the chimpanzee.
Figure 6.
Length comparison of R•Y tracts between human and chimpanzee. The lengths of the tracts in chimpanzee are plotted against the lengths of the orthologous tracts in human. The total R•Y tract lengths are given, including interruptions. Black dots, human pure R•Y tracts ≥250 bp in length versus orthologous tracts in chimpanzee (436 total); red dots, chimpanzee pure R•Y tracts ≥250 bp in length versus orthologous tracts in human (166 total). Inset, the 582 unique human/chimpanzee pairs of R•Y tracts from the main panel were ranked by length. Black line, human R•Y tracts (average length = 359 ± 114 bp); red line, chimpanzee R•Y tracts (average length = 260 ± 127 bp); dotted lines, curves fitted to the distributions of R•Y tracts.
Finally, the human–chimpanzee orthologous searches revealed two instances (namely, the PRKCB1 and ADAM18 genes) in which the R•Y tract and flanking sequences were present in only one of the two species. In an attempt to understand the possible mutational mechanisms involved, we queried the macaque genome (LCA ∼25 Mya) and analyzed the sequence composition and breakpoint junctions. The results of these analyses (Supplementary Figures 8 and 9 and Supplementary Text) were consistent with the R•Y tracts having been inserted and deleted as part of larger (∼3.5–8.1 kb) DNA fragments through non-homologous end joining reactions, most of which occurred at IR, suggesting that cruciform structures may have been involved in mediating these genomic rearrangements.
DISCUSSION
This report describes the distribution of the most prominent homopurine•homopyrimidine sequences in the human genome. Their preferred distribution within genes expressed in the brain and encoding membrane-bound proteins with channel and receptor activities contrasts with that of the largest IRs (44), which are mostly associated with testis-expressed genes on the X and Y chromosomes. Whereas IRs are known to form cruciform structures, these long R•Y tracts contain segments that fulfill the requirements for mirror symmetry to foster stable triplex structures, although other non-B DNA conformations, such as slipped structures and occasional tetraplexes, are also possible. Specific types of non-B DNA-forming sequences may therefore be associated with distinct gene families. The R•Y tract enrichment of the PAR1 region also differs from the distribution of the largest IR, which are absent from this portion of the sex chromosomes and are concentrated instead in downstream regions (44). This suggests that different types of non-B DNA conformations may be also functionally distinct. Whereas cruciforms have been proposed to mediate gene conversion thereby contributing to genomic integrity (44), R•Y tracts may be involved in stimulating genomic diversity and recombination by virtue of their ability to fold into triplexes and other conformations.
Length polymorphisms have been noted in R•Y tract-containing alleles in mammals (Supplementary Text). The human–mouse and human–chimpanzee comparisons are consistent with the rapid mutation of long R•Y tracts through length and sequence changes, driven by dynamic mutational mechanisms that include slippage, recombination/repair and nucleotide substitution (Supplementary Figure 7). The total length of all pairs of human–chimpanzee R•Y tracts considered amounts to 213 020 bp for human and 159 020 bp for chimpanzee, i.e. an average length divergence of ∼0.039 bases per site per My for the past 6.5 My. This value is more than an order of magnitude higher than the average rate of point mutations between these two species (∼0.0019) (60), and is likely to exceed the value of ∼0.075 reported for subtelomeric segmental duplication/transfer rates during primate evolution (61) if sequence changes and nucleotide substitutions are also included.
Large-scale analyses indicate a positive correlation between point mutation frequency and repeat sequence density (62), implying a role for dynamic mutation in genomic plasticity and hence genome divergence. Similarly, repetitive sequences increase the frequency of gross rearrangements (9,10,31,63) by engaging distant sites, whereas triplex-forming oligonucleotides increase nucleotode substitution rates (30). Genome-wide surveys have established a positive correlation between R•Y tracts and both recombination rates and nt diversity (64–67). Taken together, these findings support our contention that R•Y tracts, perhaps by virtue of their ability to fold into unconventional DNA conformations, represent hotspots for the generation of DSBs, which may then provide nucleation sites for chromatin remodeling pathways involving DNA repair and recombination (67). Studies of the relationship between genomic instabilities and the presence of repetitive sequences at breakpoint junctions suggest an active role for non-B DNA conformations (including slipped structures, cruciforms and triplexes) in promoting DSBs leading to rearrangements both in the context of human disease [reviewed in (8) and (16,35,68)] and evolution (69,70).
