Abstract
Here, we present a novel method for the directed genetic manipulation of the Bacillus subtilis chromosome free of any selection marker. Our new approach employed the Escherichia coli toxin gene mazF as a counter-selectable marker. The mazF gene was placed under the control of an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible expression system and associated with a spectomycin-resistance gene to form the MazF cassette, which was flanked by two directly-repeated (DR) sequences. A double-crossover event between the linearized delivery vector and the chromosome integrated the MazF cassette into a target locus and yielded an IPTG-sensitive strain with spectomycin-resistance, in which the wild-type chromosome copy had been replaced by the modified copy at the targeted locus. Another single-crossover event between the two DR sequences led to the excision of the MazF cassette and generated a strain with IPTG resistance, thereby realizing the desired alteration to the chromosome without introducing any unwanted selection markers. We used this method repeatedly and successfully to inactivate a specific gene, to introduce a gene of interest and to realize the in-frame deletion of a target gene in the same strain. As there is no prerequisite strain for this method, it will be a powerful and universal tool.
INTRODUCTION
Bacillus subtilis and several of its close relatives have long been exploited for industrial and biotechnological applications (1). The completion of the sequencing and annotation of the B.subtilis 168 strain genome supplied a complete view of the B.subtilis protein machinery, and this knowledge stimulated new approaches to the analysis of its biochemical pathways (2). Post-genomic studies require simple and highly efficient tools to enable genetic manipulations. The most prominent and widely used systems are delivery plasmids, which allow the insertion of any type of genetic information into the bacterial chromosome. Classically, these chromosomal modifications have been achieved by a method using a positive selection marker, usually an antibiotic-resistance marker generated by the insertion of a selection marker gene into the chromosome. According to this strategy, the introduction of a second chromosomal modification requires a second resistance gene; alternatively, if the same resistance gene is used, the eviction of the gene by a single-crossover event prior to further genetic manipulation is required. In the first case, the number of chromosomal modifications is limited by the number of available resistance genes; moreover, the multi-antibiotic pressure could modify the physiology of the manipulated strain. In the second case, selection of the strain that has lost resistance is time-consuming due to the relatively low frequencies and the absence of positive selection. Counter-selectable markers are often instrumental in the construction of clean and unmarked mutations in bacteria (3). Under appropriate growth conditions, a counter-selectable gene can promote the death of the microorganisms that harbor it. Hence, transformants that have integrated a suicide vector containing a counter-selectable marker through a single-crossover or double-crossover event retain a copy of the counter-selectable marker in the chromosome, and are therefore eliminated in the presence of the counter-selective compound.
Until now, only two similar methods have been described that allow the subsequent excision of the selection marker coupled with positive selection in B.subtilis; these approaches use the upp gene and the blaI gene, respectively, as a counter-selectable marker (4,5). However, for the two abovementioned methods to be effective, it is essential to have a strain with a mutation for a specific gene in the chromosome. When these methods are applied to different strains, new mutants must also be prepared. The two existing methods can therefore only be used in strains that have a clear genetic background for preparing mutations of a specific gene, which limits their application. The novel method described here can satisfy the strong demand for a universal unmarked delivery system that can be applied in any Bacillus species without requiring any prior modification to the host.
Toxin–antitoxin (TA) systems comprise pairs of adjacent genes in which a stable toxic peptide is neutralized by an unstable antitoxin (6). The mazEF cassette in the Escherichia coli chromosome is a well-characterized TA locus; ectopic expression of the MazF toxin inhibits cell growth and the MazE antitoxin neutralizes the cognate toxin (7). The latest research has revealed that MazF is an endoribonuclease that specifically cleaves free mRNAs at ACA sequences (8). Here, we describe a procedure based on the use of mazF as a novel counter-selectable maker in B.subtilis, which has enabled us to inactivate a single gene, to introduce a gene of interest and to realize the in-frame deletion of a target gene into the B.subtilis chromosome. In these cases, the resulting strains were free of selection markers, thus allowing the repeated use of the method for further manipulations of the Bacillus chromosome. No prerequisite strain is needed for this newly developed method, so it will have wide applications.
MATERIALS AND METHODS
Bacterial strains, plasmids and oligonucleotides
The bacterial strains and plasmids used in this study are listed in Table 1. The specific primers (Table 2) used for the PCR amplification were synthesized by Invitrogen Biotechnology Co., Ltd.
Table 1.
