Abstract
Cells deficient in the Werner syndrome protein (WRN) or BRCA1 are hypersensitive to DNA interstrand cross-links (ICLs), whose repair requires nucleotide excision repair (NER) and homologous recombination (HR). However, the roles of WRN and BRCA1 in the repair of DNA ICLs are not understood and the molecular mechanisms of ICL repair at the processing stage have not yet been established. This study demonstrates that WRN helicase activity, but not exonuclease activity, is required to process DNA ICLs in cells and that WRN cooperates with BRCA1 in the cellular response to DNA ICLs. BRCA1 interacts directly with WRN and stimulates WRN helicase and exonuclease activities in vitro. The interaction between WRN and BRCA1 increases in cells treated with DNA cross-linking agents. WRN binding to BRCA1 was mapped to BRCA1 452–1079 amino acids. The BRCA1/BARD1 complex also associates with WRN in vivo and stimulates WRN helicase activity on forked and Holliday junction substrates. These findings suggest that WRN and BRCA1 act in a coordinated manner to facilitate repair of DNA ICLs.
INTRODUCTION
Maintenance of genomic integrity requires efficient responses to DNA damage. This involves the activation of signaling pathways that delay cell cycle progression and recruit factors to facilitate repair of DNA lesions (1). DNA interstrand cross-links (ICLs) are toxic lesions because they are strong blocks to DNA replication and transcription. The toxicity of ICLs has led to wide use of DNA cross-linking agents for cancer chemotherapy. Repair of DNA ICLs involves homologous recombination (HR) and nucleotide excision repair (NER), both of which are relatively error-free DNA repair pathways. However, DNA ICLs are also repaired by a mutagenic error-prone pathway (2). In Saccharomyces cerevisiae, genetic studies implicate a large number of genes in ICL repair (3); however, the precise mechanisms of ICL repair have not been established and little is known about ICL repair in mammals.
WRN and BRCA1 are both involved in genome surveillance and participate in repair of DNA double strand breaks (DSB). BRCA1 germline heterozygotes with one functional allele are predisposed to breast and ovarian cancer, and tumor progression is associated with loss of heterozygosity at BRCA1 by somatic mutation. In contrast, WRN germline mutations cause the segmental progeroid Werner syndrome (WS) and WS patients are predisposed to sarcomas. WRN and BRCA1 interact with the MRN complex (MRE11, RAD50 and NBS1) and with RAD51, both of which play critical roles in HR (4–8). Cells from WS patients are defective in the repair of DNA ICLs (9). BRCA1 interacts with Fanconi anemia proteins, which protect against DNA ICLs (10,11), and is required for RAD51 focus formation in response to cisplatin, a DNA cross-linking agent (12). Cells deficient in WRN or BRCA1 are defective in HR-dependent DNA repair reactions. WRN prevents defective mitotic recombination resolution, whereas BRCA1 promotes DNA DSB repair by HR (13–15). In addition, BRCA1 and WRN are both implicated in the G2/M-checkpoint response (16,17). Thus, there is indirect evidence suggesting that WRN and BRCA1 cooperate in the cellular response to DNA ICLs, and in HR-mediated repair of DNA ICLs.
Recent evidence suggests that BRCA1 regulates HR-dependent aspects of ICL repair. For example, BRCA1 is required for formation of cisplatin-induced RAD51 foci but not for formation of γ-irradiation induced RAD51 foci (18). Moreover, BRCA1 is necessary for RAD51-mediated gene conversion, crossover and sister chromatid replication slippage events (19). Lastly, recent studies identified a BRCA1-interacting protein, BACH1, that participates in the Fanconi anemia pathway of DNA ICL repair (20,21).
In spite of recent advances that implicate BRCA1 and WRN in the cellular response to DNA ICLs, the biochemical and cellular bases for their roles in the repair of DNA ICLs have remained obscure. This study demonstrates and characterizes physical and functional interactions between WRN and BRCA1. Importantly, processing of DNA ICLs in cells requires both BRCA1 and the helicase activity of WRN. BRCA1 stimulates WRN helicase and exonuclease activities and the interaction between BRCA1 and WRN increases in cells exposed to DNA cross-linking agents. Together with other results presented here, these data suggest that WRN and BRCA1 act in a coordinated manner to facilitate processing of DNA ICLs.
MATERIALS AND METHODS
Proteins, cell lines and siRNA
Purification of WRN, BLM, BRCA1/BARD1 complex, and BRCA1 fragment proteins and maintenance of HeLa cells were described previously (4,22,23). We purchased 6× His-tagged BRCA1 from Jena Biosciences (>95% pure by SDS–PAGE, Jena, Germany) and 6× His-tag peptide from Abcam (Cambridge, MA). Generation and maintenance of the telomerase-immortalized 03141 WS cells complemented with wild-type WRN, exonuclease-inactive E84A WRN (E-), helicase-inactive K577M WRN (H-), or exonuclease- and helicase-inactive WRN (E-H-) were described previously (24). The short hairpin RNA (shRNA) targeted against WRN mRNA was cloned into the pSilencer™ 3.1-H1 hygro vector (Ambion Inc.) for a stable transfection into U-2 OS cells. The WRN RNAi target sequence is: TGAAGAGCAAGTTACTTGA. A pSilencer vector expressing a shRNA that is not homologous to any known human genes (Ambion) was used as a negative control. The WRN and control shRNA cells were selected and maintained in the presence of hygromycin B. The siRNAs targeted against BRCA1 mRNA and its control siRNA (Upstate Inc.) were transiently transfected into the WRN and control shRNA knockdown cells using the Lipofectamine™ 2000 transfection reagent (Invitrogen) according to the manufacturer's instructions.
Cell proliferation assay
One day after siRNA transfection for BRCA1 knockdown, cells were seeded into 6 cm dishes and ICLs were generated in vivo by treatment with 8-methoxypsoralen (8-MOP, Sigma) for 10 min and 365 nm ultraviolet (UV) light at 1.8 J/cm2 for 3 min (25). Cells were counted using the Beckman Z1 Coulter® Particle Counter (Beckman Coulter, Inc.) each day for 3 days following treatment. Because cells transfected with BRCA1 siRNA showed decreased proliferation (data not shown), viability was corrected for cell numbers with the respective time points in the absence of photoactivated psoralen treatment. Then, values were normalized to the cell count on day 0.