Genes involved in ion transport, synaptic transmission and brain-related functions display accelerated rates of evolution in hominids when compared with murids (60). Concomitantly, human, chimpanzee and other non-human primates exhibit accelerated changes in gene expression in brain tissues, with increased expression specifically in the human lineage (71–73). Such acceleration has been suggested to be ‘caused by positive selection that changed the functions of genes expressed in the brains of humans more than in the brains of chimpanzees’ (71). The presence of R•Y tracts within large brain-expressed genes may have synergized with increased mutation rates, thereby contributing to accelerated sequence divergence; these tracts may also have potentiated the acquisition of novel transcriptional activities.
The number of RNA transcripts in the mammalian genome is estimated to be at least one order of magnitude greater than the number of annotated genes (currently ∼20 000), implying that the majority of the mammalian genome is transcribed (74). Transcription on both strands, generating sense and antisense transcripts with a role in RNA interference and transcriptional regulation, is over-represented in genes encoding cytoplasmic proteins but under-represented in genes encoding membrane and extracellular proteins (75). Expansion of a (GAA•TTC)n triplex-forming motif is associated with gene silencing in Friedreich's ataxia as a result of strong secondary structure formation (2). It is therefore conceivable that R•Y tracts, particularly those with mirror symmetry such as the highly recurrent (GAAA•TTTC)n repeats and other MR within the tracts, may also contribute to sense/antisense transcriptional regulation (76).
Several R•Y tract-containing genes encode proteins that function at convergent nodes in signaling pathways at synapses and also represent susceptibility genes for schizophrenia (77,78). This association and the realization that non-B DNA conformations induce genetic instabilities may underscore the delicate balance that exists between the benefits of accelerated evolution and the risks associated with acquiring deleterious mutations.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
This research was supported by grants from the National Institutes of Health (NS37554 and ES11347), the Robert A. Welch Foundation, the Friedreich's Ataxia Research Alliance, the Muscular Dystrophy Foundation (Seek-a-Miracle Foundation) to R.D.W., and in part by the Intramural Research Program of the NIH, NCI and Federal funds from the NCI, NIH (contract number N01-CO-12400). D.N.C. acknowledges the financial support of BIOBASE GmbH. Certain commercial equipment, instruments, materials, or companies are identified in this paper to specify adequately the experimental procedure. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the materials or equipment identified are the best available for the purpose. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government. Funding to pay the Open Access publication charges for this article was provided by the NIH and the Robert A. Welch Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Sinden R.R. DNA Structure and Function. San Diego, CA: Academic Press; 1994. [Google Scholar]
- 2.Wells R.D., Dere R., Hebert M.L., Napierala M., Son L.S. Advances in mechanisms of genetic instability related to hereditary neurological diseases. Nucleic Acids Res. 2005;33:3785–3798. doi: 10.1093/nar/gki697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mirkin S.M., Frank-Kamenetskii M.D. H-DNA and related structures. Annu. Rev. Biophys. Biomol. Struct. 1994;23:541–576. doi: 10.1146/annurev.bb.23.060194.002545. [DOI] [PubMed] [Google Scholar]
