Abstract
The acid-sensitive ion channel 1 (ASIC1) is a neuronal Na+ channel insensitive to changes in membrane potential but is gated by external protons. Proton sensitivity is believed to be essential for the role of ASIC1 in modulating synaptic transmission and nociception in the mammalian nervous system. To examine the structural determinants that confer proton sensitivity, we cloned and functionally characterized ASIC1 from different species of the chordate lineage: lamprey, shark, toadfish and chicken. We observed that ASIC1s from early vertebrates (lamprey and shark) were proton insensitive in spite of a high degree of amino acid conservation (66–67%) with their mammalian counterparts. Sequence analysis showed that proton-sensitive ASIC1s could not be distinguished from proton-insensitive channels by any signature in the protein sequence. Chimeras made with rat ASIC1 (rASIC1) and lamprey or shark indicated that most of the ectodomain of rASIC1 was required to confer proton sensitivity and the distinct kinetics of activation and desensitization of the rat channel. Proton-sensitive chimeras contained the segment D78–E136, together with residues D351, Q358 and E359 of the rat sequence. However, none of the functional chimeras containing only part of the rat extracellular domain retained the kinetics of activation and desensitization of rASIC1, suggesting that residues distributed in several regions of the ectodomain contribute to allosteric changes underlying activation and desensitization. The results also demonstrate that gating by protons is not a feature common to all ASIC1 channels. Proton sensitivity arose recently in evolution, implying that agonists different from protons activate ASIC1 in lower vertebrates.
The acid-sensitive ion channels (ASICs) belong to the large class of ENaC/Degenerin channels, which is distinguished by the presence of two transmembrane domains and a single large extracellular loop. Human and rat ASIC2a were the first cloned members of this family (Waldmann et al. 1996; Price et al. 1996); since then, four genes and six spliced forms have been identified in mammals. Later, Waldmann et al. observed that external protons activated these channels (Waldmann et al. 1997), providing the molecular identity of the proteins that carry acid-activated currents in sensory neurones first reported by Krishtal more than two decades ago (Krishtal & Pidoplichko, 1981) and later found in most neurones of the mammalian brain.
Not all of the functional roles of ASICs have been definitively established, but several putative functions have been proposed. Expression in nociceptors and activation by protons suggest that the ASICs may act as pain receptors (Chen et al. 1998; Immke & McCleskey, 2001), mainly because ASIC1 and ASIC3 exhibit the highest proton affinity among the ASICs. In the central nervous system, ASIC1 is the most abundant and most ubiquitously expressed of all the ASICs. ASIC1 has been implicated in modulating synaptic transmission, memory and fear conditioning (Wemmie et al. 2002, 2003). Most recently, it has been shown that ASIC1 mediates cell injury induced by ischaemia, inflammation and other conditions associated with acidosis in the mammalian nervous system (Xiong et al. 2004). Although the precise mechanisms underlying these effects have not been elucidated, the prevailing notion is that protons released by synaptic vesicles activate ASIC1, enhancing depolarization of postsynaptic membranes.
The goals of this work were, first, to define structural determinants in ASIC1 that render the channel sensitive to protons and, second, to investigate whether proton sensitivity is conserved in the chordate lineage. For that purpose we cloned and characterized ASIC1 from lamprey (Lampetra fluviatilis) as representative of cyclostomes, the dogfish (Squalus acanthias) from chondrichthyes, and additional channels from the teleost Opsanus tau, and from chicken (Gallus gallus). We report the sequences and results of functional studies of the recombinant proteins expressed in Xenopus oocytes, and of chimeras made with shark or lamprey and rat ASIC1. The data indicate that proton sensitivity is not a universal feature of ASIC1. In the evolution of the chordate lineage, proton sensitivity was acquired with the rise of fishes. The determinants of proton sensitivity are located in noncontiguous regions of the ectodomain, and are tightly linked to the kinetics of activation and desensitization, indicating that an allosteric mechanism underlies gating by protons.
Methods
Cloning of shark, fish, lamprey and chicken ASIC cDNAs
Prior to removal of brain or spinal cord from shark, toadfish and lamprey, the animals were anaesthetized with 0.5% tricaine added to the water. Chicken was first anaesthetized by ether inhalation, followed by decapitation. Total RNA was extracted using the Trizol reagent (Invitrogen). First strand cDNA synthesis was conducted with 5 μg of total RNA using oligodT primers and SuperScript RT II reverse transcriptase (Invitrogen). Screening for ASIC message was performed by PCR using degenerate primers from highly conserved sequences corresponding to NCNCRMVHMPG (sense: AA(C/T)TG(C/T)AG(A/G)ATGGTICA(C/T)ATGCCIGG) and GDIGGQMG (reverse: CCCAT(C/T)TGICC-ICCIAT(A/G)TCICC), (where I represents inosine). PCR was performed with Taq polymerase (Invitrogen) with the following parameters: 20 s denaturing at 94°C, 30 s annealing at 55°C, and 30 s extension at 72°C, repeated for 30 cycles. PCR products were sequenced and used to design specific primers for 5′ and 3′ RACE (Rapid Amplification of cDNA Ends). Whole-length cDNAs were obtained using the GeneRacer System for full-length, RNA ligase-mediated rapid amplification of 5′ and 3′ cDNA ends (Invitrogen). Lamprey ASIC1 cDNA was tagged with FLAG epitope at the carboxy terminus, and subcloned in pCRIITOPO vector (Invitrogen). Shark ASIC1α and ASIC1β cDNAs were subcloned in pcDNA3.1V5/HisTOPO vector (Invitrogen), which provides the V5 epitope at the carboxy terminus of the protein.
To search for other ASIC genes, different from toadfish ASIC1 (fASIC1), we treated the 360 bp PCR fragment obtained with the above pair of degenerated primers with DNA restriction enzymes StuI, StyI or MluI (sites present in fASIC1). After digestion, the DNA was amplified again by PCR using the same degenerated primers. Template digested with StuI or StyI did not yield reamplification products in contrast to template digested with MluI. DNA sequencing of products digested with MluI identified two additional fASICs. Based on the new sequences, we designed specific primers for 5′ RACE and 3′ RACE to amplify the complete coding region of the cDNAs. We introduced a FLAG epitope in the reverse primer. The PCR products were subcloned in pCRII TOPO Dual Promoter Vector (Invitrogen).