Determination of ICL by comet assay
The comet assay was performed under alkaline conditions to detect the presence of ICLs as previously reported (26) with minor modifications. This assay measures the unhooking of DNA ICLs, the first step for the repair of DNA ICLs. Under alkaline conditions, denaturation of DNA containing ICLs is repressed, such that DNA migration is retarded upon electrophoresis. Therefore, DNA without ICLs forms tails while cross-linked DNA shows no tails. We used this approach to quantify the induction and incision of DNA ICLs induced by photoactivated psoralen (8-MOP plus UVA). Angelicin (purchased from Sigma) is a psoralen mimetic compound that forms DNA monoadducts but cannot form DNA ICLs. Forty hours after siRNA transfection for BRCA1 knockdown, cells were treated with photoactivated psoralen. At various intervals after 8-MOP treatment (1 µg/ml), cells were scraped and washed with phosphate-buffered saline (PBS), mixed with 75 µl of 0.5% low-melting point agarose at 37°C (104 cells, 10 µl), and spread onto a cold slide precoated with agarose for 5 min. Another 75 µl of melted agarose was added and slides were chilled for 5 min. Slides were placed in cold lysis buffer for 1 h [2.5 M NaCl, 100 mM EDTA, 10 mM Tris (pH 10), 1% Triton X-100 and 10% dimethyl sulfoxide (DMSO)], washed 3× with PBS for 5 min and incubated in lysis buffer containing 1 mg/ml proteinase K for 2 h at 37°C. Slides were washed again and then incubated for 30 min in alkaline denaturation buffer [300 mM NaOH, 1 mM EDTA (pH 13)] at 4°C, followed by electrophoresis in the same buffer at 25 V. Tail moments were quantified using the Komet 5.5 software (Kinetic Imaging, Durham, NC) according to the manufacturer's instructions. We normalized tail moments to that of the 12 h time point of control RNAi cells (as 100%).
Dot blot assay
Recombinant proteins (1–10 ng) were spotted onto PVDF membranes. The membrane was blocked in 5% non-fat milk in T-TBS buffer [0.75% Tween-20, 25 mM Tris (pH 7.4), 150 mM NaCl] for 1 h at room temperature and incubated in T-TBS with or without WRN (1 µg/ml) overnight at 4°C. After extensive washing, the membrane was analyzed by immunoblotting with anti-WRN antibodies (clone 30; BD Biosciences, Lexington, KY) followed by chemiluminescent detection with SuperSignal substrates (Pierce, Rockland, IL).
Immunoprecipitation, western analysis, enzyme-linked immunosorbent assay (ELISA) and catalytic activity assays
Methods for immunoprecipitation, immunoblot and ELISA were described previously (27). Relative densities of protein bands were quantified using an Gel Doc™ XR imaging system (Bio-Rad, Hercules, CA); the values were normalized with the controls (lamin B in Figure 1A and H2AX in Supplementary Figure S1) and expressed as ratios to those in the cells without RNAi or psoralen treatment. The antibodies used were: anti-WRN (H-300, Santa Cruz Biotechnology; clone 30, Transduction Laboratories; Ab200, Novus) and anti-BRCA1 (Ab-4, Oncogene). WRN helicase activity was measured using a 22 bp forked duplex substrate (28) or a Holliday junction substrate (29). WRN exonuclease activity was measured using a 34 bp forked duplex substrate (28). Experimental procedures, conditions and quantification were described previously (28).
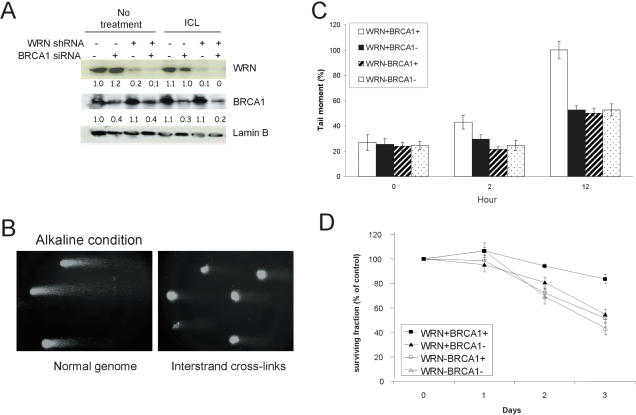
Figure 1.
Roles of WRN and BRCA1 in response to psoralen ICLs. (A) RNAi was used to knock down expression of WRN and/or BRCA1 in U-2 OS cells as described in Materials and Methods. Forty hours after BRCA1 siRNA trasnfection, knockdown or control cells were treated with psoralen ICLs (0.1 µg 8-MOP/ml +UVA) and harvested 12 h thereafter for western blot analyses. (B) Alkaline comet assay with untreated cells or psoralen-treated cells. (C) Comet assay was performed at the indicated intervals after cells were exposed to psoralen (1 µg 8-MOP/ml +UVA). Knockdown phenotype for cells in each set of assays is indicated. Tail moments are normalized to the 12 h time point for control siRNA transfected cells. Data are averages of three independent assays, from each of which at least 50 cells were randomly selected. Values are means ± SD. (D) One day after BRCA1 siRNA transfection, control (+) or RNAi (−) treated cells were treated with 0.01 µg 8-MOP/ml +UVA, and cells were counted each day for 3 days (n = 4). Numbers shown here were corrected for cell number with the respective time points in the absence of photoactivated psoralen treatment.