- 4.Rich A., Zhang S. Timeline: Z-DNA: the long road to biological function. Nature Rev. Genet. 2003;4:566–572. doi: 10.1038/nrg1115. [DOI] [PubMed] [Google Scholar]
- 5.Neidle S., Parkinson G.N. The structure of telomeric DNA. Curr. Opin. Struct. Biol. 2003;13:275–283. doi: 10.1016/s0959-440x(03)00072-1. [DOI] [PubMed] [Google Scholar]
- 6.Soyfer V.N., Potaman V.N. Triple-Helical Nucleic Acids. New York: Springer-Verlag; 1996. [Google Scholar]
- 7.Hurley L.H. DNA and its associated processes as targets for cancer therapy. Nature Rev. Cancer. 2002;2:188–200. doi: 10.1038/nrc749. [DOI] [PubMed] [Google Scholar]
- 8.Bacolla A., Wells R.D. Non-B DNA conformations, genomic rearrangements, and human disease. J. Biol. Chem. 2004;279:47411–47414. doi: 10.1074/jbc.R400028200. [DOI] [PubMed] [Google Scholar]
- 9.Bacolla A., Jaworski A., Larson J.E., Jakupciak J.P., Chuzhanova N., Abeysinghe S.S., O'Connell C.D., Cooper D.N., Wells R.D. Breakpoints of gross deletions coincide with non-B DNA conformations. Proc. Natl Acad. Sci. USA. 2004;101:14162–14167. doi: 10.1073/pnas.0405974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wojciechowska M., Bacolla A., Larson J.E., Wells R.D. The myotonic dystrophy type 1 triplet repeat sequence induces gross deletions and inversions. J. Biol. Chem. 2005;280:941–952. doi: 10.1074/jbc.M410427200. [DOI] [PubMed] [Google Scholar]
- 11.Kuroda-Kawaguchi T., Skaletsky H., Brown L.G., Minx P.J., Cordum H.S., Waterston R.H., Wilson R.K., Silber S., Oates R., Rozen S., et al. The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nature Genet. 2001;29:279–286. doi: 10.1038/ng757. [DOI] [PubMed] [Google Scholar]
- 12.Repping S., Skaletsky H., Lange J., Silber S., Van Der Veen F., Oates R.D., Page D.C., Rozen S. Recombination between palindromes P5 and P1 on the human Y chromosome causes massive deletions and spermatogenic failure. Am. J. Hum. Genet. 2002;71:906–922. doi: 10.1086/342928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gotter A.L., Shaikh T.H., Budarf M.L., Rhodes C.H., Emanuel B.S. A palindrome-mediated mechanism distinguishes translocations involving LCR-B of chromosome 22q11.2. Hum. Mol. Genet. 2004;13:103–115. doi: 10.1093/hmg/ddh004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abeysinghe S.S., Chuzhanova N., Krawczak M., Ball E.V., Cooper D.N. Translocation and gross deletion breakpoints in human inherited disease and cancer I: Nucleotide composition and recombination-associated motifs. Hum. Mutat. 2003;22:229–244. doi: 10.1002/humu.10254. [DOI] [PubMed] [Google Scholar]
- 15.Chuzhanova N., Abeysinghe S.S., Krawczak M., Cooper D.N. Translocation and gross deletion breakpoints in human inherited disease and cancer II: Potential involvement of repetitive sequence elements in secondary structure formation between DNA ends. Hum. Mutat. 2003;22:245–251. doi: 10.1002/humu.10253. [DOI] [PubMed] [Google Scholar]
- 16.Kato T., Inagaki H., Yamada K., Kogo H., Ohye T., Kowa H., Nagaoka K., Taniguchi M., Emanuel B.S., Kurahashi H. Genetic variation affects de novo translocation frequency. Science. 2006;311:971. doi: 10.1126/science.1121452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wells R.D., Collier D.A., Hanvey J.C., Shimizu M., Wohlrab F. The chemistry and biology of unusual DNA structures adopted by oligopurine.oligopyrimidine sequences. FASEB J. 1988;2:2939–2949. [PubMed] [Google Scholar]
- 18.Frank-Kamenetskii M.D., Mirkin S.M. Triplex DNA structures. Annu. Rev. Biochem. 1995;64:65–95. doi: 10.1146/annurev.bi.64.070195.000433. [DOI] [PubMed] [Google Scholar]