Chicken ASIC1 was cloned by RT-PCR using the following sense and reverse primers: CGGACCATGAT-GGACCTGAAGGTGGACGAGG and GCAGGTGAAGT-CCTCAAAGGTGCCCCGG located in the 5′ and 3′ ends of the predicted coding sequence of ASIC1 (NCBI accession no. XM-424489). DNA samples of at least three independent clones of each of the new ASIC reported here were sequenced with an automatic sequencer at the Keck Facility at Yale University.
Construction of chimeras and site directed mutagenesis
Chimeras were made with rat and lamprey ASIC1 cDNAs, and with rat and shark ASIC1α. To make lamprey–rat CH1, we introduced a HindIII site at amino acid position 78 and a StuI site at position 424 of lamprey ASIC1 protein. Lamprey cDNA was digested with HindIII and StuI, and ligated with the corresponding HindIII and StuI fragment from rat ASIC1 cDNA. For chimeras CH3 and CH4, XbaI and NheI sites were introduced in positions 187 and 346 of lamprey and the corresponding site in the rat protein. The sites were returned to the original sequences by a second round of site-directed mutagenesis using Quickchange (Stratagene). For CH5, CH6 and CH7, SacII sites were introduced at positions 150, 136 and 128 in lamprey and rat. The restriction sites SacII did not change the sequence of the proteins. To make shark–rat CH1, we introduced HindIII and StuI sites in the shark ASIC1α cDNA at positions corresponding to 78 and 424, and for CH2, SacII and NheI sites at positions corresponding to 150 and 346.
Western blotting and surface biotinylation of oocytes
After cRNA injection, oocytes were incubated at 18°C for 3 days prior to experiments. Twenty oocytes from each group were washed with ice-cold buffer A (mm): 90 NaCl, 3 KCl, 1 CaCl2, 5 triethanolamine, pH 8.0, and then transferred to 200 μl of biotinylation solution, buffer A containing 1.5 mg ml−1 of freshly prepared sulfo-NHS-SS-Biotin (Pierce, Rockford, IL, USA). Cells were incubated on a rocker at 4°C for 20 min. Fresh biotinylation solution was added a second time after which cells were transferred to a 5 ml container with ice-cold quenching solution (buffer A with 50 mm glycine) and incubated for 10 min. Oocytes were transferred to a microfuge tube containing 400 μl of buffer B (mm): 90 NaCl, 20 Tris, pH 7.4, 1% Triton X-100. Homogenates were cleared by centrifugation (8000 g) for 10 min. The supernatant was mixed with 50 μl of streptavidin bead slurry (Pierce) and incubated on a rocker overnight at 4°C. Beads were separated by centrifugation and washed three times with buffer B, and three times with buffer B supplemented with 300 mm NaCl. A 50 μl volume of loading buffer containing DTT was added to the beads and heated at 90°C for 5 min. The eluted proteins were loaded on 10% SDS-PAGE, together with 10 μl of supernatant recovered after incubation of lysate with streptavidin beads. Proteins were transferred to PVDF membranes (Immobilon P, Millipore). Membranes were blocked with 5% nonfat dry milk in TBST (mm: 20 Tris, pH 7.6, 120 NaCl, 0.1% Tween) for 30 min, then incubated for 2 h at room temperature with anti-FLAG or V5 monoclonal antibodies at a dilution 1:5000. After three 15 min washes with TBST, they were incubated for 1 h at RT in a 1:10000 dilution of horseradish-peroxidase-labelled antimouse secondary antibody (Sigma). Membranes were washed three times with TBST, signals were developed with ECL+ (Amersham Pharmacia Biotech) according to the manufacturer's protocol and exposed to BioMax MR film (Eastman Kodak, New Haven, CT, USA).
Co-immunoprecipitation
After electrophysiological measurements, oocytes were homogenized in nondenaturing lysis buffer (mm): 50 Tris pH 7.4, 150 NaCl, 1 EDTA, 1% Triton X-100, and COMPLETE protease inhibitors (Roche), cleared by centrifugation at 11 000 g for 10 min in a microcentrifuge followed by 10-fold dilution with homogenization buffer and incubation with 5 μl of anti-HA polyclonal antibody (Covance, Inc.) for 3 h at 4°C. Immune complexes were recovered with protein A linked to sepharose beads (Sigma). After several washes with homogenization buffer, complexes were eluted from the beads with Laemmli loading buffer containing β-mercaptoethanol and resolved in 7.5% SDS-PAGE. Proteins were transferred to Immobilon membranes (Millipore) and were blotted with monoclonal anti-FLAG (Sigma).
Expression in Xenopus oocytes and two-electrode voltage clamp (TEVC)
Stage V and VI oocytes were harvested from female Xenopus laevis, which were anaesthetized using 0.17% tricaine prior to the operation. Oocytes were injected with 4 ng of single cRNA or combinations of cRNAs, as indicated in the experiments. Plasmids containing the coding sequence of the ASIC genes were linearized at the 3′ end to serve as a template for synthesis of capped cRNAs with T7 polymerase, mMessagemMachine kit (Ambion, Austin, TX, USA). Injected oocytes were examined with the TEVC after 2–4 days. Oocytes were placed in a recording chamber (400 μl) perfused by gravity at a rate of 2–3 ml min−1. Oocytes were impaled with two glass microelectrodes filled with 3 m KCl having resistance lower than 1 MΩ. Whole-cell currents were recorded with a Clamp OC-725B (Warner Instrument Corp., Hamden, CT, USA), digitized at a sampling rate of 2 kHz (PowerLab/200, ADInstruments) and the data stored in a computer. Composition of the standard bath solution was (mm): 150 NaCl, 2 KCl, 5 CaCl2, 10 Hepes-Mes adjusted to pH 7.4. Activation solutions were administered by a perfusion system positioned directly in front of the oocyte. Composition of activating solutions was (mm): 150 NaCl, 2 KCl, 1 CaCl2, 10 Hepes-Mes or 10 Mes adjusted to pH 7.0–4.0.