RESULTS
Proliferation and DNA repair defects in WRN- and/or BRCA1-deficient cells treated with psoralen ICLs
To test whether WRN and BRCA1 act cooperatively in an ICL-specific DNA response pathway, sequences for RNA interference (RNAi) were designed to selectively knock down expression of WRN or BRCA1 in human osteosarcoma cells (U-2 OS). WRN RNAi was achieved by hygromycin B selection for stable expression of WRN shRNA, whereas BRCA1 RNAi was achieved by transient transfection of siRNA. Depletion of WRN by shRNA exhibited continuous low levels of WRN protein level as it was a stable knockdown (Supplementary Figure S1). To test the efficacy of BRCA1 knockdown, we first analyzed the kinetics of BRCA1 depletion. Supplementary Figure S1 shows that BRCA1 protein levels were reduced by 73, 98 and 89% at 2, 3 or 4 days after the transient BRCA1 siRNA transfection, respectively. Next, we determined the possibility that psoralen treatment had an effect on the protein levels of WRN and BRCA1. Figure 1A shows that BRCA1 protein levels were reduced by 60% in the control and WRN shRNA cells 40 h after BRCA1 siRNA transfection. Thus, cells were treated with psoralen at this time point. We found that the BRCA1 protein levels were reduced by 70 to 80% 12 h after the psoralen treatment. WRN protein levels were not affected by psoralen treatment. Lamin B protein levels were not affected by the WRN or BRCA1 knockdown, or by the psoralen treatment. After verifying the levels of WRN and BRCA1 knockdown during the course of these experiments, the cells were assessed for the extent of DNA cross-linking by the comet assay under alkaline conditions (26). Psoralen forms DNA monoadducts that are converted to ICLs by photoactivation. Tail moment, a measure of relative DNA electrophoretic mobility, was lower in psoralen-treated cells, consistent with the expected slower mobility of cross-linked DNA (Figure 1B). In contrast, there was no delay in electrophoretic mobility in (i) cells treated with photoactivated angelicin that forms DNA monoadduct only, but not ICLs, (ii) cells treated with psoralen alone without UVA exposure and (iii) control or WRN shRNA cells treated with UVA alone (Supplementary Figure S2), indicating that these three conditions did not induce DNA ICLs in cells. Poot et al. (30,31) have shown that WS cells are not more sensitive than wild-type cells to exposure to UVA treatment alone or psoralen monoadducts. The conclusion that WS cells are not particularly sensitive to monoadduct forming agents was also reached by Fujiwara et al. (32). Consequently we conclude that the sensitivity of the WS cells to psoralen/UVA and the reduced incision of psoralen treated cells reflect a role for the WRN in cross-link metabolism.
The comet assay results showed similar tail moments in cells knocked down for WRN, BRCA1, or both at all time points (Figure 1C). Moreover, the tail moment in wild-type cells was not different from cells knocked down for WRN, BRCA1 or both immediately after exposure to psoralen (0 h), indicating that psoralen-induced ICL formation is independent of the level of expression of WRN and BRCA1. The tail moment gradually increased in wild-type cells from 0 to 12 h after exposure to psoralen, presumably due to ongoing ICL repair. The rate at which the tail moment increased was slower in cells knocked down for WRN, BRCA1 or both. This suggests that substantial DNA ICLs remain in the genomes of the WRN and BRCA1 knockdown cells 12 h after treatment with photoactivated psoralen. The effects of WRN and BRCA1 knockdown in ICL removal were not additive, suggesting that WRN and BRCA1 may participate in a common pathway for efficient repair of psoralen-induced ICLs.
To address if the WRN and BRCA1 genes functioned in the same or different pathways, we next measured the effect of reduced WRN or BRCA1 expression on cell proliferation in response to psoralen-induced ICLs. In the absence of ICLs, cells deficient in WRN, BRCA1 or both, grew slower compared to control cells (data not shown). Therefore, the data presented in Figure 1D and in Supplementary Figure S3 were corrected for cell growth rate prior to photoactivated psoralen treatment. The proliferation of cells deficient in WRN or BRCA1 was reduced (P < 0.05) by low to moderate levels of psoralen-induced ICLs (0.01 or 0.1 µg 8-MOP/ml) compared to that of control cells expressing wild-type levels of both WRN and BRCA1 during a 3 day interval. In addition, the reduction in cell proliferation was similar whether cells were deficient in WRN, BRCA1 or both. Consistent with the comet assay data (Figure 1C), the results suggest that WRN and BRCA1 are involved in a common DNA ICL response pathway.
Direct interaction between WRN and BRCA1
If WRN and BRCA1 participate in the same ICL DNA response pathway, they may interact physically and functionally. This possibility was tested by co-immunoprecipitation of WRN and BRCA1 in psoralen-treated and untreated control HeLa cells. In untreated control cells, there was little or no evidence for an interaction between WRN and BRCA1. In contrast, 12 h after exposure to psoralen or 3 h after γ-irradiation (10 Gy), a significant amount of BRCA1 co-immunoprecipitated with WRN (Figure 2A). This WRN-BRCA1 association persisted in the presence of ethidium bromide (data not shown), indicating that the interaction is not mediated by DNA. The amount of BRCA1 brought down by the WRN antibodies is not trivial, since its level is comparable to that of other proteins associated with WRN (4,27). Figure 2B showed that the majority of HeLa cells (86%) were arrested in S phase 12 h after the psoralen treatment. In contrast, there was only a slight increase in cell population in S phase compared to the untreated cells (60 versus 45%) 3 h after cellular exposure to γ-irradiation treatment (10 Gy). Also, the cell cycle profiles were similar between untreated and γ-irradiation-treated cells. Further, it is known that BRCA1 expression is most abundant during S phase. Therefore, it is likely that both photoactivated psoralen and γ-irradiation contributes to the increased WRN-BRCA1 association, and such interaction is strengthened when DNA damage takes place in S phase.
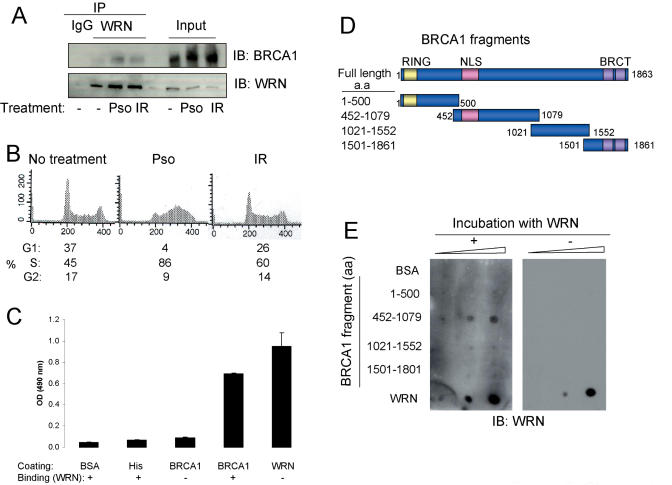
Figure 2.
Interaction between WRN and BRCA1. (A and B) HeLa cells were treated with psoralen (0.1 µg/ml +UVA, 12 h) or γ-irradiation (10 Gy, 3 h). WRN immunoprecipitates were analyzed by western blot with the indicated antibodies. Input shows 10% of cell lysate used for immunoprecipitation. DNA content and cell cycle distributions were determined by flow cytometry. (C) Purified full-length His-tagged BRCA1 or WRN, His peptides or BSA was coated onto ELISA plates and incubated with purified WRN. A rabbit antibody against WRN (Ab200, Novus) was used to detect WRN bound to the ELISA plate. (D) Schematic diagram of human BRCA1 protein is shown including the N-terminal RING domain, the NLS and two C-terminal BRCT domains. (E) Regions of BRCA1 that bind WRN. BSA, WRN and GST-fusion proteins containing different regions of BRCA1 (1, 3 or 10 ng/dot) were immobilized on a membrane surface. The membrane was incubated with WRN (1 µg/ml) and then probed with anti-WRN antibody.