- 19.Chan P.P., Glazer P.M. Triplex DNA: fundamentals, advances, and potential applications for gene therapy. J. Mol. Med. 1997;75:267–282. doi: 10.1007/s001090050112. [DOI] [PubMed] [Google Scholar]
- 20.Inman R.B. Transitions of DNA Homopolymers. J. Mol. Biol. 1964;9:624–637. doi: 10.1016/s0022-2836(64)80171-6. [DOI] [PubMed] [Google Scholar]
- 21.Wells R.D., Larson J.E. Buoyant density studies on natural and synthetic deoxyribonucleic acids in neutral and alkaline solutions. J. Biol. Chem. 1972;247:3405–3409. [PubMed] [Google Scholar]
- 22.Jaishree T.N., Wang A.H. NMR studies of pH-dependent conformational polymorphism of alternating (C-T)n sequences. Nucleic Acids Res. 1993;21:3839–3844. doi: 10.1093/nar/21.16.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morgan A.R., Wells R.D. Specificity of the three-stranded complex formation between double-stranded DNA and single-stranded RNA containing repeating nucleotide sequences. J. Mol. Biol. 1968;37:63–80. doi: 10.1016/0022-2836(68)90073-9. [DOI] [PubMed] [Google Scholar]
- 24.Krasilnikov A.S., Panyutin I.G., Samadashwily G.M., Cox R., Lazurkin Y.S., Mirkin S.M. Mechanisms of triplex-caused polymerization arrest. Nucleic Acids Res. 1997;25:1339–1346. doi: 10.1093/nar/25.7.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patel H.P., Lu L., Blaszak R.T., Bissler J.J. PKD1 intron 21: triplex DNA formation and effect on replication. Nucleic Acids Res. 2004;32:1460–1468. doi: 10.1093/nar/gkh312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohshima K., Montermini L., Wells R.D., Pandolfo M. Inhibitory effects of expanded GAA•TTC triplet repeats from intron I of the Friedreich ataxia gene on transcription and replication in vivo. J. Biol. Chem. 1998;275:14588–14595. doi: 10.1074/jbc.273.23.14588. [DOI] [PubMed] [Google Scholar]
- 27.Kohwi Y., Panchenko Y. Transcription-dependent recombination induced by triple-helix formation. Genes Dev. 1993;7:1766–1778. doi: 10.1101/gad.7.9.1766. [DOI] [PubMed] [Google Scholar]
- 28.Faruqi A.F., Datta H.J., Carroll D., Seidman M.M., Glazer P.M. Triple-helix formation induces recombination in mammalian cells via a nucleotide excision repair-dependent pathway. Mol. Cell. Biol. 2000;20:990–1000. doi: 10.1128/mcb.20.3.990-1000.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo Z., Macris M.A., Faruqi A.F., Glazer P.M. High-frequency intrachromosomal gene conversion induced by triplex-forming oligonucleotides microinjected into mouse cells. Proc. Natl Acad. Sci. USA. 2000;97:9003–9008. doi: 10.1073/pnas.160004997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vasquez K.M., Narayanan L., Glazer P.M. Specific mutations induced by triplex-forming oligonucleotides in mice. Science. 2000;290:530–533. doi: 10.1126/science.290.5491.530. [DOI] [PubMed] [Google Scholar]
- 31.Wang G., Vasquez K.M. Naturally occurring H-DNA-forming sequences are mutagenic in mammalian cells. Proc. Natl Acad. Sci. USA. 2004;101:13448–13453. doi: 10.1073/pnas.0405116101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bacolla A., Jaworski A., Connors T.D., Wells R.D. PKD1 unusual DNA conformations are recognized by nucleotide excision repair. J. Biol. Chem. 2001;276:18597–18604. doi: 10.1074/jbc.M100845200. [DOI] [PubMed] [Google Scholar]
- 33.Pandolfo M., Koenig M. Freidreich's Ataxia. In: Wells R.D., Warren S.T., editors. Genetic Instabilities and Hereditary Neurological Diseases. San Diego, CA: Academic Press; 1998. pp. 373–398. [Google Scholar]