Sequence analysis of ASIC1
clustal_x program (Thompson et al. 1997) was used to align and calculate conservation score (c-score) of the protein sequences of human, rat, chicken, zebrafish, toadfish, shark and lamprey ASIC1. c-scores were computed for each position in the multiple sequence alignment, separately for proton-sensitive and -insensitive groups. These scores were then averaged over a sliding window of seven amino acids with a step of four amino acids and plotted on the y-axis, and residue position was plotted on the x-axis. Differences in evolutionary rate between the two ASIC1 groups were inferred by computing the ratio between the nonsynonymous versus synonymous rates of substitution (Ka/Ks) using the YN00 program within the paml evolutionary package (Nei & Gojobori, 1986; Yang, 1997). Analysis was performed on cDNA sequences using a window of 90 nt (30 codons) with a step of 12 nt.
Results
Cloning and functional expression of chicken ASIC1
Chicken ASIC1 was cloned and characterized to determine the degree of protein conservation, and to compare functional properties with another tetrapod different from the mammalian one. The sequence obtained from chicken ASIC1 (see supplemental data) differed from the predicted sequence by automated computational analysis (NCBI accession no. XM-424489). The protein shares 89% amino acid identity with rat, mouse and human ASIC1. Functional expression in oocytes indicated that chicken ASIC1 is proton sensitive (calculated apparent EC50 for activation by external protons, pH 5.8), but it generates channels with rapid kinetics of activation and desensitization very different from the mammalian channels, but indistinguishable from fish ASIC1 (data not shown). This is an unexpected finding given that the human and chicken proteins are more similar than chicken is to fish (89 versus 77% identity). The rapid kinetics could be attributed to presence of residues T84R85 in chicken versus S83Q84 in rat, which we had previously identified as important in determining the kinetics of desensitization (Coric et al. 2003).
Cloning of ASIC1.2 and ASIC2 channels from toadfish
We previously cloned and characterized ASIC1.1 from toadfish (Coric et al. 2003). Here we searched for additional ASIC1 genes using the approach described in methods. We cloned two additional closely related cDNAs: one clone showed high sequence homology to mammalian ASIC1 and was labelled fASIC1.2 (it is a paralogue of fASIC1.1); whereas the other sequence was most closely related to mammalian ASIC2, and this clone was labelled fASIC2. The three proteins exhibit 73–78% conservation and 51–54% overall amino acid identity. The regions of highest homology are M2 and the ectodomain, whereas the cytoplasmic amino and carboxy termini, as well as M1, are less conserved.
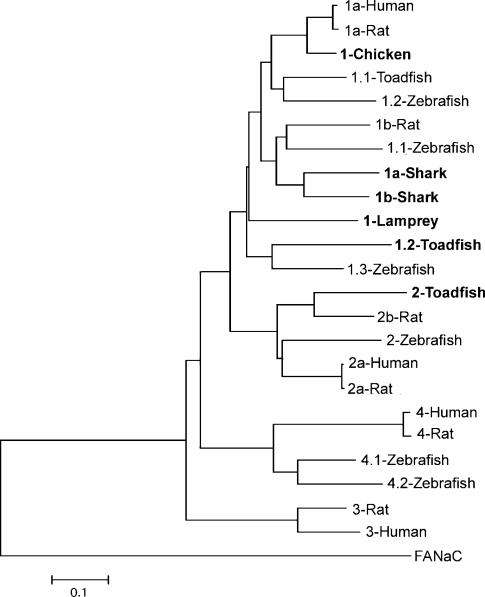
The relation of the three toadfish proteins with all the other ASICs cloned to date is shown in the dendrogram in Fig. 1. The degree of amino acid identity of fish and rat ASICs is 75% for fASIC1.1, 61% for fASIC1.2, and 56% for fASIC2.
Figure 1. Phylogenetic analysis of ASIC gene family generated using mega version 2.1 (Kumar et al. 2001).
The tree for the phylogram was established by Neighbour-Joining, with the alignment form clustal_x. The tree was rooted with FaNaC as an outgroup. The bar represents genetic distance in substitutions per amino acid. New sequences reported in this work are in bold. The corresponding nucleotide sequences have been submitted to GenBank: the accession numbers are AY278028, AY275841, AY275840, AY956390, AY956391, AY956392 and AY956393 for ASIC 1-toadfish, 2-toadfish, 1.2-toadfish, 1-lamprey, 1a-shark, 1b-shark and 1-chicken, respectively.
Functional characterization of fish ASIC1.2 and ASIC2 in Xenopus oocytes
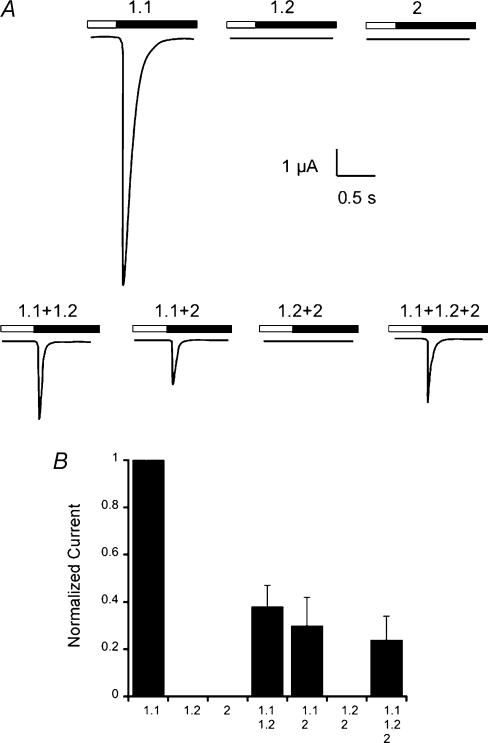
To examine the functional properties of fASIC1.2 and fASIC2, we injected oocytes with cRNAs of these clones alone or in combination with toadfish ASIC1.1. Upper traces in Fig. 2A show that fASIC1.1 induced currents that activate and inactivate rapidly, as we previously described (Coric et al. 2003). In contrast, oocytes injected with fASIC1.2 or fASIC2 did not exhibit proton-sensitive channels. Co-injection of fASIC1.1 with the other fish clones produced currents with properties similar to those of fASIC1.1 but of smaller magnitude, whereas coinjection of fASIC1.2 with fASIC2 did not generate proton-activated currents (lower traces in Fig. 2A). A summary of the data from two independent experiments normalized to the values obtained with fASIC1.1 is shown in Fig. 2B. All the conditions exhibited statistically significant smaller currents than fASIC1.1 alone (P = 0.001).