ELISA with His-tagged purified full-length proteins confirmed that BRCA1 and WRN interact, and demonstrated that WRN binds directly and specifically to BRCA1 (Figure 2C). The specificity of binding was confirmed by the observations that WRN did not bind to BSA or the 6× His peptide. The purity of His-tagged WRN and glutathione S-transferase (GST)-tagged BRCA1 fragments was shown in the Supplementary Figure S4. The region of BRCA1 required for this interaction was mapped using GST fusion proteins containing different BRCA1 domains (Figure 2D), which were immobilized on a membrane and probed with purified WRN in vitro. The results indicate that the fragment containing BRCA1 452–1079 amino acids interacts with WRN (Figure 2E), suggesting that BRCA1 interacts with WRN via this region. This region of BRCA1 contains the nuclear localization signals (NLSs) and the DNA binding motifs (23). Because BARD1 binds to the N and C terminal domains of BRCA1 (33), our results imply that WRN and BARD1 bind to different regions of BRCA1 and thus may simultaneously interact with BRCA1.
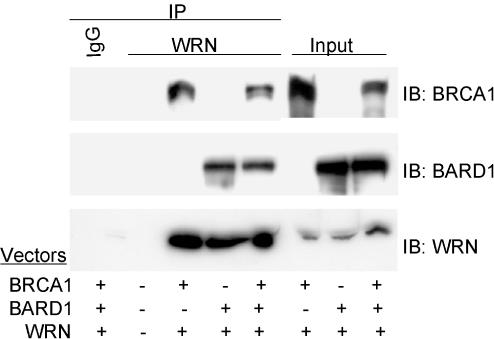
To confirm a direct interaction of WRN and BRCA1 in vivo, Sf9 insect cells were infected with both WRN and BRCA1. We also tested a potential WRN-BARD1 association, as BARD1 is considered an important protein partner of BRCA1 on the basis of the observation that BRCA1 and BARD1 protein levels are reduced in the absence of BARD1 and BRCA1, respectively (34,35). Analyses of anti-WRN immunoprecipitates by immunoblotting showed that WRN associated not only with BRCA1 but also BARD1 (Figure 3). When the amount of BRCA1 and BARD1 brought down by anti-WRN immunoprecipitates in cells expressing all three proteins was compared with those co-expressing WRN and BRCA1 or WRN and BARD1, we did not observe a noticeable difference. Whatever the case, we conclude that WRN is likely to associate with both BRCA1 and BARD1 in vivo.
Figure 3.
Direct interaction of BRCA1 and BARD1 with WRN in vivo. Lysates from the insect Sf9 cells infected with the indicated baculoviruses were immunoprecipitated with antibodies against WRN or IgG, followed by immunoblotting using the indicated antibodies. Input shows 10% of the lysate used for immunoprecipitation.
BRCA1 stimulates WRN exonuclease and helicase activities
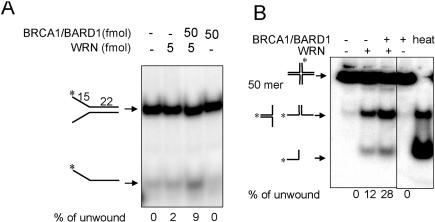
BRCA1 is expressed during S phase, where HR activity is high. To explore the significance of the physical interaction between WRN and BRCA1, we assayed WRN catalytic activities on DNA substrates that are intermediates in HR. Also, HR is essential for the repair of DNA ICLs. WRN helicase activity may play a role in resolving recombination intermediates formed during HR-mediated recombination or repair reactions (14,15). We tested WRN helicase activity in the presence or absence of the BRCA1/BARD1 complex. We used a sub-optimal amount of WRN in order to determine whether WRN helicase activity is promoted by its protein partners (4). We observed that pre-incubation with BRCA1/BARD1 significantly (P < 0.05) increased the extent of strand displacement on a forked substrate from 2 to 9% (Figure 4A) and on a Holliday (four-way) junction substrate 12 to 28% (Figure 4B). The BRCA1/BARD1 complex alone had no intrinsic DNA unwinding activity on either substrate, supporting the view that the BRCA1/BARD1 complex stimulates WRN helicase activity via its interaction with WRN instead of with DNA.
Figure 4.
Effect of BRCA1 on WRN helicase activities. Purified WRN (5 fmol) was pre-incubated with the indicated concentrations of BRCA1/BARD1 complex in 20 µl assay buffer. WRN helicase activity was measured using a 22 bp forked duplex substrate (A) or a 50mer Holliday junction substrate (B). Product ssDNA molecules are fast migrating species at the bottom of the gels. The mean percentage DNA strand displacement (n ≥ 2) is shown. Amount of DNA used per reaction was 10 fmol for forked substrates and 30 fmol for the Holliday junction substrate. Heated controls show 100% strand displacement.
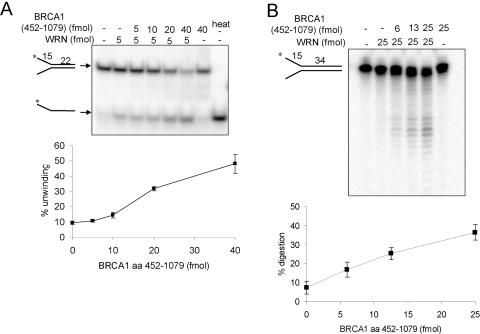
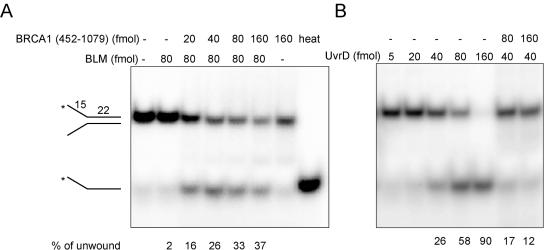
Because only the 452–1079 amino acids fragment of BRCA1 binds WRN (Figure 2C), we tested whether this fragment affected WRN catalytic activities. WRN helicase activity was stimulated 5-fold (P < 0.05) by pre-incubation with the 452–1079 amino acids fragment of BRCA1 (Figure 5A), whereas other regions of BRCA1 had no effect on WRN helicase activity (Supplementary Figure S5). None of the BRCA1 fragments had intrinsic DNA unwinding activity. The specificity of the helicase-stimulating function of BRCA1 was tested by assaying other helicases in the presence or absence of the BRCA1 452–1079 amino acids fragment. The Bloom syndrome protein (BLM) is another member of the RecQ helicase family. The BRCA1 fragment also stimulated BLM helicase activity; it did not, however, significantly stimulate the bacterial UvrD helicase (Figure 6). Thus, the BRCA1 helicase-stimulating function appears to be specific for the RecQ members of helicases, at least the ones tested here.
