Abstract
Integrons are genetic elements that acquire and exchange exogenous DNA, known as gene cassettes, by a site-specific recombination mechanism. Characterized gene cassettes consist of a target recombination sequence (attC site) usually associated with a single open reading frame coding for an antibiotic resistance determinant. The affiliation of multiresistant integrons (MRIs), which contain various combinations of antibiotic resistance gene cassettes, with transferable elements underlies the rapid evolution of multidrug resistance among diverse Gram-negative bacteria. Yet the origin of MRIs remains unknown. Recently, a chromosomal super-integron (SI) harboring hundreds of cassettes was identified in the Vibrio cholerae genome. Here, we demonstrate that the activity of its associated integrase is identical to that of the MRI integrase, IntI1. We have also identified equivalent integron superstructures in nine distinct genera throughout the γ-proteobacterial radiation. Phylogenetic analysis revealed that the evolutionary history of the system paralleled that of the radiation, indicating that integrons are ancient structures. The attC sites of the 63 antibiotic-resistance gene cassettes identified thus far in MRIs are highly variable. Strikingly, one-fifth of these were virtually identical to the highly related yet species-specific attC sites of the SIs described here. Furthermore, antimicrobial resistance homologues were identified among the thousands of genes entrapped by these SIs. Because the gene cassettes of SIs are substrates for MRIs, these data identify SIs as the source of contemporary MRIs and their cassettes. However, our demonstration of the metabolic functions, beyond antibiotic resistance and virulence, of three distinct SI gene cassettes indicates that integrons function as a general gene-capture system for bacterial innovation.
The impact of lateral gene transfer on bacterial evolution is underscored by the realization that foreign DNA can represent up to one-fifth of a given bacterial genome (1). Perhaps the most striking embodiment of its affect on microbial adaptation has been the rapid and widespread emergence of similar antibiotic-resistance profiles among phylogenetically diverse Gram-negative clinical and environmental isolates over the last half-century (2). The localization of antibiotic-resistance determinants to mobile entities such as plasmids and transposons readily explained this phenomenon (3–6). Closer examination revealed that in many cases a new type of genetic element, termed an integron, harbored the resistance determinants. Integrons are natural cloning and expression systems that incorporate open reading frames and convert them to functional genes (for review see refs. 7 and 8). The integron platform codes for an integrase (intI) that mediates recombination between a proximal primary recombination site (attI) and a secondary target called an attC site [or 59-base element (59be)]. The attC site is normally found associated with a single open reading frame (ORF), and the attC-ORF structure is termed a gene cassette (9–12). Insertion of the cassette at the attI site, which is located downstream of a resident promoter internal to the intI gene, drives expression of the encoded proteins (13). Most of the attC sites of the integron cassettes identified to date are unique. Their length and sequence vary considerably (from 57 to 141 bp) and their similarities are primarily restricted to their boundaries, which correspond to the inverse core site (RYYYAAC) and the core site [G↓TTRRRY; ↓, recombination point (11, 14)]. More than 60 different antibiotic-resistance genes, covering most antimicrobials presently in use, have been characterized in cassette structures (15). The stockpiling of exogenous genetic loci to create multiresistant integrons (MRIs) has contributed substantially to the current dilemma in the treatment of infectious disease (2, 8, 15, 16), as MRIs harboring up to five different resistance cassettes have been described (17). However, the origin of integrons and their cassettes has remained obscure.
Recently a distinct type of integron was discovered in the Vibrio cholerae genome (18). Located on the smaller of the two circular chromosomes (19), this element spans 126 kb and gathers at least 179 cassettes (20, 21) into a single structure termed a super-integron (SI), dwarfing previously identified MRIs. Of particular interest were the observations that: (i) the cassette-associated attC sites, termed VCRs [for Vibrio cholerae repeats (22); other repeated sequences are named similarly] displayed a strikingly high degree of sequence relatedness, unlike their counterparts of MRIs; (ii) the attC site of two MRI antibiotic-resistance gene cassettes, CARB4 and dfrVI, was similar to the VCRs; and (iii) the V. cholerae SI cassettes were substrates for the class 1 integrase of MRIs (18).
These relationships led us to hypothesize that MRIs evolved from SIs through the entrapment of intI genes and their cognate attI sites into highly mobile structures like transposons. Subsequent harvesting of cassettes from various SI sources then led to the establishment of contemporary MRIs. In this scenario, each distinct attC site of MRI cassettes would represent a specific SI cluster containing related attC sites, as in V. cholerae. If SIs are the progenitors of MRIs and their cassettes, then the SI integrases should be active, the cassettes should encode adaptive functions, and SIs should be numerous in the bacterial kingdom. Here, we have demonstrated that the V. cholerae SI integrase is active for site-specific cassette recombination, and we have determined the metabolic function of three SI cassettes: a sulfate-binding protein, a psychrophilic lipase, and a restriction enzyme. Furthermore, a systematic search for SI structures beginning with Vibrio and closely related species and extending to distantly related genera has revealed that this gene acquisition machinery is an ancient system that is widespread among the proteobacteria.