Figure 2. Proton-gated currents of fish ASICs.
Whole-cell currents elicited by a change in pHo from 7.4 to 5.0 (bars above the current traces) were recorded with two-electrode voltage clamp (TEVC) from injected oocytes. The bath solution contained 150 mm NaCl and the membrane potential was held at –60 mV. A, representative examples of current trances from oocytes injected with single cRNAs corresponding to each of the toadfish ASICs, and oocytes injected with the indicated combinations of cRNAs. B, relative currents induced by pHo 5.0. The data represent the summary of five independent experiments where the peak currents were normalized to the values of fASIC1. Error bars are the standard deviation, n = 8 oocytes for each column. All conditions were statistically different from fASIC1 (P < 0.001).
Mutations of residue G430 located in the outer pore of the mammalian ASIC2a render the channel constitutively active and more sensitive to external protons (Huang & Chalfie, 1994; Champigny et al. 1998). We thus substituted the corresponding amino acids in fASIC1.2 (G458F) and fASIC2 (G431F); however, these mutations did not induce gain of function whether the mutant channels were expressed alone or in combination with fASIC1.1.
Fish ASICs form heteromeric complexes
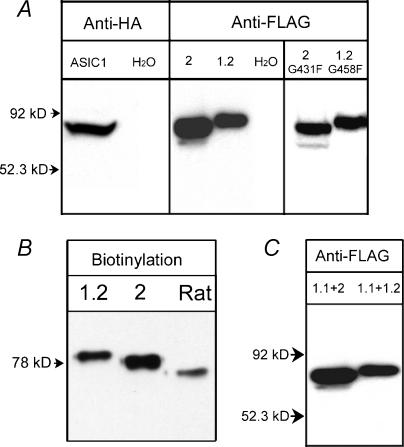
The inability to induce proton-gated currents was not due to the absence of protein expression because Western blots revealed abundant expression of all three subunits. Figure 3A shows fASIC1.1 detected with anti-HA, and fASIC1.2 and fASIC2 with anti-FLAG antibodies. fASIC1.2G458F and fASIC2G431F mutants were also expressed in oocytes that did not exhibit proton-gated currents (Fig. 3A, right panel). Moreover, using surface biotinylation, we demonstrated that fASIC1.2 and fASIC2 reach the plasma membrane (Fig. 3B).
Figure 3. Expression of fish ASICs Xenopus oocytes.
A, identification of fASIC1-HA, fASIC2-FLAG and fASIC1.2-FLAG from injected oocytes by Western blotting using HA and FLAG monoclonal antibodies. fASIC2G431F and fASIC1.2G458F mutants are also shown. B, Western blotting of surface biotinylated proteins of cells expressing toadfish ASIC1.2 or ASIC2, or rat ASIC1, revealed with anti-FLAG monoclonal. C, co-immunoprecipitation of fish ASICs from injected Xenopus oocytes. Oocytes were injected with equal amounts of cRNAs from fASIC1 + fASIC2 or fASIC1 + fASIC1.2. Two days after injection, oocytes were examined for proton-gated currents and saved for co-immunoprecipitation with anti-HA rabbit polyclonal antibody under non-denaturing conditions. Immunocomplexes were resolved on SDS-PAGE and then transferred to membranes for Western blotting. Signals corresponding to fASIC2 and fASIC1.2 were detected with anti-FLAG monoclonal antibody. Arrows on the left of the gels indicate molecular mass markers.
The decrease in fASIC1.1 current induced by coexpression with the other fish ASICs raised the possibility that fASIC1.1 associates with ASIC1.2 and/or ASIC2 to form heteromeric channels that are insensitive to external protons. We therefore conducted co-immunoprecipitations from oocytes expressing fASIC1.1 +1.2 or fASIC1.1 +2. Non-denaturing immunoprecipitations were performed with rabbit anti-HA antibody that recognizes fASIC1.1, followed by detection of fASIC1.2 and fASIC2 with anti-FLAG monoclonal antibody by Western blotting. The Western blot in Fig. 3C shows the bands corresponding to ASIC1.2 and ASIC2, indicating assembly with fASIC1.1. Therefore, reduction of the magnitude of fASIC1.1 proton-induced currents by coexpression with ASIC1.2 and ASIC2 is most likely to reflect the formation of non proton-gated heteromeric channels.
Expression of lamprey and shark ASICs in Xenopus oocytes
We cloned two cDNAs from shark brain and one from lamprey spinal cord. The shark proteins differed only in the first amino-terminal third (intracellular amino terminus, TM1 and first segment of the extracellular domain), but were identical in the distal two-thirds. The site of identity starts in the same position as the rat ASIC1 and ASIC2 spliced forms. Comparison with other sequences in GenBank indicated that the two shark clones share closest identity with ASIC1, 66% amino acid identity with rat ASIC1; therefore, these cDNAs correspond to two splice forms of the shark ASIC1 gene: sASIC1α and sASIC1β.
The cDNA from lamprey also corresponded to ASIC1; it shared highest identity (66.7%) with rat ASIC1. Alignment of sASIC1α, sASIC1β and lASIC1 is shown in supplemental data, and their relation with other ASICs is shown in the dendrogram of Fig. 1.
Functional properties of lamprey and shark ASIC1 channels were examined in Xenopus oocytes by the TEVC technique. Oocytes expressing single cRNAs of lASIC1, sASIC1α or sASIC1β, or coexpressing sASIC1α and sASIC1β, did not respond to a rapid change of pH in the bathing solution (from 7.4 to 5.0). Additional manoeuvres reported in the literature to increase or modify the response to protons were also tested: increase in Ca2+ concentration in the preconditioning solution to 6 mm (Babini et al. 2002; Coric et al. 2003), removal of Ca2+ from the low pH solution (0 mm) (Immke & McCleskey, 2004), pretreatment of oocytes with 200 μg ml−1 of trypsin (Poirot et al. 2004), and introduction of the degenerin's gain-of-function mutation, G410F (Champigny et al. 1998). However, none of these interventions rendered the channels sensitive to protons.