Figure 5.
Effect of the BRCA1 452–1079 amino acids fragment on WRN helicase and exonuclease activities. (A) WRN helicase activity was measured as described in Figure 4 using a 22 bp forked duplex substrate. (B) WRN exonuclease activity was measured on a 34 bp forked duplex substrate. Purified WRN was pre-incubated with BRCA1 452–1079 amino acids at the indicated concentrations in 10 µl assay buffer. The mean percent strand displacement or digestion (n = 2) on the substrate is shown.
Figure 6.
BRCA1 452–1079 amino acids fragment stimulates BLM helicase activity (A), but not UvrD helicase activity (B). Purified BLM helicase and UvrD helicase were pre-incubated with the BRCA1 fragment in 20 µl assay buffer and helicase activity was measured using the 34 bp forked duplex substrate as described in Figure 4.
BRCA1 452–1079 amino acids also stimulated WRN exonuclease activity, increasing product formation from 4 to 32% (Figure 5B). The BRCA1 fragment had no intrinsic exonuclease activity, and heat-inactivated BRCA1 did not stimulate WRN exonuclease activity (data not shown). Thus, BRCA1 and the KU heterodimer (36) share the ability to stimulate WRN exonuclease activity. Collectively, these data indicate that the 452–1079 amino acids fragment of BRCA1 stimulates both WRN catalytic activities, linking the physical and functional significance of the WRN-BRCA1 interaction.
Processing of DNA ICLs requires the helicase, but not the exonuclease, activity of WRN
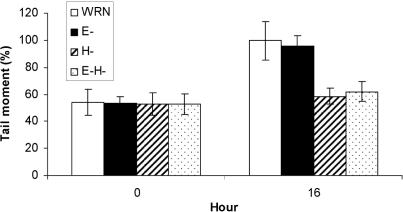
Because WRN functions with BRCA1 in the cellular response to DNA ICLs, and WRN helicase and exonuclease activities are stimulated by BRCA1, we asked whether both activities of WRN were important for the removal of DNA ICLs. For this purpose, we used AG03141 WS cells immortalized by telomerase and complemented with either wild-type WRN, or WRN defective in exonuclease activity (E-), helicase activity (H-), or both activities (E-H-) (24). Although telomerase immortalization has been shown to suppress the growth defects of WS due to telomere dysfunction (37), it does not affect sensitivity to ionizing radiation (38,39), and its effects on 4-nitroquinoline-1-oxide sensitivity is confined to SV-40 transformed WS cells (data not shown). The cells were treated with photoactivated psoralen. Western analyses using anti-WRN antibody demonstrated comparable levels of WRN expression in the four cells [(24), and data not shown]. The comet assay results showed decreased tail moments in wild-type WRN, E-WRN, H-WRN, and E-H-WRN complemented cells immediately after exposure to psoralen (0 h) and there was no difference among them, indicating that psoralen-induced ICL formation is independent of WRN catalytic activities (Figure 7). However, the comet assay results showed increased tail moments in the wild-type WRN and E-WRN complemented cells 16 h after exposure to psoralen. In contrast, tail moment recovery was not observed in the H-WRN and E-H-WRN complemented cells after exposure to psoralen. Tail moments in the wild-type and the E-WRN complemented cells were comparable 16 h after exposure to psoralen, and significantly greater in the H-WRN and the E-H-WRN complemented cells. Collectively, these data indicate that WRN may participate in the processing of psoralen-induced ICLs via its helicase activity, but not its exonuclease activity in vivo.
Figure 7.
Role of WRN helicase activity in the processing of DNA ICLs. Comet assays were performed between 0 and 16 h after cells were exposed to psoralen (1 µg 8-MOP/ml +UVA) as described in Figure 1B. The AG 03141 WS cells were complemented with wild-type WRN, exonuclease-inactive WRN (E-), helicase-inactive WRN (H-), or exonuclease- and helicase-inactive WRN (E-H-). Tail moments are normalized to the 16 h time point of wild-type WRN complemented cells. Data are averages of three independent assays, from each of which at least 50 cells were randomly selected. Values are means ± SD (n = 4).
DISCUSSION
This study provides the first evidence that WRN and BRCA1 coordinately participate in the response to DNA ICLs. BRCA1 directly binds to WRN and stimulates WRN helicase and exonuclease activities, and the interaction between WRN and BRCA1 increases in cells exposed to DNA cross-linking agents. Efficient processing of DNA ICLs in vivo required WRN helicase, but not exonuclease, activities. These observations suggest that WRN and BRCA1 may coordinately facilitate repair of DNA ICLs.
ICLs form a covalent attachment between the two strands of the DNA helix, thereby generating a strong block to DNA replication and inducing DSBs (40). Because WRN associates with BRCA1 after DNA ICLs and DSBs are formed in S phase, the WRN–BRCA1 interaction may prevent aberrant HR-mediated recombination during S phase (14). In the absence of BRCA1 or WRN, the frequency of DSBs during S phase might increase and induce additional DNA damage response pathways. Although it has recently been shown that WRN localizes to γH2AX foci (41,42), the role of WRN in repair of DNA DSBs associated with ICLs during DNA replication remains unclear.
Previous studies indicate that BLM may also participate in ICL repair in mammalian cells. In particular, cellular deficiency in BLM causes hypersensitivity to DNA cross-links (43) and a defect in BRCA1 focus formation after replication stress (44). Consistent with this notion, we found that BRCA1 also stimulates BLM helicase activity. However, the BLM helicase activity is not required for formation of BRCA1 foci after replication stress (44). Therefore, BRCA1 may recruit BLM to DNA lesions and stimulates BLM helicase, promoting efficient resolution of recombinational intermediates for the repair of DNA ICLs.