Materials and Methods
Bacterial Strains, PCR, Sequencing, and Phylogenetic Analysis.
Bacterial strains were provided by the Collection de l'Institut Pasteur (CIP). All genomic DNA was isolated by using the Qiagen DNEasy Kit. PCR-amplified genes were cloned by using the TA TOPO cloning kit (Invitrogen) and verified by sequencing of the corresponding genomic clones [performed by MWG-Biotech (Ebersberg, Germany), on a Pharmacia 4 × 4 sequencing system or an Applied Biosystems 377 sequencer]. Primers were obtained from Genset (Evry, France) or MWG-Biotech. Alignments were generated with the clustalX (23) software program (version 1.8) and dendrograms were compiled by using the neighbor-joining method (computed from 1,000 independent trials) of clustalW and phylip (24) or treeview (25). 16S rRNA sequences were retrieved from GenBank.
Integrase Activity and Cassette Function Assays.
The VchintIA gene (Vch indicating the V. cholerae source; other genes are identified similarly) was digested with EcoRI and BamHI after PCR amplification with the primers VINTup (CCGGAATTCTGTACAAATAACCAGTTAAAT) and VINTdw (CGCGGATCCGTGTTATTAAGAAAGGGGAG) and cloned into pUC18 digested with the same enzymes to create pVC3. The substrate plasmids pSU38∷CARB4 and pSU38∷ORF1-cat have been previously described (18). Escherichia coli DH5α (recA−) containing the indicated plasmids was incubated overnight at 37°C with the appropriate antibiotics. Plasmid DNA was prepared and digested with either XbaI (for pSU38∷ORF1-cat) or MfeI (for pSU38∷CARB4), which cut specifically within the respective cassettes. Samples of both the original and digested plasmid preparations were used to transform E. coli DH5α. The recombination frequency is the number of ampicillin-sensitive (ω32 and ω34) or chloramphenicol-sensitive (ω33 and ω35) transformants among the number of kanamycin-resistant transformants (average of two independent trials; Table 1). The precise location of the recombination event was established by PCR and sequencing of 25 random clones per test set to determine the VchIntIA-dependent recombination frequency.
Table 1.
VchIntIA-dependent recombination frequencies for the CARB4 and ORF1∷cat cassette substrates
| Strain | Plasmid | Recombination frequency |
|---|---|---|
| ω32 | pSU38∷CARB4, pVC3 | 5.0 × 10−3 |
| ω34 | pSU38∷CARB4 | <10−6 |
| ω33 | pSU38∷ORF1-cat, pVC3 | 1.6 × 10−2 |
| ω35 | pSU38∷ORF1-cat | <10−5 |
The sulfate-binding protein cassette of the V. cholerae SI was amplified by PCR with the primers VCR1/SBP (CGTGAATTCTTGAGGCGTTTGTTAGGTTT) and SBP3′ (ATAGGATCCTCATTTTTAAACTACCTTTTAC), digested with EcoRI and BamHI, and cloned into pUC18 digested with the same enzymes to create pVchSBP. Complementation of the sulfate auxotroph E. coli sbp/cysP double mutant, EC2402 (26), was carried out in liquid and solid minimal media containing 0.1 mM sodium sulfate as the sole source of sulfate. The lipase cassette of the Vibrio marinus SI was amplified by PCR with the primers Vmarlip1 (TCTAGACAAACGTGTTATGCAGGCGTT) and Vmarlip3 (GAATTCTTATGGTTTTAATGCGAAAGTTAAAAAG). The product was digested with NdeI and BamHI and cloned into pET3a digested with the same enzymes to give pET3aVml. T7 RNA polymerase-dependent lipase expression in recombinant E. coli BL21(DE3)pLysS was induced by the addition of isopropyl β-d-thiogalactoside (IPTG) to a final concentration of 0.5 mM on solid LB medium containing 100 μg/ml ampicillin, 25 μg/ml chloramphenicol, and 0.5% tributyrin as previously described (27). Positive clones produced a zone of clearing caused by hydrolysis of the lipid in the surrounding medium. Control strains carried the vector without insert.
Identification of the Integrase Genes.