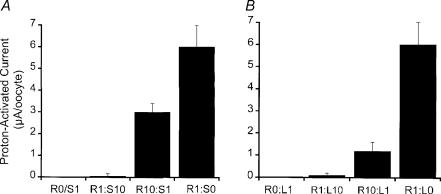
Only when lamprey or shark ASIC1 was coexpressed with rat ASIC1 did we detect proton-induced currents, but the magnitude of the currents was smaller than in oocytes expressing rASIC1 alone. Figure 4A shows the results of injections with rat and shark cRNAs. Most of the oocytes injected with a rat:shark cRNA ratio of 1:10 expressed very small currents upon stimulation by protons (<0.3 μA oocyte−1). When the ratio rat:shark was changed to 10:1, the average proton-induced current was 3.1 ± 0.5 μA oocyte−1, which represents 50% of the current expressed by oocytes injected with the same amount of only rat ASIC1 cRNA. Injection of rat:lamprey cRNAs in ratios 1:10 and 10:1 produced similar results; although, the combination rat:lamprey in the ratio 10:1 expressed an average current of 1.3 ± 0.6 μA oocyte−1, which represents 25% of the current expressed by oocytes injected with rat ASIC1 alone (Fig. 4B). Thus, shark and lamprey ASIC1 channels seem to assemble with rASIC1, but the heteromeric channels are not proton sensitive.
Figure 4. Functional expression of homomeric shark or lamprey channels and heteromeric rat–shark and rat–lamprey ASIC1 channels in Xenopus oocytes.
A, 3 days after injection of oocytes with cRNA of rat and/or sASIC1α in ratios indicated below the columns, the oocytes were examined for expression of proton-induced currents with the TEVC. Oocytes were exposed to a preconditioning solution of pHo 7.4 and activated with pHo 4.0 for 5 s. The mean of the values ±s.d. of whole-cell currents (μA oocyte−1) are shown in the ordinate (n = 8). B, similar experiments conducted with oocytes injected with rat and/or lamprey shark cRNAs.
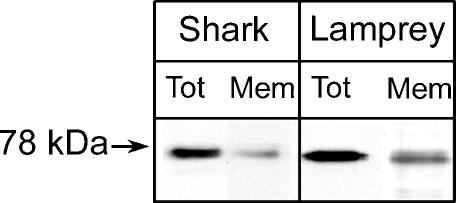
Because the absence of proton-induced currents with shark or lamprey ASIC1 may be due to the inability of assembly or delivery to the plasma membrane, these possibilities were examined by surface biotinylation of oocytes expressing shark or lamprey ASIC1s tagged in the carboxy terminus with the FLAG or V5 epitopes, respectively. Western blots in Fig. 5 show the presence of shark and lamprey ASIC1 in whole-cell lysates and in the plasma membrane. Therefore, homomeric lamprey and shark ASIC1 reach the plasma membrane, but do not conduct upon stimulation by protons.
Figure 5. Expression of shark and lamprey ASIC1 in the plasma membrane of oocytes.
Western blot of oocytes injected with shark or lamprey ASIC1 cRNA. Oocytes were first treated with sulfo-NHS-SS-Biotin and the biotinylated proteins were isolated with streptavidin beads. Total cell lysate (Tot) and biotinylated proteins (Mem) were resolved in 10% SDS-PAGE, transferred to membranes and processed for Western blotting with V5 (shark) or FLAG (lamprey) monoclonal antibodies, respectively.
Proton sensitivity of rat–lamprey chimeric channels
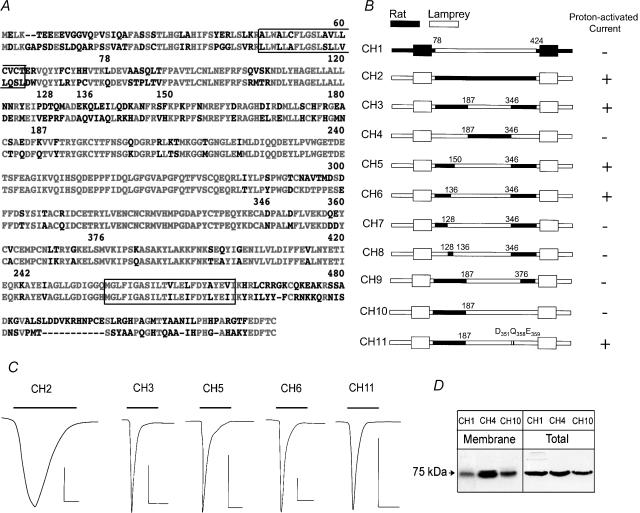
The difference in response to protons while maintaining a high degree of amino acid identity prompted us to generate chimeras between lamprey and rat ASIC1 in order to identify sequences important for conferring proton sensitivity to the channels. An alignment of the amino acid sequences of rat (top) and lamprey (bottom) ASIC1 is shown in Fig. 6A. Red and black indicate identical and different residues (numbers correspond to the positions indicated in Fig. 6B) and the boxes enclose TM1 and TM2. The sequence comparison makes evident a high degree of identity of TM2 and the distal two-thirds of the extracellular domain, whereas the intracellular amino and carboxy termini, and the first part of the extracellular domain, are less conserved. Figure 6B shows a schematic representation of the chimeric constructs that were tested in oocytes. Represented in black are sequences from rat, and in white sequences from lamprey; the numbers indicate the amino acid position in the rat sequence. Response with a transient inward current to a test pulse of pHo 4.0 is indicated on the right column.
Figure 6. Rat–lamprey chimeras.
A, alignment of rat and lamprey ASIC1. Identical residues are shown in red and different ones in black. Numbers above the sequence correspond to the amino acid positions in the lamprey sequence. Boxes enclose TM1 and TM2. B, schematic representation of rat–lamprey CHs. Sequences corresponding to rat and lamprey are indicated in black and white, respectively. The names of the chimeras are on the left. The numbers over each chimera are the same as in A. On the right is indicated the response to a pulse of pHo 4.0. C, representative current traces of oocytes expressing proton-sensitive CHs activated by pHo 4.0 indicated by the bars over the current traces. The time and current scale bars under each trace correspond to 1 s and 1 μA. D, Western blots of surface biotinylated CHs (Membrane) and a fraction of cell lysate (Total) revealed by anti-FLAG monoclonal antibody.