Because BRCA1 stimulates the helicase activity of WRN, another implication from this study is that BRCA1 and WRN act cooperatively in the resolution stages of HR. The process of HR generates recombination intermediates including Holliday junctions and other forked or branched DNA structures. These structures are preferred substrates for WRN, and WRN is likely to play a role in their resolution (45,46). Because BRCA1 stimulates WRN unwinding and digestion of forked duplexes, BRCA1 and WRN may also play a role in replication fork maintenance. Recent studies show that SGS1, a yeast ortholog of WRN, plays a role in maintaining replication fork stability (47).
Identification of a functional and physical interaction between WRN and BRCA1 may have some relevance for novel therapies for breast and ovarian cancer. Recent studies show that BRCA-deficient cells are susceptible to killing by an inhibitor of poly(ADP-ribose) polymerase (PARP) (48,49), and previous studies in our laboratory demonstrate that WRN interacts with PARP-1. Furthermore, WS cells have less poly(ADP-ribosyl)ation than wild-type cells (50), suggesting a functional interaction WRN and PARP. Because BRCA1 stimulates WRN exonuclease and helicase, BRCA mutant cells may have lower WRN catalytic activities than wild-type cells. In contrast, PARP-1 inhibits WRN helicase and exonuclease activities (51), so inhibition of PARP-1 should increase WRN catalytic activities. In BRCA1 mutant cells, WRN may not be stimulated by PARP inhibitors, while in wild-type cells, PARP inhibitors may stimulate WRN by a BRCA1-dependent pathway. We believe it is potentially critical to understand this complex interaction between PARP, BRCA1 and WRN, and that this understanding may stimulate development of therapeutic approaches to breast and ovarian cancer based on PARP inhibitors.
Lastly, it should be pointed out that both BRCA1 and WRN are likely to play roles in human aging. For example, murine BRCA1 hypomorphs manifest many features of accelerated aging (52), which may reflect an increased frequency of DSBs (53). If this is true, any factor that inhibits the interaction between WRN and BRCA1 could potentially contribute to aging-like pathologies in mammalian species. This hypothesis is consistent with the notion that BRCA1 and WRN contribute significantly to genomic stability and thus prevent both aging and cancer (54).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
The authors thank L. Brunner, L. Dawut, C. Kasmer and A. May for technical support, I. Hickson for BLM protein, S. Matson for UvrD protein, R. Wersto and C. Morris for FACS analysis, A. Majumdar, R. Brosh,Jr, P. Mason and M. Hedayati for suggestion and critical reading of this manuscript. This research was supported by the Intramural Research Program of the NIH, National Institute on Aging. Funding to pay the Open Access publication charges for this article was provided by the Intramural Research Program of the NIH, National Institute on Aging.
Conflict of interest statement. None declared.
REFERENCES
- 1.West S.C. Molecular views of recombination proteins and their control. Nature Rev. Mol. Cell Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 2.Majumdar A., Puri N., Cuenoud B., Natt F., Martin P., Khorlin A., Dyatkina N., George A.J., Miller P.S., Seidman M.M. Cell cycle modulation of gene targeting by a triple helix-forming oligonucleotide. J. Biol. Chem. 2003;278:11072–11077. doi: 10.1074/jbc.M211837200. [DOI] [PubMed] [Google Scholar]
- 3.Dronkert M.L., Kanaar R. Repair of DNA interstrand cross-links. Mutat. Res. 2001;486:217–247. doi: 10.1016/s0921-8777(01)00092-1. [DOI] [PubMed] [Google Scholar]
- 4.Cheng W.H., von Kobbe C., Opresko P.L., Arthur L.M., Komatsu K., Seidman M.M., Carney J.P., Bohr V.A. Linkage between werner syndrome protein and the Mre11 complex via Nbs1. J. Biol. Chem. 2004;279:21169–21176. doi: 10.1074/jbc.M312770200. [DOI] [PubMed] [Google Scholar]
- 5.Zhong Q., Chen C.F., Li S., Chen Y., Wang C.C., Xiao J., Chen P.L., Sharp Z.D., Lee W.H. Association of BRCA1 with the hRad50-hMre11-p95 complex and the DNA damage response. Science. 1999;285:747–750. doi: 10.1126/science.285.5428.747. [DOI] [PubMed] [Google Scholar]
- 6.Dong Y., Hakimi M.A., Chen X., Kumaraswamy E., Cooch N.S., Godwin A.K., Shiekhattar R. Regulation of BRCC, a holoenzyme complex containing BRCA1 and BRCA2, by a signalosome-like subunit and its role in DNA repair. Mol. Cell. 2003;12:1087–1099. doi: 10.1016/s1097-2765(03)00424-6. [DOI] [PubMed] [Google Scholar]
- 7.Scully R., Chen J., Plug A., Xiao Y., Weaver D., Feunteun J., Ashley T., Livingston D.M. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell. 1997;88:265–275. doi: 10.1016/s0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- 8.Sakamoto S., Nishikawa K., Heo S.J., Goto M., Furuichi Y., Shimamoto A. Werner helicase relocates into nuclear foci in response to DNA damaging agents and co-localizes with RPA and Rad51. Genes Cells. 2001;6:421–430. doi: 10.1046/j.1365-2443.2001.00433.x. [DOI] [PubMed] [Google Scholar]
- 9.Bohr V.A., Souza P.N., Nyaga S.G., Dianov G., Kraemer K., Seidman M.M., Brosh R.M., Jr DNA repair and mutagenesis in Werner syndrome. Environ. Mol. Mutagen. 2001;38:227–234. doi: 10.1002/em.1076. [DOI] [PubMed] [Google Scholar]
- 10.Folias A., Matkovic M., Bruun D., Reid S., Hejna J., Grompe M., D'Andrea A., Moses R. BRCA1 interacts directly with the Fanconi anemia protein FANCA. Hum. Mol. Genet. 2002;11:2591–2597. doi: 10.1093/hmg/11.21.2591. [DOI] [PubMed] [Google Scholar]