A primer to rpmI (L35), R16–1 (CCAAACGCCATTCTGCCTAAG), and VCR1 (18) were used to amplify VmiintIA from Vibrio mimicus genomic DNA. Primers VCR1 and H7–1 (GGACTATGGCAAGTTCCTCT), which targeted infC (IF3), were used to amplify the entire VmeintIA gene from Vibrio metschnikovii chromosomal DNA. The VmeintIA gene was used to probe a Listonella pelagia HindIII library in pNot218R [modified from the phagemid pTZ18R vector (Pharmacia) to include in-frame NotI sites on both sides of the multiple cloning site]. Single-strand DNA was prepared after infection with M13K07 helper phage. This library was used in a PCR with the M13 forward primer and a set of degenerate intI primers, 17–2C [C(AC)A(AG)(CAT)AC(AG)TG(CTA)GTGTAGAT]. The PCR product, corresponding to LpeintIA, was cloned and sequenced. This gene was in turn used as a probe to clone a 6-kb Vibrio parahaemolyticus fragment containing VpaintIA from an XbaI library in pNot218R.
On the basis of estimates of the size of the Vibrio SIs, it was assumed that the Vibrio fischeri SI would be more or less equal in length (ca. 3% of the genome). Fifty random clones of a V. fischeri BglII genomic DNA library (in pNot218R) were selected for sequencing, and one was found to contain a short cassette array plus chromosomal DNA with homology to general housekeeping genes, indicative of a clone corresponding to the 3′ end of the SI. Primers VFR1 (GCAAATCGTGTTGATGCATTTGTTA) and VFR2 (CAAACGTGTTATGCAGGCGTTA), designed from this sequence, were used in a PCR screen to determine the consensus sequence of the VFRs from several independent clones containing nonoverlapping cassette arrays. A unique conserved restriction enzyme site, NsiI, was identified among the VFRs near the 5′ end. V. fischeri genomic DNA was digested with NsiI, with the expectation that the enzyme would cut regularly between most of the SI cassettes but less frequently in the rest of the genome. The majority of DNA fragments that hybridized with a VFR-specific probe were less than 2.5 kb, but three larger hybridizing fragments of 3, 4.5, and 6 kb, containing at least one VFR were observed. Rehybridization with the 3′ end of the SI as a probe gave an intense signal corresponding to the 3-kb fragment. We surmised that the remaining two fragments likely corresponded to either the 5′ end of the SI or cassette arrays in which the enzyme did not cut. NsiI partial libraries corresponding to the 4.5- and 6-kb regions were constructed in NsiI-digested pMTL22 (28). Because the NsiI site was located close to the 5′ end of the VFR, unique fusion primers, NsiFUS1 (GTACCATGGCATGCATTTGTT) and NsiFUS2 (GAGATCTCTCGATGCATTTGTT), corresponding to a region of the multiple cloning site of pMTL22 immediately adjacent to and including the NsiI site plus an additional 5 nt specific to the 5′ end of the VFR sequence were created. These were used in a PCR with VFR2 and the respective libraries as matrix to identify the proper clones. Subcloning and sequencing of the 6-kb fragment unveiled VfiintIA.
Analysis of the available sequencing data [The Institute for Genomic Research (TIGR)] revealed an integron-integrase gene in the genome of Shewanella oneidensis MR-1 (formerly Shewanella putrefaciens MR1). However, mobile elements [insertion sequences (ISs) or transposons (Tns)] had disrupted the SI structure. A native SI structure in strain 69.34 was identified as follows: Primers SpintI-1 (GACAACCAGCAAGGGATAGGT) and SpintI-3 (TTTATAATGTGTCTAACGGGC), based on the integrase gene of S. oneidensis MR1 (SonintIA), were used to amplify the 3′ end of SpuintIA of S. putrefaciens strain 80.40T. Again, the SI of this strain was also disrupted by IS or Tn elements. Using SpuintIA as a probe, we identified SpuintIB of the S. putrefaciens strain 69.34 SI by Southern analysis and sequenced it from a BamHI library in pNot218R.
A search of the GenBank database with the attC site of the aadA7 aminoglycoside-resistance gene cassette (29) revealed that the XbaI restriction–modification locus of Xanthomonas campestris pv. badrii was coded in a cassette-like structure. SIs were identified in the genomes of several X. campestris pathovars and Xanthomonas spp. as follows: The primers XCR1 (CGTCGGCTTGAATGAATTGTTA) and XCR2 (GCGCGGCTTAACTCAGGTGTT), designed from the attC site of aadA7 were used to PCR amplify several cassettes in X. campestris pv. campestris 100069T. These cassettes were used as probes to clone a 26-kb EcoRI genomic fragment, containing a large portion of the SI cassette array less the integrase gene, in pUC18. The XcaintIA gene was obtained by sequencing of an overlapping 12-kb PstI fragment. The intI gene of Xanthomonas sp. 102397 (XspintIA) and X. campestris pv. badrii (XcaintIB) were cloned by PCR with the primers XCR1 and intI9RHS (ATGCGTGGCGAACGAATGCCG) designed from the XcaintIA gene.