Strains and plasmids
| Strain or plasmid | Characteristicsa | Reference |
|---|---|---|
| E.coli JM109 | recA1, supE44 endA1 hsdR17 (, ) gyrA96 relA1 thi (lac-proAB) F′[traD36 proAB+lacIqlacZΔM15] | (9) |
| B.subtilis | ||
| 1A751 | his nprR2 nprE18 ΔaprA3 ΔeglS102 ΔbglT bglSRV | (10) |
| BS752 | 1A751 derivate, Spcr, amyE::(DR, lacI, mazF, spc, DR) | This work |
| BS752S | 1A751 derivate, amyE::DR | This work |
| BS753 | 1A751 derivate, Spcr, amyE::(mpd, DR, lacI, mazF, spc, DR) | This work |
| BS753S | 1A751 derivate, amyE::(mpd, DR) | This work |
| BS754 | 1A751 derivate, Spcr, amyE::(mpd, DR), bpr::(DR, lacI, mazF, spc) | This work |
| BS754S | 1A751 derivate, amyE::(mpd, DR), Δbpr | This work |
| Brevibacillus brevis B15 | Wild-type | Stored in this lab |
| Plasmids | ||
| pDG148 | Ampr, Kmr, B.subtilis–E.coli shuttle expression vector | (11) |
| pDG1730 | Ampr, Spcr, Ermr, B.subtilis amyE integration vector | (12) |
| pDGIEF | Ampr, Spcr, B.subtilis amyE integration vector | This work |
| pP43NMK | Ampr, Kmr, B.subtilis–E.coli shuttle expression vector containing an expression cassette including ytkA promoter, NprB signal peptide-encoding sequence and mpd gene | (13) |
| pP15MK | Ampr, Kmr, B.subtilis–E.coli shuttle expression vector | This work |
| pDGIEF-mpd | pDGIEF with mpd expression cassette inserted in NotI/NheI site | This work |
| pBluescript II SK(−) | Ampr, E.coli cloning vector | Stratagene |
| pIEFBPR | Ampr, Spcr, B.subtilis bpr integration vector | This work |
| pIEFBPR-ID | Ampr, Spcr, B.subtilis bpr integration vector | This work |
aSpcr: Spectinomycin-resistance, Ampr: Ampicillin-resistance, Kmr: Kanamycin resistance, Ermr: Erythromycin resistance.
Table 2.
Oligonucleotides used for construction of plasmids
| Primer | Sequence |
|---|---|
| P1 | 5′-CGAGCTCCGATAACCAGAAGCAATA-3′ |
| P2 | 5′-ACTGTGGATCATTATTAATACCCTCT-3′ |
| P3 | 5′-AGAGGGTATTAATAATGATCCACAGT-3′ |
| P4 | 5′-CATCGATTTACCAGACTTCCTTATC-3′ |
| P5 | 5′-GAAGCTTAAGGAGGAAGCAGGTATGGTAAGCCGATAC-3′ |
| P6 | 5′-GCGCATGCCTACCCAATCAGTACGTT-3′ |
| P7 | 5′-CACGGATCCGCGGCTAGCCATGGTCGACGCGGCCGCAAAATTGAAAAAATGGTGG-3′ |
| P8 | 5′-CGAGGATCCGATCCCCCTATGCAAGGG-3′ |
| P9 | 5′-CCTGGATCCATCTTCAACTTGGCTGTCGTA-3′ |
| P10 | 5′-CTCCTGCAGCGAAAGCCATGGGAGCAACAG-3′ |
| P11 | 5′-GACGCGGCCGCATCTTCAACTTGGCTGTC-3′ |
| P12 | 5′-GACGCTAGCTCATCATCACTTGGGGTTG-3′ |
| P13 | 5′-CTCGAGTTAACCTGCAGGCGCGCCTTTGATGGGTATCGG-3′ |
| P14 | 5′-GTCGACCATGGCTAGCATCGATAGATCTTTAAATTTAATCAGAAAAG-3′ |
| P15 | 5′-CCCGGGCCGGCCAATTGATGCATCAGTTCAAAAGGG-3′ |
| P16 | 5′-GACTAGTGCGATCGCCCTAGGACCGGTATTTTCTGTGTTC-3′ |
| P17 | 5′-GACACTAGTCGATAACCAGAAGCAATA-3′ |
| P18 | 5′-GCTGAGCTCTTACCAGACTTCCTTATC-3′ |
| P19 | 5′-CGAGGATCCGCGGTTTAAACTCCGGAGATCCCCCTATGCAAGGG-3′ |
| P20 | 5′-CAGATCTCAGTTCAAAAGGG-3′ |
| P21 | 5′-GGATCCACTGCCAGTTGCGG-3′ |
| P22 | 5′-TCATCAATCATACCACCAG-3′ |
| P23 | 5′-GTGCCTGTCAGTTTACCA-3′ |
Culture and growth conditions
All organisms were grown in Luria–Bertani (LB) (9) or LBG medium (LB medium containing 1% glucose). When required, the final concentrations of antibiotics were as follows: 100 µg/ml ampicillin and 100 µg/ml spectinomycin.