- 34.Vetcher A.A., Napierala M., Iyer R.R., Chastain P.D., Griffith J.D., Wells R.D. Sticky DNA, a long GAA.GAA.TTC triplex that is formed intramolecularly, in the sequence of intron 1 of the frataxin gene. J. Biol. Chem. 2002;277:39217–39227. doi: 10.1074/jbc.M205209200. [DOI] [PubMed] [Google Scholar]
- 35.Raghavan S.C., Chastain P., Lee J.S., Hegde B.G., Houston S., Langen R., Hsieh C.L., Haworth I.S., Lieber M.R. Evidence for a triplex DNA conformation at the bcl-2 major breakpoint region of the t(14;18) translocation. J. Biol. Chem. 2005;280:22749–22760. doi: 10.1074/jbc.M502952200. [DOI] [PubMed] [Google Scholar]
- 36.Raghavan S.C., Lieber M.R. Chromosomal translocations and non-B DNA structures in the human genome. Cell Cycle. 2004;3:762–768. [PubMed] [Google Scholar]
- 37.Raghavan S.C., Swanson P.C., Wu X., Hsieh C.L., Lieber M.R. A non-B-DNA structure at the Bcl-2 major breakpoint region is cleaved by the RAG complex. Nature. 2004;428:88–93. doi: 10.1038/nature02355. [DOI] [PubMed] [Google Scholar]
- 38.Knauert M.P., Lloyd J.A., Rogers F.A., Datta H.J., Bennett M.L., Weeks D.L., Glazer P.M. Distance and affinity dependence of triplex-induced recombination. Biochemistry. 2005;44:3856–3864. doi: 10.1021/bi0481040. [DOI] [PubMed] [Google Scholar]
- 39.Hoffman-Liebermann B., Liebermann D., Troutt A., Kedes L.H., Cohen S.N. Human homologs of TU transposon sequences: polypurine/polypyrimidine sequence elements that can alter DNA conformation in vitro and in vivo. Mol. Cell. Biol. 1986;6:3632–3642. doi: 10.1128/mcb.6.11.3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Christophe D., Cabrer B., Bacolla A., Targovnik H., Pohl V., Vassart G. An unusually long poly(purine)-poly(pyrimidine) sequence is located upstream from the human thyroglobulin gene. Nucleic Acids Res. 1985;13:5127–5144. doi: 10.1093/nar/13.14.5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Behe M.J. The DNA sequence of the human beta-globin region is strongly biased in favor of long strings of contiguous purine or pyrimidine residues. Biochemistry. 1987;26:7870–7875. doi: 10.1021/bi00398a050. [DOI] [PubMed] [Google Scholar]
- 42.Schroth G.P., Ho P.S. Occurrence of potential cruciform and H-DNA forming sequences in genomic DNA. Nucleic Acids Res. 1995;23:1977–1983. doi: 10.1093/nar/23.11.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ussery D., Soumpasis D.M., Brunak S., Staerfeldt H.H., Worning P., Krogh A. Bias of purine stretches in sequenced chromosomes. Comput. Chem. 2002;26:531–541. doi: 10.1016/s0097-8485(02)00013-x. [DOI] [PubMed] [Google Scholar]
- 44.Warburton P.E., Giordano J., Cheung F., Gelfand Y., Benson G. Inverted repeat structure of the human genome: the X-chromosome contains a preponderance of large, highly homologous inverted repeats that contain testes genes. Genome Res. 2004;14:1861–1869. doi: 10.1101/gr.2542904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Collins J.R., Stephens R.M., Gold B., Long B., Dean M., Burt S.K. An exhaustive DNA micro-satellite map of the human genome using high performance computing. Genomics. 2003;82:10–19. doi: 10.1016/s0888-7543(03)00076-4. [DOI] [PubMed] [Google Scholar]
- 46.Ming Y., Horton D., Cohen J.C., Hobbs H.H., Stephens R.M. WholePathwayScope: a comprehensive pathway-based analysis tool for high-throughput data. BMC Bioinformatics. 2006;7:30. doi: 10.1186/1471-2105-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karlin S., Macken C. Some statistical problems in the assessment of inhomogenesis of DNA sequence data. J. Am. Statist. Assoc. 1991;86:27–35. [Google Scholar]
- 48.Gusev V.D., Nemytikova L.A., Chuzhanova N.A. On the complexity measures of genetic sequences. Bioinformatics. 1999;15:994–999. doi: 10.1093/bioinformatics/15.12.994. [DOI] [PubMed] [Google Scholar]