SI Signature Detection Among Various Bacterial Genera.
SI structures were identified in various species by PCR amplification of VCR-like cassettes with primers designed to detect the XXRs of closely related species as described above (Vibrio marinus, Vibrio anguillarum, Vibrio salmonicida, and Photobacterium phosphoreum with VCR1 and VCR2 or VFR1 and VFR2) or by Southern hybridization with intI or cassette specific probes (Vibrio hollisae, Vibrio harveyi, Alteromonas macleodii, Pseudomonas pseudoalcaligenes). All PCR products were verified as sharing the characteristics of typical integron gene cassettes (8, 21).
Cloning of the rplT Genes.
The rplT genes of the following species were cloned after PCR amplification with the indicated set of primers. Degenerate positions are bracketed: V. mimicus and V. metschnikovii with primers VM20.SOS (CGTCAACTACGCCCTAACTCA) and VM20.TPT (CTTACGACGATCTCGATAGAT); S. putrefaciens strains 80.40T and 69.34 with primers SpL20–1 (ATATGCTTACCGTGACCGTCG) and SpL20–2 [(AT)(AG)CTTTGTCGAATAC(N)GCGAT]; V. marinus and A. macleodii with primers SputL20–1 [ATA(CT)GCTTACCGTGACCGTCG] and SputL20–3 [(AT)(AG)CTTTGTCAAATAC(N)GCGAT]; P. phosphoreum, Photobacterium profundum, L. pelagia, V. fischeri, V. harveyi, V. parahaemolyticus, and V. hollisae with primers DRL20U (GCCTCGCGTAAAACGTGGTGT) and DRL20D (TTTGTCGAATACTGCGATATC); X. campestris with primers XcL20–2 [TTTGTCGAATAC(GT)GCGATATC] and XcL20–4 [(TAG)TGGAT(CT)GC(TAG)CGTATCAACGC].
Deposition of Nucleic Acid Sequences.
All nucleic acid sequences have been deposited at the National Center for Biotechnology Information (NCBI) GenBank Database. Accession numbers can be found on our website: http://www.pasteur.fr/recherche/unites/pmtg/.
Results
A Conserved Activity for the Integron-Integrases.
To verify the evolutionary link between MRIs and the V. cholerae SI, an activity for the SI integrase gene, intI4 (renamed VchintIA, see below), had to be demonstrated. We monitored integrase activity through deletion of a selectable marker caused by VchintIA-dependent site-specific recombination. We used as substrates pSU38∷ORF1-cat, which harbors a chloramphenicol-resistance gene bordered by two VCRs, and pSU38∷CARB4, which contains a carbenicillinase-resistance gene cassette between two attC sites of an MRI (18). The junctions of 25 recombinant plasmids from each test set were sequenced to ensure that recombination occurred specifically within the GTT triplet of the core site. The results, presented in Table 1, clearly indicated the presence of a VchintIA-dependent site-specific recombination activity for both cassette substrates that was congruent with the activity of IntI1 (18), the only other integron-integrase activity demonstrated to date. Furthermore, the residues identified as critical for the recombination activity of IntI1 (30) were absolutely conserved in VchIntIA (see Fig. 3, which is published as supplemental data on the PNAS web site, www.pnas.org), suggesting that both catalyze recombination by the same mechanism.
SI Distribution.
Among the Vibrionaceae and their close relatives (Fig. 1), we have identified SI structures in V. mimicus, V. metschnikovii, V. parahaemolyticus, L. pelagia and V. fischeri. Estimates by DNA hybridization revealed that each of the Vibrio species SI were at least 100 kb, collectively equaling a small genome (for example, Mycoplasma genitalium). Sequencing of a portion of these SI (≈10%) indicated that, as seen for V. cholerae (21), the majority of the cassette-borne genes had no counterpart in the database, or the sole homologues were unassigned ORFs of viral, bacterial, or eukaryotic origins. Several distinct genera of the γ-proteobacteria also harbor SI structures, including Shewanella and Xanthomonas. In addition, SIs were recently characterized in Pseudomonas alcaligenes and Pseudomonas mendocina.§ SI signatures were obtained by PCR for a gamut of γ-proteobacteria, including V. harveyi, V. marinus, V. hollisae, V. anguillarum, V. salmonicida, Alteromonas macleodii, Photobacterium phosphoreum, Moritella marina, several Xanthomonas spp. (Collection de l'Institut Pasteur nos. 102336, 102338, and 105155), and Pseudomonas pseudoalcaligenes. Notably, integron-integrase genes were also identified by blast analysis in the genome of Nitrosomonas europaea, a member of the β-proteobacterial radiation (U.S. Dept. of Energy sequencing program).