Amylase activity detection
Amylase expression by Bacillus colonies was detected by growing colonies overnight on an LB plate containing 1% starch and then staining the plate with iodine.
DNA-manipulation techniques
The isolation and manipulation of recombinant DNA was performed using standard techniques. All enzymes were commercial preparations, and were used as specified by the suppliers (TaKaRa). E.coli transformation was performed as described by Sambrook et al. (9). B.subtilis transformation was performed by the competent-cell method (14).
Construction of pDGIEF
Using E.coli JM109 chromosomal DNA as the template, the promoter functional region of the E.coli lpp gene was PCR amplified using the primer pair P1/P2, with the SacI site introduced by the forward primer; the mazE gene was amplified using the primer pair P3/P4, with the ClaI site introduced by the reverse primer. Primers P2 and P3 were designed as completely reverse complements, and the two PCR fragments of the lpp promoter and the mazE gene were spliced by overlap extension; this generated the mazE over-expression cassette, which was cloned into the corresponding sites of the integration vector pDG1730, thereby generating pDGE. The EcoRI/BamHI fragment excised from the vector pDG148, which contained the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible Pspac promoter, and the lac-repressor-encoding gene lacI, were inserted into the corresponding sites of the vector pDGE, thereby yielding pDGIE. The mazF gene was amplified using the primer pair P5/P6, with the HindIII site and the Shine–Dalgarno sequence (UAAGGAGG) introduced by the forward primer, and the SphI site introduced by the reverse primer; the HindIII/SphI-digested PCR product was then cloned into the corresponding site of pDGIE to yield pDGIEFO. Finally, a 125 bp DNA sequence between the spc gene and the amyE-back homologous arm in pDGIEFO was amplified using the primer pair P7/P8; the PCR product was flanked with the BamHI site and then cloned into the corresponding site of pDGIEFO. The fact that the inserted sequence ran in the same direction as that seen between spc and amyE-back was confirmed by DNA sequencing, and the resulting vector was designated as pDGIEF.
Construction of pDGIEF-mpd
The multiple and tandem promoters, and the signal peptide-encoding sequence of the cell-wall protein-encoding gene (cwp) (GenBank accession no. AY956423), were amplified from Brevibacillus brevis B15 chromosomal DNA, using the primer pair P9/P10, with the BamHI and PstI sites introduced by the forward and reverse primers, respectively. The PCR product was cloned into the corresponding site of pP43NMK to generate pP15MK, which made the promoters and peptide-encoding sequence fused in-frame with the mpd gene in pP43NMK. The mpd expression cassette containing the promoters, signal peptide-encoding sequence and mpd gene in pP15MK was then amplified using the primer pair P11/P12, with the NotI and NheI sites being introduced by the forward and reverse primers, respectively. The PCR product of the mpd expression cassette was digested with NotI/NheI and cloned into pDGIEF, yielding pDGIEF-mpd.
Construction of pIEFBPR
The two primer pairs P13/P14 and P15/P16 were used to amplify the upstream (bpr-front) and downstream (bpr-back) DNA sequences of the bpr gene (encoding bacillopeptidase F) from B.subtilis 1A751 chromosomal DNA; the XhoI-HpaI-SbfI-AscI/ClaI-NheI-NcoI-SalI and XmaI-FseI-MfeI-NsiI/AgeI-AvrII-AsiSI-SpeI sites were introduced by the two primer pairs, respectively. The Plpp-mazE cassette was amplified from pDGIEF using the primer pair P17/P18, which introduced the SpeI and SacI sites, respectively. The MazF cassette and the two DR sequences were excised from pDGIEF with SalI/XmaI. The fragment of the MazF cassette flanked by the DR sequences and the three PCR fragments described above, which were digested with the appropriate restriction enzymes (the bpr-front and bpr-back fragments were digested with XhoI/SalI and XmaI/SpeI, respectively), were cloned into the corresponding sites of pBluescript II SK(−), yielding pIEFBPRO. To introduce a multiple cloning site (MCS) at the 3′ end of the upstream DR sequence in pIEFBPRO, the DR sequence was amplified using the primer pair P7/P19, which, along with an MCS (BspEI-PmeI-SacII-BamHI), was introduced by primer P19. The new DR sequence was digested with NotI/BamHI and cloned into the corresponding sites of pIEFBPRO, in order to replace the original DR sequence, thereby resulting in the new vector pIEFBPR.