The first two chimeras, CH1 and CH2, have the whole extracellular domain exchanged between rat and lamprey, while maintaining TM1, TM2 and the amino and carboxy termini from the parent protein. CH1 was not functional, but CH2 exhibited properties indistinguishable from those of the rat ASIC1: EC50 for proton activation of pHo 6.6, activation rate of 2 s−1, and desensitization rate of 0.58 s−1 (Fig. 6C). To prove that CH1 was expressed at the plasma membrane by surface biotinylation, all the biotinylated proteins (Membrane) and 1/10 of the cytosolic fraction (Total) were revealed with anti-FLAG monoclonal (Fig. 6D). This result indicates that the extracellular domain of rat contains the elements that confer proton sensitivity and also determines the kinetics of activation and desensitization.
Other chimeras were made to replace smaller segments of the rat extracellular domain. CH3, but not CH4, responded to pHo 4.0; however, the currents of CH3 differed from rat ASIC1 and CH2 by having very rapid rise and decay rates, 10 s−1 and 2.9 s−1, and by a smaller magnitude of response, mean peak current of 2.2 ± 0.4 versus 5.3 ± 0.8 μA oocyte−1. CH5 and CH6 exhibited properties indistinguishable from CH3, whereas CH7 was not functional suggesting that the short segment of amino acids between positions 128 and 136 may be essential. To test this latter idea, we generated CH8, which contains the segment 128–136 from rat, while the other sequences were replaced by lamprey. CH8 did not respond to protons, indicating that the rat segment 128–136, although necessary, is not sufficient to confer proton sensitivity.
Thus far, the above chimeras demonstrate that rat sequences in the extracellular domain following TM1 are necessary for the function of the chimeras. Additional chimeras were designed to examine the contribution of the region between residues 346 and 424 in the most distal part of the extracellular domain. CH9 replaced rat residues 346–376 with lamprey, which resulted in loss of function. This is a highly conserved area with only a few different residues. Substitutions of these residues with the ones corresponding to rat restored function as indicated by CH11. Three residues were required for full recovery: D351, Q358 and E359.
Proton sensitivity of rat–shark chimeric channels
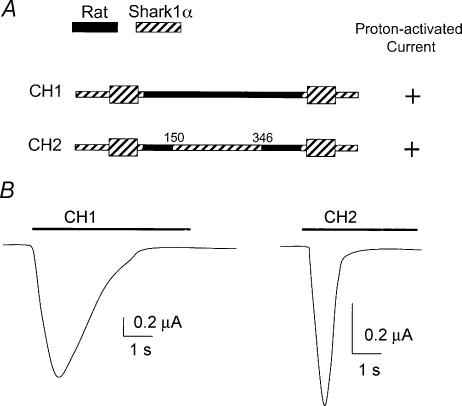
To test whether the segments of the extracellular domain in rASIC1 identified by rat–lamprey chimeras are important, we made two rat–shark chimeras using the sASIC1α cDNA comparable to rat–lamprey CH2 and CH5. Figure 7 shows in black and hatched the sequences corresponding to rat and shark in the chimeras. The two constructs were functional: CH1 produced currents similar to rASIC1, but CH2 exhibited kinetics of activation and desensitization much faster than rASIC1.
Figure 7. Rat–shark1α chimeras.
A, schematic of rat–shark1α chimeras. Black and hatched represent sequences from rat and shark, respectively. Numbers are amino acid positions in the shark sequence. B, representative examples of whole-cell currents elicited by changes in pHo from 7.4 to 4.0 (bar above traces) obtained with TEVC. Bars indicate current and time scales.
Sequence analysis of ASICs
Functional studies of ASIC1 from different species have made it apparent that these channels can be separated into proton-sensitive and proton-insensitive types. This raises the question of whether specific residues and/or motifs are responsible for distinguishing between the two groups. Such motifs may form the binding sites for protons or they may mediate conformational changes for channel gating in response to variations in local pH. To identify such residues, we constructed multiple sequence alignment of 12 known ASIC1 sequences (human-1; rat-1α and -1β, chicken-1; toadfish-1.1 and -1.2; zebrafish-1.1, -1.2 and -1.3; shark-1α and -1β; and lamprey-1) and examined the amino acid compositions at individual positions in the alignment. The aim was to find residues that are either conserved or selected for some amino acids in proton-sensitive, but not in proton-insensitive, channels, or vice versa. Even though several methods were used (see supplemental data), our analyses did not identify residues with amino acids selected exclusively for the proton-sensitive group. We only found positions significantly enriched with certain types of amino acids in one group, but not in the other (red and blue in the sequence alignments in supplemental data). These residues were not only distributed in the extracellular domain, but also in the amino and carboxy terminus.
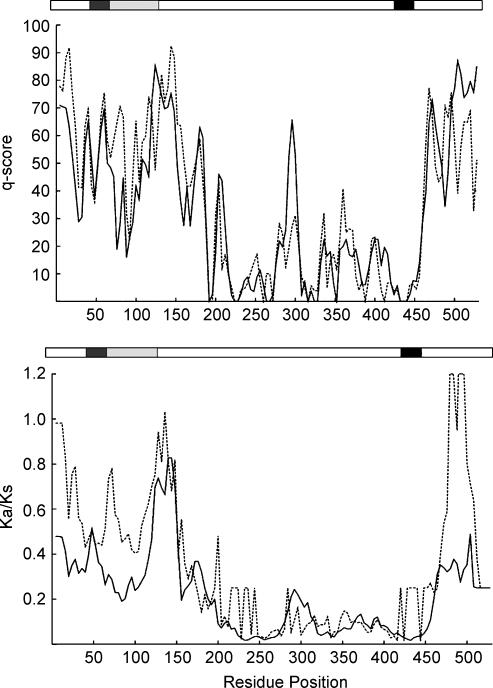
Subsequently, we examined the sequence conservation of ASIC1 by calculating c-scores, as explained in Methods. The continuous and dashed lines in Fig. 8A represent proton-sensitive and -insensitive channels; a low c-score indicates a high degree of sequence conservation, and a high score indicates high variability within the group. Overall the pattern of sequence conservation is similar between the two groups. TM2 and many segments of the ectodomain are identical, whereas the first third of the protein and the carboxy terminus are the most variable (c-score 50–90).