- 11.Garcia-Higuera I., Taniguchi T., Ganesan S., Meyn M.S., Timmers C., Hejna J., Grompe M., D'Andrea A.D. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 12.Bhattacharyya A., Ear U.S., Koller B.H., Weichselbaum R.R., Bishop D.K. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J. Biol. Chem. 2000;275:23899–23903. doi: 10.1074/jbc.C000276200. [DOI] [PubMed] [Google Scholar]
- 13.Venkitaraman A.R. Tracing the network connecting BRCA and Fanconi anaemia proteins. Nature Rev. Cancer. 2004;4:266–276. doi: 10.1038/nrc1321. [DOI] [PubMed] [Google Scholar]
- 14.Prince P.R., Emond M.J., Monnat R.J., Jr Loss of Werner syndrome protein function promotes aberrant mitotic recombination. Genes Dev. 2001;15:933–938. doi: 10.1101/gad.877001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moynahan M.E., Chiu J.W., Koller B.H., Jasin M. Brca1 controls homology-directed DNA repair. Mol. Cell. 1999;4:511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 16.Yarden R.I., Pardo-Reoyo S., Sgagias M., Cowan K.H., Brody L.C. BRCA1 regulates the G2/M checkpoint by activating Chk1 kinase upon DNA damage. Nature Genet. 2002;30:285–289. doi: 10.1038/ng837. [DOI] [PubMed] [Google Scholar]
- 17.Franchitto A., Oshima J., Pichierri P. The G2 phase decatenation checkpoint is defective in Werner syndrome cells. Cancer Res. 2003;63:3289–3295. [PubMed] [Google Scholar]
- 18.Zhou C., Huang P., Liu J. The carboxyl-terminal of BRCA1 is required for subnuclear assembly of RAD51 after treatment with cisplatin but not ionizing radiation in human breast and ovarian cancer cells. Biochem. Biophys. Res. Commun. 2005;336:952–960. doi: 10.1016/j.bbrc.2005.08.197. [DOI] [PubMed] [Google Scholar]
- 19.Cousineau I., Abaji C., Belmaaza A. BRCA1 regulates RAD51 function in response to DNA damage and suppresses spontaneous sister chromatid replication slippage: implications for sister chromatid cohesion, genome stability and carcinogenesis. Cancer Res. 2005;65:11384–11391. doi: 10.1158/0008-5472.CAN-05-2156. [DOI] [PubMed] [Google Scholar]
- 20.Levran O., Attwooll C., Henry R.T., Milton K.L., Neveling K., Rio P., Batish S.D., Kalb R., Velleuer E., Barral S., et al. The BRCA1-interacting helicase BRIP1 is deficient in Fanconi anemia. Nature Genet. 2005;37:931–933. doi: 10.1038/ng1624. [DOI] [PubMed] [Google Scholar]
- 21.Levitus M., Waisfisz Q., Godthelp B.C., de V.Y., Hussain S., Wiegant W.W., Elghalbzouri-Maghrani E., Steltenpool J., Rooimans M.A., Pals G., et al. The DNA helicase BRIP1 is defective in Fanconi anemia complementation group J. Nature Genet. 2005;37:934–935. doi: 10.1038/ng1625. [DOI] [PubMed] [Google Scholar]
- 22.Orren D.K., Brosh R.M., Jr, Nehlin J.O., Machwe A., Gray M.D., Bohr V.A. Enzymatic and DNA binding properties of purified WRN protein: high affinity binding to single-stranded DNA but not to DNA damage induced by 4NQO. Nucleic Acids Res. 1999;27:3557–3566. doi: 10.1093/nar/27.17.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paull T.T., Cortez D., Bowers B., Elledge S.J., Gellert M. Direct DNA binding by Brca1. Proc. Natl. Acad. Sci. USA. 2001;98:6086–6091. doi: 10.1073/pnas.111125998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen L., Huang S., Lee L., Davalos A., Schiestl R.H., Campisi J., Oshima J. WRN, the protein deficient in Werner syndrome, plays a critical structural role in optimizing DNA repair. Aging Cell. 2003;2:191–199. doi: 10.1046/j.1474-9728.2003.00052.x. [DOI] [PubMed] [Google Scholar]
- 25.Majumdar A., Khorlin A., Dyatkina N., Lin F.L., Powell J., Liu J., Fei Z., Khripine Y., Watanabe K.A., George J., et al. Targeted gene knockout mediated by triple helix forming oligonucleotides. Nature Genet. 1998;20:212–214. doi: 10.1038/2530. [DOI] [PubMed] [Google Scholar]
- 26.Rothfuss A., Grompe M. Repair kinetics of genomic interstrand DNA cross-links: evidence for DNA double-strand break-dependent activation of the Fanconi anemia/BRCA pathway. Mol. Cell Biol. 2004;24:123–134. doi: 10.1128/MCB.24.1.123-134.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng W.H., von Kobbe C., Opresko P.L., Fields K.M., Ren J., Kufe D., Bohr V.A. Werner syndrome protein phosphorylation by abl tyrosine kinase regulates its activity and distribution. Mol. Cell Biol. 2003;23:6385–6395. doi: 10.1128/MCB.23.18.6385-6395.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Opresko P.L., Laine J.P., Brosh R.M., Jr, Seidman M.M., Bohr V.A. Coordinate action of the helicase and 3′ to 5′ exonuclease of Werner Syndrome protein. J. Biol. Chem. 2001;276:44677–44687. doi: 10.1074/jbc.M107548200. [DOI] [PubMed] [Google Scholar]
- 29.Mohaghegh P., Karow J.K., Brosh J.R., Jr, Bohr V.A., Hickson I.D. The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poot M., Yom J.S., Whang S.H., Kato J.T., Gollahon K.A., Rabinovitch P.S. Werner syndrome cells are sensitive to DNA cross-linking drugs. FASEB J. 2001;15:1224–1226. doi: 10.1096/fj.00-0611fje. [DOI] [PubMed] [Google Scholar]
- 31.Poot M., Gollahon K.A., Emond M.J., Silber J.R., Rabinovitch P.S. Werner syndrome diploid fibroblasts are sensitive to 4-nitroquinoline-N-oxide and 8-methoxypsoralen: implications for the disease phenotype. FASEB J. 2002;16:757–758. doi: 10.1096/fj.01-0906fje. [DOI] [PubMed] [Google Scholar]
- 32.Fujiwara Y., Higashikawa T., Tatsumi M. A retarded rate of DNA replication and normal level of DNA repair in Werner's syndrome fibroblasts in culture. J. Cell Physiol. 1977;92:365–374. doi: 10.1002/jcp.1040920305. [DOI] [PubMed] [Google Scholar]
- 33.Simons A.M., Horwitz A.A., Starita L.M., Griffin K., Williams R.S., Glover J.N., Parvin J.D. BRCA1 DNA-binding activity is stimulated by BARD1. Cancer Res. 2006;66:2012–2018. doi: 10.1158/0008-5472.CAN-05-3296. [DOI] [PubMed] [Google Scholar]