Construction of pIEFBPR-ID
A 125 bp DNA sequence, which was identical to the sequence of the 5′ end of the bpr-back homologous arm, was amplified using the primer pair P20/P21. The PCR product was flanked with the BglII and BamHI sites, and was cloned into the corresponding site of pIEFBPR; the resulting vector was designated as pIEFBPR-ID.
Methyl parathion hydrolase-activity assay
A methyl parathion hydrolase-activity assay was performed as described previously (15).
RESULTS
Construction of the selection–eviction MazF cassette
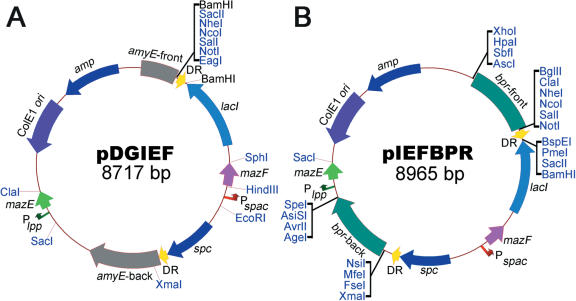
In order to test the feasibility of using mazF as a counter-selectable marker in B.subtilis, we preliminarily tried to clone mazF into an IPTG-inducible expression vector pDG148, and attempted to place the toxin gene under the control of the Pspac promoter for the regulated expression of the toxin gene. Although the IPTG-inducible expression system of pDG148 has been optimized to achieve the tightly-regulated expression of foreign protein in B.subtilis (11), there is some leakage when using this system in E.coli. For this reason, we failed to clone the toxin gene mazF into pDG148, even in E.coli JM109, which is a lacIq genotype strain. To resolve this problem, the antitoxin gene mazE was placed under the control of the E.coli lpp promoter, which is considered to be one of the strongest promoters in E.coli (16), in order to counteract the toxicity of the leaked MazF. This mazE over-expression cassette was transferred to a B.subtilis-integration vector, pDG1730, which can stably insert the DNA sequences at the amyE locus into the B.subtilis chromosome via a double-crossover event (12). The EcoRI/BamHI fragment excised from pDG148, containing the IPTG-inducible expression cassette, was also cloned into the corresponding sites of pDG1730. With MazE counteracting the toxicity of the leaked MazF, the mazF gene was successfully cloned into the integration vector under the control of the IPTG-inducible Pspac promoter. To complete the construction of the selection–eviction MazF cassette, a 125 bp directly-repeated (DR) sequence was generated by PCR and inserted into the BamHI site, which is identical to the sequence between the spc gene and the amyE-back homologous arm; an MCS was also introduced by the forward primer of the DR sequence. The recombinant vector was designated as pDGIEF (Figure 1A). The genetic elements between the two DR sequences were designated as the MazF cassette (Figure 1A). This design allowed the self eviction of the MazF cassette, which was integrated into the host chromosome, through a single-crossover event between the two DR sequences.
Figure 1.
Relevant features of the delivery vector pDGIEF (A) and its derivative pIEFBPR (B). ColE1 ori, ColE1 replication origin; amp, ampicillin-resistance marker; spc, spectinomycin-resistance marker; lacI, lac-repressor-encoding gene; Pspac, Pspac promoter; mazF, E.coli toxin gene; Plpp, promoter of E.coli lpp gene; mazE, E.coli antitoxin gene; DR, directly-repeated sequence; amyE-front and amyE-back, upstream and downstream sequences of the B.subtilis amyE gene, respectively; bpr-front and bpr-back, upstream and downstream sequences of the B.subtilis bpr gene, respectively. All of the DNA sequences located between the amyE-front and amyE-back or the bpr-front and bpr-back allow integration into the chromosome of B.subtilis via a double-crossover event, resulting in selection for spectinomycin-resistance. The unique restriction enzyme sites are highlighted in blue.