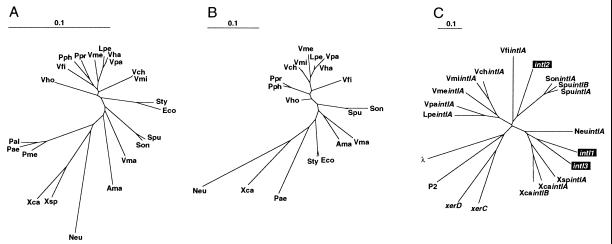
Figure 1.
Phylogenetic relationship of the proteobacteria. Unrooted dendrograms based on the 16S rRNA (A), rplT (ribosomal L20) (B), and intI (C) genes among the proteobacteria used in this study. The integrases from the three classes of MRIs are boxed in C. Organism abbreviations are as follows: Vch, Vibrio cholerae; Vme, V. metschnikovii; Vpa, V. parahaemolyticus; Vma, V. marinus; Vha, V. harveyi; Vho, V. hollisae; Vfi, V. fischeri; Lpe, Listonella pelagia; Pph, Photobacterium phosphoreum; Ppr, P. profundum; Eco, Escherichia coli; Ave, Aeromonas veronii; Son, Shewanella oneidensis; Spu, S. putrefaciens; Xca, Xanthomonas campestris; Xsp, Xanthomonas sp.; Neu, Nitrosomonas europaea. For the integrase and recombinase proteins: λ (of phage lambda), P2 (of phage P2), XerC/D (of E. coli). For intI nomenclature see Table 2.
For each SI identified (Table 2), an intI gene was found to be divergently transcribed from an upstream gene-XXR cluster (in which the first X of XXR denotes the genus and the second X denotes the species in which the repeat occurs). This structure was identical to previously identified MRIs and the V. cholerae SI (8). Because of the large number of integrases that were found, a species-letter nomenclature system for the intI genes was adopted. For example, the integrase gene of V. cholerae 569B was designated VchintIA and the integrase genes of S. putrefaciens strains 80.40T and 69.34 were designated SpuintIA and SpuintIB, respectively.
Table 2.
Attributes of the chromosomal SIs
| Strain | Size of SI, kb* | Integrase gene | Length of intI, nt | XXR
|
|||
|---|---|---|---|---|---|---|---|
| Name | Length, nt | Similarity
|
|||||
| % identity | No. cassettes† | ||||||
| V. cholerae 569B | >100 | VchintIA | 963 | VCR | 121–123 | 74 | 23 |
| V. mimicus 101888T | >100 | VmiintIA | 963 | VMiR | 121–123 | 80 | 3 |
| V. metschnikovii A267 | >100 | VmeintIA | 963 | VMeR | 117–120 | 83‡ | 20‡ |
| V. parahaemolyticus 75.2T | >100 | VpaintIA | 963 | VPR | 119–120 | 75‡ | 4‡ |
| V. fischeri 103206T | >100 | VfiintIA | 951 | VFR | 119–120 | 71‡ | 16‡ |
| L. pelagia 10276.2T | >100 | LpeintIA | 963 | LPR | 117–120 | 86 | 3 |
| S. oneidensis MR1 | ND | SonintIA | 960 | ND | ND | ND | ND |
| S. putrefaciens 80.40T | ND | SpuintIA | 665§ | ND | ND | ND | ND |
| S. putrefaciens 69.34 | ND | SpuintIB | 954 | SPR | 68–94 | ND | ND |
| X. campestris pv. campestris 100069T | ND | XcaintIA | 1,014 | XCcR | 53–54 | 70 | 10 |
| X. campestris pv. badrii 11672 | ND | XcaintIB | 285§ | XCbR | 53–54 | — | — |
| X. sp. 102397 | ND | XspintIA | 858§ | XSR | 53–54 | 80 | 7 |
| N. europaea 103999 | ND | NeuintIA | 975 | ND | ND | ND | ND |
ND, not determined.
> signs denote estimations by Southern blot with the respective XXR-specific probes.
Number of cassettes sequenced.
Two distinct subgroups with at least 60% identity between them were present in the total number of cassettes examined. The percent similarity is among XXR within the respective subgroups.
Partial sequence of intI gene.
Integrons Are Ancient Structures.