- 49.Van Raay T.J., Burn T.C., Connors T.D., Petri L.R., Germino G.G., Klinger K.W., Landes G.M. A 2.5 kb polypyrimidine tract in the PKD1 gene contains at least 23 H-DNA-forming sequences. Microb. Comp. Genomics. 1996;1:317–327. doi: 10.1089/mcg.1996.1.317. [DOI] [PubMed] [Google Scholar]
- 50.Vasquez K.M., Wang G., Havre P.A., Glazer P.M. Chromosomal mutations induced by triplex-forming oligonicleotides in mammalian cells. Nucleic Acids Res. 1999;27:1176–1181. doi: 10.1093/nar/27.4.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang G., Seidman M.M., Glazer P.M. Mutagenesis in mammalian cells induced by triple helix formation and transcription-coupled repair. Science. 1996;271:802–805. doi: 10.1126/science.271.5250.802. [DOI] [PubMed] [Google Scholar]
- 52.Filatov D.A., Gerrard D.T. High mutation rates in human and ape pseudoautosomal genes. Gene. 2003;317:67–77. doi: 10.1016/s0378-1119(03)00697-8. [DOI] [PubMed] [Google Scholar]
- 53.Perry J., Palmer S., Gabriel A., Ashworth A. A short pseudoautosomal region in laboratory mice. Genome Res. 2001;11:1826–1832. doi: 10.1101/gr.203001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Napierala M., Dere R., Vetcher A., Wells R.D. Structure-dependent recombination hot spot activity of GAA.TTC sequences from intron 1 of the Friedreich's ataxia gene. J. Biol. Chem. 2004;279:6444–6454. doi: 10.1074/jbc.M309596200. [DOI] [PubMed] [Google Scholar]
- 55.Sakamoto N., Chastain P.D., Parniewski P., Ohshima K., Pandolfo M., Griffith J.D., Wells R.D. Sticky DNA: self-association properties of long GAAaTTC repeats in R R Y triplex structures from Friedreich's ataxia. Molecular Cell. 1999;3:465–475. doi: 10.1016/s1097-2765(00)80474-8. [DOI] [PubMed] [Google Scholar]
- 56.Subramanian S., Mishra R.K., Singh L. Genome-wide analysis of microsatellite repeats in humans: their abundance and density in specific genomic regions. Genome Biol. 2003;4:R13. doi: 10.1186/gb-2003-4-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sandstrom K., Warmlander S., Graslund A., Leijon M. A-tract DNA disfavours triplex formation. J. Mol. Biol. 2002;315:737–748. doi: 10.1006/jmbi.2001.5249. [DOI] [PubMed] [Google Scholar]
- 58.Roberts R.W., Crothers D.M. Prediction of the stability of DNA triplexes. Proc. Natl Acad. Sci. USA. 1996;93:4320–4325. doi: 10.1073/pnas.93.9.4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.James P.L., Brown T., Fox K.R. Thermodynamic and kinetic stability of intermolecular triple helices containing different proportions of C+*GC and T*AT triplets. Nucleic Acids Res. 2003;31:5598–5606. doi: 10.1093/nar/gkg782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mikkelsen T.S., Hillier L.W., Eichler E.E., Zody M.C., Jaffe D.B., Yang S.-P., Enard W., Hellmann I., Lindblad-Toh K., Altheide T.K., et al. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 61.Linardopoulou E.V., Williams E.M., Fan Y., Friedman C., Young J.M., Trask B.J. Human subtelomeres are hot spots of interchromosomal recombination and segmental duplication. Nature. 2005;437:94–100. doi: 10.1038/nature04029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chiaromonte F., Yang S., Elnitski L., Yap V.B., Miller W., Hardison R.C. Association between divergence and interspersed repeats in mammalian noncoding genomic DNA. Proc. Natl Acad. Sci. USA. 2001;98:14503–14508. doi: 10.1073/pnas.251423898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meservy J.L., Sargent R.G., Iyer R.R., Chan F., McKenzie G.J., Wells R.D., Wilson J.H. Long CTG tracts from the myotonic dystrophy gene induce deletions and rearrangements during recombination at the APRT locus in CHO cells. Mol. Cell. Biol. 2003;23:3152–3162. doi: 10.1128/MCB.23.9.3152-3162.