Integration and eviction of the MazF cassette in B.subtilis 1A751
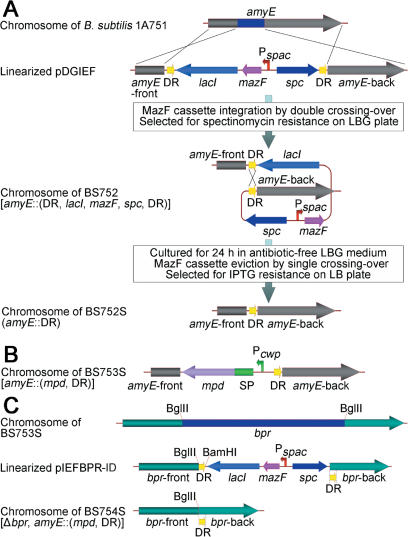
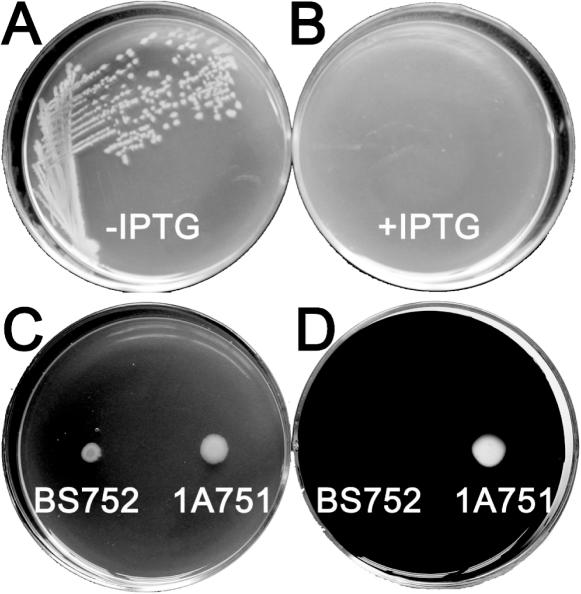
To probe the feasibility of the eviction of the MazF cassette after its integration into the B.subtilis 1A751 chromosome amyE locus, competent cells of this strain were transformed with SacI-linearized pDGIEF and plated on LBG medium supplemented with 100 µg/ml spectinomycin. The selected spectinomycin-resistant B.subtilis 752 (BS752) strain (Figure 2A) could not form colonies on the LB medium supplemented with 1 mM IPTG (Figure 3B), and the occurrence of a double-crossover event (Figure 2A) was confirmed by the lack of α-amylase activity visualized by a starch-plate assay (Figure 3D). The integration of the mazF gene into the host chromosome was confirmed by PCR (Figure 4A). The fact that BS752 was unable to grow on the medium containing IPTG suggests that mazF can be used as a counter-selectable marker during the eviction of the MazF cassette. The eviction of the MazF cassette by a single-crossover event (Figure 2A) was achieved by growing BS752 for 24 h in 10 ml antibiotic-free LBG medium. After dilution of the culture, the clones isolated on LB medium with added IPTG were analyzed for their ability to grow on LB medium supplemented with spectinomycin. The eviction of the MazF cassette from the spectinomycin-sensitive strain BS752S chromosome (Figure 2A) was confirmed by PCR, which detected the existence of the mazF gene (Figure 4A). After the single-crossover event between the two DR sequences, a scar of the DR sequence was left behind at the amyE locus of the BS752S chromosome (Figure 2A), which replaced the amyE partial sequence and disrupted the amyE open reading frame (ORF). Notably, during the selection of the integrants and the culture of the BS752 strain to achieve the self eviction of the MazF cassette, complex mediums containing tryptone should be supplemented with 1% glucose, because the unintended induction of mazF expression caused by small amounts of lactose in tryptone (17) will result in the accumulation of spontaneous mazF mutants. The addition of 1% glucose to the complex medium can prevent the unintended induction by well-studied mechanisms (17,18).
Figure 2.
Construction of BS752S (amyE::DR), BS753S [amyE::(mpd, DR)] and BS754S [Δbpr, amyE::(mpd, DR)]. (A) Flow scheme for the construction of BS752S. X indicates one crossover event. (B) Schematic representation of the BS753S chromosome. Pcwp, promoter of cwp gene; SP, cwp signal peptide-encoding sequence; mpd, methyl parathion hydrolase-encoding gene. (C) Schematic representation of linearized pIEFBPR-ID, and the BS753S and BS754S chromosomes.
Figure 3.
A test of the feasibility of the mazF gene as a counter-selectable marker. B.subtilis BS752 was streaked on an LB plate in the absence (A) and presence (B) of 1 mM IPTG. (C and D) B.subtilis 1A751 and BS752 were grown on LB plates containing 1% starch; one plate (D) was stained with iodine to detect the α-amylase activity, which was indicated by the transparent plaque.
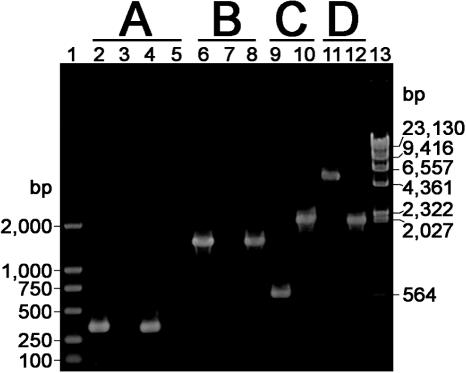
Figure 4.