Dendrograms were constructed to probe the evolutionary relationship between the extent of species and intI gene divergence. These were based on two essential chromosomal markers unlikely to be subject to horizontal transfer (31), the 16S rRNA gene and the rplT gene (coding for the ribosomal L20 protein). Comparison of the 16S rRNA, rplT, and intI trees revealed a robust phylogenetic congruence for the distribution of the species used in this study (Fig. 1). The extent of divergence between the intI genes adhered to the line of descent among the bacterial species. Consequently, the integrases partitioned in genus-specific clades. The V. cholerae and V. mimicus integrases clustered together, whereas the integrases of V. metschnikovii, V. parahaemolyticus, L. pelagia, and V. fischeri, species on separate lines of descent from V. cholerae according to their 16S rRNA sequences, were more distantly related. This superimposable dispersal pattern was also evident for the integrase genes of the Shewanella and Xanthomonas genera. SonintIA, SpuintIA, and SpuintIA formed a clade, whereas the integrase genes of X. campestris pv. campestris, X. campestris pv. badrii, and Xanthomonas sp. CIP 102397 partitioned in a separate clade. The congruence of the three dendrograms argued for the presence of an integron in the ancestral organism of each genus, and perhaps the γ-proteobacterial radiation itself, over the independent acquisition of integron platforms (i.e., the intI gene and the attI site) within each lineage. Furthermore, no evidence was found that the SI platforms were mobile. Thus integrons are ancient structures that have been steering the evolution of bacterial genomes for hundreds of millions of years, as the substitution rates calculated by Ochman and Wilson estimate that the Vibrio and Pseudomonas/Xanthomonas genera separated some 300–800 million years ago (32).
Despite their divergence, the integron-integrase genes clearly formed a related but distinct group from the λ family of site-specific recombinases (Fig. 1C). Furthermore, the Shewanella integrase genes clustered with intI2 and the Xanthomonas integrases grouped with intI1 and intI3, suggesting that the root for the three classes of MRIs may be within these divisions. Because the catalytic residues were absolutely conserved among all of the SI integrases identified, it can be presumed that they all use the same mechanics for cassette recombination. Thus, we have opted to group all of the integrase genes within a single, functionally defined class.
Coevolution of the SI Integrase Genes and XXRs.
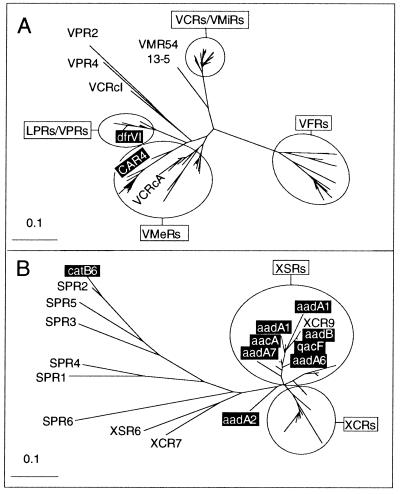
Comparison of the XXR and intI dendrograms revealed an extraordinary symmetry of divergence among the integron-integrase genes and their cognate SI repeats. The XXRs of closely related intI genes shared extensive similarity. For example, VchintIA and VmiintIA share 83% identity (94% at the amino acid level) and their respective XXRs, the VCRs and VMiRs, converged within a clade. However, VmeintIA was only 69% identical to VchintIA (76% at the amino acid level) and its repeat sequences, the VMeRs, were similarly divergent and formed a separate clade. This pattern of decreasing similarity between the XXRs continued as the divergence between the intIs increased (Fig. 2A). This resulted in species-specific clustering of the repeated sequences. However, several repeats were found to group with the bulk of those from another species, such as the VCRcA repeat with the VMeRs. The XXRs of genera remote from the Vibrionaceae were compiled in a separate dendrogram because of their extreme disparity to the VXRs; a meaningful alignment could not be achieved. As seen with the VCRs, tight clustering among the XCRs and XSRs was observed (Fig. 2B). The anomalous variability among the SPRs is reminiscent of MRIs and may reflect the recent acquisition of exogenous cassettes in this strain. The frequency of cassette genesis, capture, and turnover as well as the overall stability of integron cassette arrays remains to be established. However, it is important to note that the SPRs were distinct from the other XXRs. Thus, the parallel divergence between the intI genes and the XXRs suggests that they are coevolving.
Figure 2.