Figure 8. Sequence analysis of proton-sensitive and -insensitive ASIC1s.
A, conservation of amino acid composition within proton-sensitive (continuous line) and -insensitive (dashed line) ASIC1s assessed by conservation score (q-scores). A low score indicates that a region is conserved. B, sliding-window analysis of Ka/Ks ratios performed on proton-sensitive (continuous line) and proton-insensitive (dashed line) ASIC1s. Window of 90 nt with 12 nt step was used for the study. The x-axis indicates the residue numbered according to the rat ASIC1α sequence. Protein domains are drawn schematically above each graph, with transmembrane segments shown in black. Regions that were found important for proton sensitivity in functional studies are shaded in grey.
Two small regions exhibit different levels of variability between the groups. The region of residues 76–88 in the proton-sensitive type is highly conserved (c-score ∼27), whereas in the proton-insensitive type it is more variable (c-score ∼56). This former region coincides with the segment we identified as important for proton sensitivity. In contrast, the region 289–295 is more conserved in the proton-insensitive (c-score ∼20) than in the -sensitive group (c-score ∼48). However, functional studies did not implicate this segment as important for proton sensitivity.
To further investigate whether the differences in protein sequence observed in the two groups of ASIC1 reflect changes in natural selection that gave rise to a new functional specialization of the channel, we examined the pace of protein evolution as scaled to neutral divergence approximated by the ratio between nonsynonymous (Ka) and synonymous (Ks) substitution rates in the coding region of the cDNA (Li, 1997). Two processes have been identified that increase the Ka/Ks ratio (>1): one is positive selection in favour of a change in gene function; the other is relaxed selection.
The average of Ka/Ks ratios in pair-wise comparison is 0.13 (0.02–0.19) for the proton-sensitive type, and 0.1 (0.07–0.13) for the proton-insensitive type, indicating high conservation and functional constraint within each group. Pair-wise comparison between proton-sensitive and proton-insensitive ASIC1s gives the average Ka/Ks value 0.14 (0.02–0.55), which also indicates a high level of conservation between the two groups. However, if gain of proton sensitivity requires changes only in a small domain of the protein, the Ka/Ks analysis using the entire sequence would miss such differences. We thus applied a sliding-window analysis of the Ka/Ks ratio (Fig. 8B). Residues between 184 and 450 show smaller Ka/Ks ratios than residues in other regions, and thereby they are under strong functional constraint in both groups. A Ka/Ks value >1 in 480–500 could reflect that the carboxy terminus is under positive selection in proton-insensitive channels, whereas a value of ∼0.4 in proton-sensitive channels is indicative of moderate constraint. The Ka/Ks ratios of residues 64–112 following TM1 are also different between the two groups, with a value of ∼0.26 for proton-sensitive and 0.53 for proton-insensitive, which is indicative of stronger functional constrains in the proton-sensitive group than in the proton-insensitive group. Together, sequence analyses suggest that the highly variable segment after TM1 has become ‘fixed’ in proton-sensitive ASIC1, which in turn may reflect adaptation to a new function.
Discussion
Determinants of proton sensitivity and gating of ASIC1
Of particular interest is the question regarding what structural elements underlie the differences between proton-sensitive and proton-insensitive ASIC1 channels, and how differences in proton sensitivity of ASIC1 affect brain functions. Here, we addressed the first question by examining chimeras derived from proton-sensitive and proton-insensitive ASIC1 channels. The studies identified noncontiguous regions in the extracellular domain of rASIC1 that are required to confer proton sensitivity. The region encompassed by residues 78–136 together with a few more distal residues, D351, Q358 and E359, from rASIC1 rendered lamprey and shark ASIC1 responsive to protons. However, with the exception of chimeras that contained the whole extracellular domain of rat, all other functional constructs induced channels with kinetics of activation and desensitization different from rASIC1.
A proposed mechanism for ASIC gating is the release of a blocking Ca2+ ion from the outer pore (Immke & McCleskey, 2004). Protons compete for this high-affinity Ca2+-binding site promoting the release of Ca2+, and thereby opening the channel. To interpret our results in light of this hypothesis, the first question is whether the regions we identified constitute a Ca2+-binding site. Residues D351, Q358 and E359 could potentially make a high-affinity Ca2+-binding site. However, they are not conserved in all proton-sensitive ASIC1s and their substitution by alanine did not abolish proton sensitivity (data not shown) indicating that, although required, they are not sufficient to confer proton sensitivity to rat ASIC1. On the other hand, the segment 78–136 is essential, although it also does not contain a canonical Ca2+-binding site. Moreover, the variability of amino acid composition of segment 78–136 in both proton-sensitive and proton-insensitive groups makes it unlikely to encode for the Ca2+-binding site. On the other hand, proton-sensitive chimeras exhibited marked differences in kinetics of activation and desensitization, suggesting that the regions identified as functionally important do not form a Ca2+-binding site, but rather are involved in conformational changes elicited upon proton binding.
Evolution of proton sensitivity of ASIC1
Gating by external protons is considered to be the most distinct feature of the ASICs. Indeed, it was this property that gave to ASIC1 the name of acid-sensing ion channel. Although in most instances the physiological role of the ASICs has not been completely elucidated, the underlying notion is that protons are required for activation of the ASICs in nociceptors, synaptic terminals of central neurones and in pathological conditions such as brain injury induced by ischaemia. The results of this study challenge that notion by demonstrating that proton sensitivity is not a general property of ASIC1; it was acquired late in the evolution of vertebrates with the rise of bony fishes, some 400 million years ago. This finding has important functional implications for ASIC1, which is the prototype and the most abundant and ubiquitous channel of the ASIC family. Indeed, inactivation of the ASIC1 gene in mice abolishes proton-activated currents in many regions of the mammalian brain, indicating that it forms homomeric channels that are not substituted by channels with other subunit compositions (Wemmie et al. 2002).
It is remarkable that, in spite of the high degree of amino acid conservation of ASIC1 through the whole chordate lineage (evolutionary time spanning more than 550 million years), there is such a stark difference regarding proton sensitivity. The findings presented here imply that ASIC1 from lower vertebrates is gated by a stimulus different from protons. Whether ASIC1s of recent vertebrates still respond to the primordial agonist is not known but raises the possibility that mammalian ASIC1 may be gated in vivo by agonists other than protons.