- 34.Wu L.C., Wang Z.W., Tsan J.T., Spillman M.A., Phung A., Xu X.L., Yang M.C., Hwang L.Y., Bowcock A.M., Baer R. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nature Genet. 1996;14:430–440. doi: 10.1038/ng1296-430. [DOI] [PubMed] [Google Scholar]
- 35.Fabbro M., Savage K., Hobson K., Deans A.J., Powell S.N., McArthur G.A., Khanna K.K. BRCA1-BARD1 complexes are required for p53Ser-15 phosphorylation and a G1/S arrest following ionizing radiation-induced DNA damage. J. Biol. Chem. 2004;279:31251–31258. doi: 10.1074/jbc.M405372200. [DOI] [PubMed] [Google Scholar]
- 36.Cooper M.P., Machwe A., Orren D.K., Brosh R.M., Ramsden D., Bohr V.A. Ku complex interacts with and stimulates the Werner protein. Genes Dev. 2000;14:907–912. [PMC free article] [PubMed] [Google Scholar]
- 37.Wyllie F.S., Jones C.J., Skinner J.W., Haughton M.F., Wallis C., Wynford-Thomas D., Faragher R.G., Kipling D. Telomerase prevents the accelerated cell ageing of Werner syndrome fibroblasts. Nature Genet. 2000;24:16–17. doi: 10.1038/71630. [DOI] [PubMed] [Google Scholar]
- 38.Yannone S.M., Roy S., Chan D.W., Murphy M.B., Huang S., Campisi J., Chen D.J. Werner syndrome protein is regulated and phosphorylated by DNA-dependent protein kinase. J. Biol. Chem. 2001;276:38242–38248. doi: 10.1074/jbc.M101913200. [DOI] [PubMed] [Google Scholar]
- 39.Karmakar P., Piotrowski J., Brosh R.M., Jr, Sommers J.A., Miller S.P., Cheng W.H., Snowden C.M., Ramsden D.A., Bohr V.A. Werner protein is a target of DNA-dependent protein kinase in vivo and in vitro, and its catalytic activities are regulated by phosphorylation. J. Biol. Chem. 2002;277:18291–18302. doi: 10.1074/jbc.M111523200. [DOI] [PubMed] [Google Scholar]
- 40.Thompson L.H. Unraveling the Fanconi anemia-DNA repair connection. Nature Genet. 2005;37:921–922. doi: 10.1038/ng0905-921. [DOI] [PubMed] [Google Scholar]
- 41.Lan L., Nakajima S., Komatsu K., Nussenzweig A., Shimamoto A., Oshima J., Yasui A. Accumulation of Werner protein at DNA double-strand breaks in human cells. J. Cell. Sci. 2005;118:4153–4162. doi: 10.1242/jcs.02544. [DOI] [PubMed] [Google Scholar]
- 42.Cheng W.H., Sakamoto S., Fox J.T., Komatsu K., Carney J., Bohr V.A. Werner syndrome protein associates with gamma H2AX in a manner that depends upon Nbs1. FEBS Lett. 2005;579:1350–1356. doi: 10.1016/j.febslet.2005.01.028. [DOI] [PubMed] [Google Scholar]
- 43.Pichierri P., Franchitto A., Rosselli F. BLM and the FANC proteins collaborate in a common pathway in response to stalled replication forks. EMBO J. 2004;23:3154–3163. doi: 10.1038/sj.emboj.7600277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davalos A.R., Campisi J. Bloom syndrome cells undergo p53-dependent apoptosis and delayed assembly of BRCA1 and NBS1 repair complexes at stalled replication forks. J. Cell. Biol. 2003;162:1197–1209. doi: 10.1083/jcb.200304016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prince P.R., Ogburn C.E., Moser M.J., Emond M.J., Martin G.M., Monnat R.J., Jr Cell fusion corrects the 4-nitroquinoline 1-oxide sensitivity of Werner syndrome fibroblast cell lines. Hum. Genet. 1999;105:132–138. doi: 10.1007/s004399900078. [DOI] [PubMed] [Google Scholar]
- 46.Constantinou A., Tarsounas M., Karow J.K., Brosh R.M., Bohr V.A., Hickson I.D., West S.C. Werner's syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep. 2000;1:80–84. doi: 10.1093/embo-reports/kvd004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cobb J.A., Schleker T., Rojas V., Bjergbaek L., Tercero J.A., Gasser S.M. Replisome instability, fork collapse, and gross chromosomal rearrangements arise synergistically from Mec1 kinase and RecQ helicase mutations. Genes Dev. 2005;19:3055–3069. doi: 10.1101/gad.361805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bryant H.E., Schultz N., Thomas H.D., Parker K.M., Flower D., Lopez E., Kyle S., Meuth M., Curtin N.J., Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 49.Farmer H., McCabe N., Lord C.J., Tutt A.N., Johnson D.A., Richardson T.B., Santarosa M., Dillon K.J., Hickson I., Knights C., et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 50.von Kobbe C., Harrigan J.A., May A., Opresko P.L., Dawut L., Cheng W.H., Bohr V.A. Central role for the Werner syndrome protein/poly(ADP-ribose) polymerase 1 complex in the poly(ADP-ribosyl)ation pathway after DNA damage. Mol. Cell Biol. 2003;23:8601–8613. doi: 10.1128/MCB.23.23.8601-8613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.von Kobbe C., Harrigan J.A., Schreiber V., Stiegler P., Piotrowski J., Dawut L., Bohr V.A. Poly(ADP-ribose) polymerase 1 regulates both the exonuclease and helicase activities of the Werner syndrome protein. Nucleic Acids Res. 2004;32:4003–4014. doi: 10.1093/nar/gkh721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cao L., Li W., Kim S., Brodie S.G., Deng C.X. Senescence, aging, and malignant transformation mediated by p53 in mice lacking the Brca1 full-length isoform. Genes Dev. 2003;17:201–213. doi: 10.1101/gad.1050003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sedelnikova O.A., Horikawa I., Zimonjic D.B., Popescu N.C., Bonner W.M., Barrett J.C. Senescing human cells and ageing mice accumulate DNA lesions with unrepairable double-strand breaks. Nature Cell. Biol. 2004;6:168–170. doi: 10.1038/ncb1095. [DOI] [PubMed] [Google Scholar]
- 54.Risinger M.A., Groden J. Crosslinks and crosstalk; Human cancer syndromes and DNA repair defects. Cancer Cell. 2004;6:539–545. doi: 10.1016/j.ccr.2004.12.001. [DOI] [PubMed] [Google Scholar]