Phylogenetic relationships of the XXRs. The unrooted dendrograms were compiled from an arbitrary selection of representative repeat sequences from each species harboring a SI. For clarity, branch identifications are omitted for regions of tight clustering and instead are encircled and marked with the type of XXR. XXRs with similarity to the endogenous repeats of another species are indicated by name within the respective encircled regions. The repeat sequences associated with known antibiotic resistance cassettes are boxed and confer resistance to the following compounds: aad, streptomycin–spectinomycin; cat, chloramphenicol; qac, quaternary ammonium compounds; CAR, carbenicillin–ampicillin; dfr, trimethoprim. For XXR nomenclature, see Table 2.
The Metabolic Function of Three SI Cassettes.
Three cassettes with the highest homology to genes of known function were tested for activity. A cassette with homology to the sulfate-binding protein (SBP) of E. coli (P score of 9.3e−111) was identified in the V. cholerae SI. In E. coli, an ABC-type transporter and two periplasmic binding proteins, SBP (sbp) and a thiosulfate-binding protein (TSBP, cysP), control the uptake of sulfate and thiosulfate ions for cysteine biosynthesis and other processes. An E. coli sbp/cysP double mutant, which is sulfate auxotrophic, can be complemented by either gene (26). We have shown that this V. cholerae SI cassette can restore prototrophy to the E. coli sbp/cysP double mutant on minimal media containing sodium sulfate as the sole source of sulfur (see Fig. 4 in the supplemental data).
A cassette with homology (P score of 5e−39) to the excreted lipases of psychrophilic bacteria (27) was identified in the SI of V. marinus (a bacterium with an optimum growth temperature of 15°C). This cassette was shown to be actively secreted and functional at 10°C when expressed in E. coli (see Fig. 5 in the supplemental data). We have also shown by PCR, cloning, and sequencing that the restriction endonuclease–DNA modification methyltransferase system of X. campestris pv. badrii [composed of the xbaIR and xbaIM genes, respectively (33)] is structured as a gene cassette within the SI of this strain (see Materials and Methods).
Discussion
The pivotal role of integron-mediated gene-capture in the evolution of antibiotic resistance among Gram-negative bacteria is well established (8, 15, 16, 34, 35). However, the apparent bias toward the propagation of resistance gene cassettes by MRIs is likely due to the selective pressure of antibiotic therapy regimes driving the specific capture of resistance cassettes. It is conceivable, as indicated by the varied and unknown origins of the V. cholerae SI cassettes (21), that any ORF can be structured as a gene cassette. As such, integrons likely play a much broader role in bacterial evolution. In support of this notion, a systematic search has revealed that SIs are present throughout the γ-proteobacterial radiation. We have shown that the activity of the integrase associated with the V. cholerae SI is identical to that of the class 1 integrase of MRIs, previously the only integron-integrase activity described. We also identified the metabolic activity, other than antibiotic resistance and virulence, of three SI cassettes. Thus, integrons operate as a general gene capture system in bacterial adaptation, and the evolutionary congruence between the integron-integrases and their host bacterium indicates that they have been functioning in this capacity for hundreds of millions of years.
But how is it that genes and attC sites become associated to form cassettes? The characteristics of SIs may yield important clues. The accumulation of highly related repeat sequences within SIs could be interpreted as a substrate-preference profile for the integron-integrases. Accordingly, the variability of the attCs of MRIs could be considered as the result of forced recombination of dissimilar attCs into the integron because of the high and narrow selective pressure of antimicrobial therapy regimes. Although adaptive pressures differ in accordance with the microbial environment, they may be as selective as those of antibiotic therapy regimes. Such conditions should diminish any XXR preference and promote the acquisition of unrelated XXRs so long as they are associated with a useful gene. Furthermore, the integron-integrases can readily function in this capacity, since both IntI1 and VchIntIA have been shown to have an extremely flexible substrate recognition range. Yet, the XXRs remain remarkably uniform. This is of particular importance. It implies that the process of cassette genesis, in particular the mechanism of XXR propagation, occurs within bacteria possessing a SI. Furthermore, it implies that there is an initial directional flow of cassettes from SIs to MRIs. If SIs are the source of MRI cassettes, the gene cassette pool available to be propagated through mobile integrons is immense.
Our data suggest several ideas regarding integron-mediated lateral transfer. First, that each distinct resistance cassette attC site of MRIs represents a specific SI. Second, that MRIs have evolved from SIs through the association of intI genes and their cognate attI sites with mobile structures. Subsequently, cassettes with dissimilar attC sites were harvested from the many different kinds of SI cassette pools through multiple lateral transfers. Third, as illustrated by the grouping of VCRcA with the VMeRs, the exchange of cassettes between SIs, whether direct or indirect, must also occur. It has been noted that XXRs occur exclusively in intergenic regions. An XXR primer/reverse transcriptase model for cassette genesis has been proposed to accommodate this observation (36), but several characteristics of SIs (i.e., the apparent varied origins of the genes, promoter-associated cassettes, and inversely oriented genes) are inconsistent with this hypothesis. The homogeneous yet distinctive nature of the XXRs is an attribute that may reflect an enzymatic or gene-conversion type event governing their propagation. However, the dynamics of cassette genesis remain somewhat nebulous.