Detection and characterization of the mazF gene, the mpd expression cassette, the partial amyE gene and the bpr gene using PCR. Plasmids or B.subtilis chromosomal DNA were used as the template, and the amplified fragments were analyzed by agarose gel electrophoresis. Lanes 1 and 13, DNA markers. (A) The primer pair P5/P6 was used to amplify the mazF gene. Lane 2, pDGIEF (positive control); lane 3, 1A751 (negative control); lane 4, BS752; lane 5, BS752S. (B) The primer pair P11/P12 was used to amplify the mpd expression cassette. Lane 6, pDGIEF-mpd (positive control); lane 7, BS752S (negative control); lane 8, BS753S. (C) The primer pair P22/P23 was used to amplify the partial amyE gene. Lane 9, BS752S (negative control); lane 10, BS753S. (D) The primer pair P13/P16 was used to amplify the bpr gene. Lane 11, BS753S (negative control); lane 12, BS754S.
Integration of the mpd gene into BS752S and subsequent eviction of the MazF cassette
The mpd gene encoding methyl parathion hydrolase from Plesiomonas sp. M6 (19) was constructed under the control of the Brevibacillus brevis B15 cwp promoter and signal peptide, and inserted into pDGIEF to generate pDGIEF-mpd. The chromosomal integration of the mpd gene and the MazF cassette into the BS752S amyE gene locus, and the eviction of the MazF cassette from the chromosome, were carried out as described above. This generated the spectomycin-resistant BS753 strain and the desired spectinomycin-sensitive BS753S strain (Figure 2B). After the eviction of the MazF cassette, one copy of the DR sequence, the cwp promoter, the signal peptide-encoding sequence and the mpd gene were left behind in the chromosome of the BS753S strain, which was inserted into the amyE gene locus (Figure 2B). The BS753S chromosome was characterized by PCR, and its ability to express the mpd gene was confirmed by detecting the methyl parathion hydrolase-activity on the LB plate containing methyl parathion. A yellow halo (p-nitrophenol) around the BS753S colony (Figure 5) showed that the mpd gene was functionally expressed. The PCR amplifications obtained using the corresponding primer pairs for the mpd expression cassette and the partial amyE gene are shown in Figure 4. The mpd expression cassette-amplified fragment in BS753S (about 1.5 kb) (Figure 4B, lane 8) was the same length as that obtained when pDGIEF-mpd was used as a DNA template (Figure 4B, lane 6); this indicated that the mpd expression cassette was integrated into the BS753S chromosome. The amplified fragments for the partial amyE gene in BS752S (about 0.6 kb) and BS753S are shown in Figure 4C; in contrast to lane 9, a band of about 2.1 kb present in lane 10 indicated that the mpd expression cassette and one copy of the DR sequence were inserted into the amyE gene locus of the BS753S chromosome.
Figure 5.
Methyl parathion hydrolase-activity detection of BS752S and BS753S. The colonies were transferred to an LB plate containing 100 µg/ml methyl parathion and cultivated for 24 h.
Development of a universal integration vector using the B.subtilis bpr gene as the target
Although the MazF cassette proved to be functional in the counter-selection procedure, some restriction enzyme sites were still missing in pDGIEF flanking the two homologous arms. The introduction of new restriction sites would facilitate the replacement of the two homologous arms to create new integration vectors with the desired target sequences. To do this, the mazE over-expression cassette, MazF cassette and DR sequences were cloned into pBluescript II SK(−). Two new homologous arms (upstream and downstream sequences of the B.subtilis bpr gene encoding bacillopeptidase F) were amplified from B.subtilis 1A751 chromosomal DNA and cloned into the corresponding sites of pBluescript II SK(−) that flank the MazF cassette; all of the abovementioned genetic elements were flanked with the unique restriction enzyme sites, yielding a new integration vector pIEFBPRO that could integrate into the B.subtilis bpr gene locus. The original upstream DR sequence in pIEFBPRO was then replaced with a new one, which, along with an MCS, was introduced at the 3′ end of the DR sequence, thereby generating the new vector pIEFBPR (Figure 1B). This plasmid was constructed in a modular way, which allowed the replacement of each part by others as follows: (i) cleavage with appropriate restriction enzyme sites in MCS could release the vector backbone; (ii) the other designed homologous arms could also be replaced by the MCS sites; (iii) application of the restriction enzyme sites in MCS/SacI or MCS released the mazE over-expression cassette and the MazF cassette, respectively, which allowed this functional system to be transferred into other vectors; (iv) cleavage with MCS could release one of the DR sequences and (v) there was an MCS flanking the MazF cassette, which allowed the gene of interest to be cloned into it. This modular structure greatly improves the versatility of this vector for all possible applications.