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kong A., Gudbjartsson D.F., Sainz J., Jonsdottir G.M., Gudjonsson S.A., Richardsson B., Sigurdardottir S., Barnard J., Hallbeck B., Masson G., et al. A high-resolution recombination map of the human genome. Nature Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- 65.Jensen-Seaman M.I., Furey T.S., Payseur B.A., Lu Y., Roskin K.M., Chen C.F., Thomas M.A., Haussler D., Jacob H.J. Comparative recombination rates in the rat, mouse, and human genomes. Genome Res. 2004;14:528–538. doi: 10.1101/gr.1970304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hellmann I., Prufer K., Ji H., Zody M.C., Paabo S., Ptak S.E. Why do human diversity levels vary at a megabase scale? Genome Res. 2005;15:1222–1231. doi: 10.1101/gr.3461105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Myers S., Bottolo L., Freeman C., McVean G., Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- 68.Wells R.D., Warren S.T. Genetic Instabilities and Hereditary Neurological Diseases. San Diego, CA: Academic Press; 1998. [Google Scholar]
- 69.Kehrer-Sawatzki H., Sandig C., Chuzhanova N., Goidts V., Szamalek J.M., Tanzer S., Muller S., Platzer M., Cooper D.N., Hameister H. Breakpoint analysis of the pericentric inversion distinguishing human chromosome 4 from the homologous chromosome in the chimpanzee (Pan troglodytes) Hum. Mutat. 2005;25:45–55. doi: 10.1002/humu.20116. [DOI] [PubMed] [Google Scholar]
- 70.Szamalek J.M., Goidts V., Chuzhanova N., Hameister H., Cooper D.N., Kehrer-Sawatzki H. Molecular characterisation of the pericentric inversion that distinguishes human chromosome 5 from the homologous chimpanzee chromosome. Hum. Genet. 2005;117:168–176. doi: 10.1007/s00439-005-1287-y. [DOI] [PubMed] [Google Scholar]
- 71.Khaitovich P., Hellmann I., Enard W., Nowick K., Leinweber M., Franz H., Weiss G., Lachmann M., Paabo S. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science. 2005;309:1850–1854. doi: 10.1126/science.1108296. [DOI] [PubMed] [Google Scholar]
- 72.Preuss T.M., Caceres M., Oldham M.C., Geschwind D.H. Human brain evolution: insights from microarrays. Nature Rev. Genet. 2004;5:850–860. doi: 10.1038/nrg1469. [DOI] [PubMed] [Google Scholar]
- 73.Uddin M., Wildman D.E., Liu G., Xu W., Johnson R.M., Hof P.R., Kapatos G., Grossman L.I., Goodman M. Sister grouping of chimpanzees and humans as revealed by genome-wide phylogenetic analysis of brain gene expression profiles. Proc. Natl Acad. Sci. USA. 2004;101:2957–2962. doi: 10.1073/pnas.0308725100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 75.Katayama S., Tomaru Y., Kasukawa T., Waki K., Nakanishi M., Nakamura M., Nishida H., Yap C.C., Suzuki M., Kawai J., et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 76.Fabregat I., Koch K.S., Aoki T., Atkinson A.E., Dang H., Amosova O., Fresco J.R., Schildkraut C.L., Leffert H.L. Functional pleiotropy of an intramolecular triplex-forming fragment from the 3′-UTR of the rat Pigr gene. Physiol. Genomics. 2001;5:53–65. doi: 10.1152/physiolgenomics.2001.5.2.53. [DOI] [PubMed] [Google Scholar]
- 77.Harrison P.J., Weinberger D.R. Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Mol. Psychiatry. 2005;10:40–68. doi: 10.1038/sj.mp.4001558. [DOI] [PubMed] [Google Scholar]
- 78.Lewis D.A., Hashimoto T., Volk D.W. Cortical inhibitory neurons and schizophrenia. Nature Rev. Neurosci. 2005;6:312–324. doi: 10.1038/nrn1648. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.