One interpretation of these results is that proton sensitivity arose in evolution to provide a new function or to improve a previously existing one (such as nociception or long-term potentiation in higher vertebrates). This interpretation predicts that the residues conferring proton sensitivity would become fixed in the protein; however, the group of proton-sensitive ASIC1s could not be distinguished from the proton-insensitive group by any signature in the protein sequence. On the other hand, the relatively high Ka/Ks ratio of the 78–136 segment in the proton-insensitive group (Fig. 8B) is likely to provide conditions for emergence of amino acids combinations that support proton sensitivity. The subsequent decrease in Ka/Ks ratio of this segment is consistent with proton sensitivity conferring some evolutionary advantage. Alternatively, proton sensitivity may not be directly relevant to the functional specialization of the protein because variation in proton sensitivity might be driven by near neutral evolution, and will be tolerated because it does not hamper the survival of the species. Further characterization of the functional role of ASIC1 in early and more recent vertebrates will provide a definitive answer.
Supplemental material
The online version of this paper can be accessed at: 10.1113/jphysiol.2005.087734 http://jp.physoc.org/cgi/content/full/jphysiol.2005.087734/DC1 and contains supplemental material entitled: Sequence comparison of proton-sensitive and insensitive ASIC1s. This material can also be found as part of the full-text HTML version available from http://www.blackwell-synergy.com
References
- Babini E, Paukert M, Geisler HS, Gründer S. Alternative splicing and interaction with di- and polyvalent cations control the dynamic range of acid-sensing ion channel 1 (ASIC1) J Biol Chem. 2002;277:41597–41603. doi: 10.1074/jbc.M205877200. [DOI] [PubMed] [Google Scholar]
- Champigny G, Voilley N, Waldmann R, Lazdunski M. Mutations causing neurodegeneration in Caenorhabditis elegans drastically alter the pH sensitivity and inactivation of the mammalian H+-gated Na+ channel MDEG1. J Biol Chem. 1998;273:15418–15422. doi: 10.1074/jbc.273.25.15418. [DOI] [PubMed] [Google Scholar]
- Chen CC, England S, Akopian AN, Wood JN. A sensory neuron-specific, proton-gated ion channel. Proc Natl Acad Sci U S A. 1998;95:10240–10245. doi: 10.1073/pnas.95.17.10240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coric T, Zhang P, Todorovic N, Canessa CM. The extracellular domain determines the kinetics of desensitization in acid-sensitive ion channel 1. J Biol Chem. 2003;278:45240–45247. doi: 10.1074/jbc.M304441200. [DOI] [PubMed] [Google Scholar]
- Huang M, Chalfie M. Gene interactions affecting mechanosensory transduction in Caenorhabditis elegans. Nature. 1994;367:467–470. doi: 10.1038/367467a0. [DOI] [PubMed] [Google Scholar]
- Immke DC, McCleskey EW. Lactate enhances the acid-sensing Na+ channel on ischemia-sensing neurons. Nat Neurosci. 2001;4:869–870. doi: 10.1038/nn0901-869. [DOI] [PubMed] [Google Scholar]
- Immke DC, McCleskey EW. Protons open acid-sensing ion channels by catalyzing relief of Ca2+ blockade. Neuron. 2004;37:75–84. doi: 10.1016/s0896-6273(02)01130-3. [DOI] [PubMed] [Google Scholar]
- Krishtal OA, Pidoplichko VI. A receptor for protons in the membrane of sensory neurons may participate in nociception. Neuroscience. 1981;6:2599–2601. doi: 10.1016/0306-4522(81)90105-6. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Jakobsen IB, Nei M. MEGA2: molecular evolutionary genetcis analysis software. Bioinformatics. 2001;12:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- Li WH. Molecular Evolution. Sunderland, MA: Sinauer Associates; 1997. [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Poirot O, Vukicevic M, Boesch A, Kellenberger S. Selective regulation of acid-sensing ion channel 1 by serine proteases. J Biol Chem. 2004;279:38448–38457. doi: 10.1074/jbc.M407381200. [DOI] [PubMed] [Google Scholar]
- Price M, Snyder P, Welsh MJ. Cloning and expression of a novel human brain Na+ channel. J Biol Chem. 1996;271:7879–7882. doi: 10.1074/jbc.271.14.7879. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The clustal_x windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldmann R, Champigny G, Bassilana F, Heurteaux C, Lazdunski M. A proton-gated cation channel involved in acid-sensing. Nature. 1997;386:173–177. doi: 10.1038/386173a0. [DOI] [PubMed] [Google Scholar]
- Waldmann R, Champigny G, Voilley N, Lauritzen I, Lazdunski M. The mammalian degenerin MDEG, an amiloride-sensitive cation channel activated by mutations causing neurodegeneration in Caenorhabditis elegans. J Biol Chem. 1996;271:10433–10436. doi: 10.1074/jbc.271.18.10433. [DOI] [PubMed] [Google Scholar]
- Wemmie JA, Askwith CC, Lamani E, Cassell MD, Freeman JH, Jr, Welsh MJ. Acid-sensing ion channel 1 is localized in brain regions with high synaptic density and contributes to fear conditioning. J Neurosci. 2003;23:5496–5502. doi: 10.1523/JNEUROSCI.23-13-05496.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wemmie JA, Chen J, Askwith CC, Hruska-Hageman AM, Price MP, Nolan BC, Yoder PG, Lamani E, Hoshi T, Freeman JH, Jr, Welsh MJ. The acid-activated ion channel ASIC contributes to synaptic plasticity, learning, and memory. Neuron. 2002;34:463–477. doi: 10.1016/s0896-6273(02)00661-x. [DOI] [PubMed] [Google Scholar]
- Xiong ZG, Zhu XM, Chu XP, Minami M, Hey J, Wei WL, MacDonald JF, Wemmie JA, Price MP, Welsh MJ, Simon RP. Neuroprotection in ischemia: blocking calcium-permeable acid-sensing ion channels. Cell. 2004;18:687–698. doi: 10.1016/j.cell.2004.08.026. [DOI] [PubMed] [Google Scholar]
- Yang Z. paml: a program for package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;15:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.