In-frame deletion of the bpr gene in BS753S
To realize the in-frame deletion of the BS753S bpr gene using the newly developed system, the original DR sequence flanking the bpr-front homologous arm was replaced with a new 125 bp DR sequence that was identical to the sequence of the 5′ end of bpr-back homologous arm in the vector pIEFBPR, which generated pIEFBPR-ID (Figure 2C). In the new vector pIEFBPR-ID, the two DR sequences were located in two homologous arms and this meant that the two homologous arms overlapped. BS753S was transformed with linearized pIEFBPR-ID and the MazF cassette was integrated into the bpr gene locus through a double-crossover event; subsequently, after a single-crossover event between the two DR sequences, the desired strain BS754S (Figure 2C) was selected. In the BS754S chromosome, after the removal of the MazF cassette, the remaining ORF contained the Bpr 76 N-terminal and 346 C-terminal amino acid-encoding sequence. The internal deletion of the bpr gene was confirmed by PCR using the primer pair P13/P16 (Figure 4D); a small band of about 2.1 kb (Figure 4D, lane 12) indicated that the partial internal sequence (about 3.0 kb) of the bpr gene had been successfully deleted. The in-frame deletion event was also confirmed by DNA sequencing of the 2.1 kb PCR product of the internal deleted bpr gene.
DISCUSSION
This study developed a simple and efficient method for the directed genetic manipulation of the B.subtilis chromosome. This novel approach is based on the use of the mazF gene from the E.coli chromosome, which encodes an endoribonuclease, as a novel counter-selectable marker. The mazF gene was placed under the control of the IPTG-inducible Pspac promoter, and was associated with the LacI-repressor-encoding gene and a spectomycin-resistance gene to form the MazF cassette. Homologous integration of the linearized plasmid into the chromosome yields a strain in which the wild-type chromosome copy has been replaced by the modified copy at the targeted locus. The design of the transforming DNA molecule was such that the MazF cassette was flanked by two DR sequences; homologous recombination between these DR sequences leads to the excision of the MazF cassette, leaving only the desired alteration in the chromosome.
We used this method to inactivate a specific gene, to introduce a gene of interest and to realize the in-frame deletion of the bpr gene in the same strain. This strategy can also be used to deliver unmarked point mutations and large deletions into the chromosome. In the first case, the point mutation must be presented in the two DR sequences, and the DR sequence must be a portion of the target sequence. A deletion disrupting the ORF of a gene might have a strong effect on the expression of a downstream gene (4), and an in-frame deletion might have no polar effect (20). Our new system allows the efficient introduction of in-frame deletions, which will minimize the chance of exerting a polar effect on the expression of downstream genes. At the same time, when the MazF cassette was integrated into an operon with downstream essential genes, in the first step of the procedure, before the eviction of the cassette took place, continuous read-through transcription from the spc promoter beyond the downstream DR was able to avoid the unwanted polar effect. However, when necessary, an inducible promoter could also be inserted into the MCS positioned at the 3′ end of the MazF cassette, following the downstream DR, in order to eliminate the polar effect.
Compared with the delivery system described by Fabret et al. (4) and Brans et al. (5), our technique is well adapted to universal use, due to the fact that there is no need for a specific prerequisite strain. This will allow it to be used in wild-type Bacillus isolates for which the genetic backgrounds have not yet been clarified. So, the strategy described in this paper is highly efficient and can be used as a valuable tool to manipulate the Bacillus chromosome. Furthermore, this system should also be functional in the Gram-positive and Gram-negative bacteria when a tightly-regulated inducible expression system is employed. The latest research has revealed that MazF functions as an mRNA interferase in both lower (yeast) and higher (mouse and human) eukaryotes (8). This suggests the exciting potential for mazF to be exploited as the counter-selectable marker in these eukaryotes.
The two delivery vectors pDGIEF and pIEFBPR can be ordered from the Bacillus Genetic Stock Center (http://www.bgsc.org) and their complete sequences have been submitted to the GenBank database (accession nos: DQ358863 and DQ486035).
Acknowledgments
The authors thank Prof. John A. Buswell and You-Tao Song for the helpful comments and linguistic revision of the manuscript. The authors are also grateful to the Bacillus Genetic Stock Center and Prof. Patrick Stragier for providing bacterial strains and vectors. Many thanks to anonymous referees for their useful comments. This work was supported by grants from Chinese National Natural Science Foundation (40471073, 30300005 and 30400013). Funding to pay the Open Access publication charges for this article was provided by NSFC.
Conflict of interest statement. None declared.
REFERENCES
- 1.Schallmey M., Singh A., Ward O.P. Developments in the use of Bacillus species for industrial production. Can. J. Microbiol. 2004;50:1–17. doi: 10.1139/w03-076. [DOI] [PubMed] [Google Scholar]
- 2.Kunst F., Ogasawara N., Moszer I., Albertini A.M., Alloni G., Azevedo V., Bertero M.G., Bessieres P., Bolotin A., Borchert S., et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 3.Reyrat J.-M., Pelicic V., Gicquel B., Rappuoli R. Counter-selectable markers: untapped tools for bacterial genetics and pathogenesis. Infect. Immun. 1998;66:4011–4017. doi: 10.1128/iai.66.9.4011-4017.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fabret C., Ehrlich S.D., Noirot P. A new mutation delivery system for genome-scale approaches in Bacillus subtilis. Mol. Microbiol. 2002;46:25–36. doi: 10.1046/j.1365-2958.2002.03140.x. [DOI] [PubMed] [Google Scholar]
- 5.Brans A., Filée P., Chevigné A., Claessens A., Joris B. New integrative method to generate Bacillus subtilis recombinant strains free of selection markers. Appl. Environ. Microbiol. 2004;70:7241–7250. doi: 10.1128/AEM.70.12.7241-7250.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayes F. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science. 2003;301:1496–1499. doi: 10.1126/science.1088157. [DOI] [PubMed] [Google Scholar]
- 7.Gerdes K., Christensen S.K., Lobner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nature Rev. Microbiol. 2005;3:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y., Zhang J., Hara H., Kato I., Inouye M. Insights into the mRNA cleavage mechanism by MazF, an mRNA interferase. J. Biol. Chem. 2005;280:3143–3150. doi: 10.1074/jbc.M411811200. [DOI] [PubMed] [Google Scholar]
- 9.Sambrook J., Fritsch E.F., Maniatis T. Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 10.Wolf M., Geczi A., Simon O., Borriss R. Genes encoding xylan and beta-glucan hydrolysing enzymes in Bacillus subtilis: characterization, mapping and construction of strains deficient in lichenase, cellulase, and xylanase. Microbiology. 1995;141:281–290. doi: 10.1099/13500872-141-2-281. [DOI] [PubMed] [Google Scholar]
- 11.Stragier P., Bonamy C., Karmazyn-Campelli C. Processing of a sporulation sigma factor in Bacillus subtilis: how morphological structure could control gene expression. Cell. 1988;52:697–704. doi: 10.1016/0092-8674(88)90407-2. [DOI] [PubMed] [Google Scholar]
- 12.Guerout-Fleury A.M., Shazand K., Frandsen N., Stragier P. Antibiotic-resistance cassettes for Bacillus subtilis. Gene. 1995;167:335–336. doi: 10.1016/0378-1119(95)00652-4. [DOI] [PubMed] [Google Scholar]
- 13.Zhang X.-Z., Cui Z.-L., Hong Q., Li S.-P. High-level expression and secretion of methyl parathion hydrolase in Bacillus subtilis WB800. Appl. Environ. Microbiol. 2005;71:4101–4103. doi: 10.1128/AEM.71.7.4101-4103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anagnostopoulus C., Spizizen J. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 1961;81:741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cui Z.-L., Zhang X.-Z., Zhang Z.-H., Li S.-P. Construction and application of a promoter-trapping vector with methyl parathion hydrolase gene mpd as the reporter. Biotechnol. Lett. 2004;26:1115–1118. doi: 10.1023/B:BILE.0000035481.03854.41. [DOI] [PubMed] [Google Scholar]
- 16.Inouye S., Inouye M. Up-promoter mutations in the lpp gene of Escherichia coli. Nucleic Acids Res. 1985;13:3101–3110. doi: 10.1093/nar/13.9.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Studier F.W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 18.Grossman T.H., Kawasaki E.S., Punreddy S.R., Osburne M.S. Spontaneous cAMP-dependent derepression of gene expression in stationary phase plays a role in recombinant expression instability. Gene. 1998;209:95–103. doi: 10.1016/s0378-1119(98)00020-1. [DOI] [PubMed] [Google Scholar]
- 19.Cui Z., Li S., Fu G. Isolation of methyl parathion-degrading strain M6 and cloning of the methyl parathion hydrolase gene. Appl. Environ. Microbiol. 2001;67:4922–4925. doi: 10.1128/AEM.67.10.4922-4925.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sissler M., Delorme C., Bond J., Ehrlich S.D., Renault P., Francklyn C. An aminoacyl-tRNA synthetase paralog with a catalytic role in histidine biosynthesis. Proc. Natl. Acad. Sci. USA. 1999;96:8985–8990. doi: 10.1073/pnas.96.16.8985. [DOI] [PMC free article] [PubMed] [Google Scholar]