Strikingly, one-fifth of the antimicrobial cassettes of MRIs have attC sites virtually identical in length and sequence to the XXRs of the SIs described here. Because the XXRs of SI cassettes are substrates for the class 1 integrase of MRI (18), this implies that these resistance cassettes originated within these or closely related bacteria. Although a known antibiotic-resistance gene cassette has yet to be definitively identified within these SIs, several potential progenitor cassettes with significant homology (greater than 30% amino acid identity) to aminoglycoside, phosphinotricin, fosfomycin, streptothricin, and chloramphenicol resistance genes have been found. The function of these cassettes remains to be established. However, the phenomenal chemical diversity of naturally occurring antimicrobial compounds (37) and the effects of perpetual evolutionary changes renders precise determination of their function a daunting task.
The potency of the integron system lies in its versatility, that is, the ability to recognize highly variable target recombination sequences and an apparently limitless capacity to exchange and stockpile cassettes. Such flexibility permits rapid adaptation to the unpredictable flux of environmental niches by allowing bacteria to scavenge foreign genes that may ultimately increase their fitness. In the course of natural selection, the acquisition of sbp or lip genes may promote the colonization of new ecological niches, whereas a restriction–modification system may be a defensive measure against, for example, phage infection. Likewise, genes that fail to provide a meaningful function can be readily eliminated. In addition to the plethora of antibiotic-resistance genes, two virulence genes of V. cholerae are also structured as gene cassettes (38, 39), underscoring the potential of this system to participate in the establishment of pathogenicity islands. Although retained among many extant proteobacterial genera, this system may have been lost in others (for example, the enterobacteria). Genomes are dynamic, and ample opportunities exist for extensive changes in gene order and content to occur. These events can lead to deletion of nonessential genes. Integrons may not be essential systems, and the SI cassettes examined to date seemingly encode adaptive rather than indispensable functions. Thus, deletion of the integron platform may have occurred in certain genera or independently within species. In support of this possibility, one of the Shewanella strains we examined was found to contain only a partial intI gene. Furthermore, other elements exist that allow bacteria to survive adaptive challenges and explore new niches. The participation of bacteriophages in this regard is well documented (for review see refs. 1, 40, and 41). Even though the integron platform lacks the typical characteristics of bacteriophages (tRNA insertion preference, mobilization genes, terminal flanking direct repeats, and packaging functions), the similarity of the integron system to bacteriophages at the level of the integrase gene, the attachment sites, and the large number of ORFs of unassigned function is intriguing. It is tempting to speculate that the ancestral integron was an immobilized prophage. If so, the integron and phage systems could have diverged to the extent that their functions no longer share any overlap, except for the recombination machinery itself. Although their origin remains elusive, the presence of SIs among diverse Gram-negative bacteria establishes both an ancestry for MRIs and the quintessential role of integrons in adaptive bacterial evolution.
Supplementary Material
Acknowledgments
We especially thank Prof. Maurice Hofnung (Unité de Programmation Moléculaire et Toxicologie Génétique) for his unconditional and enthusiastic support. We also thank Stéphane Blanchard for his sequencing skills, Navith Kaim, Latefa Biskri, and Alexandre Prieur for their technical assistance, Dr. Douglas Bartlett for Photobacterium and Vibrio strains, Drs. Elizabeth Raleigh and George Feller for their gifts of X. campestris pv. badrii and pULG323, respectively. Thank you to The Institute for Genomic Research and the Department of Energy for making their genome sequence data available to all. In particular, we thank Chantal Bizet and Dominique Clermont of the Collection de l'Institut Pasteur and Dr. Martina Ochs for critical reading of the manuscript and insightful discussions. D.R.-M. was supported by a Manlio Cantarini Postdoctoral Fellowship (Institut Pasteur) and is currently a European Molecular Biology Organization Postdoctoral Fellow. This work was supported by the Institut Pasteur, the Centre National de la Recherche Scientifique, and the Program de Recherche Fondamentale en Microbiologie et Maladies Infectieuses et Parasitaires from the Ministère de l'Éducation Nationale, de la Recherche et de la Technologie.
Abbreviations
- MRI
multiresistant integron
- SI
super-integron
- VCR
Vibrio cholerae repeated sequence (repeated sequences of other species are named similarly)
- IS
insertion sequence
- Tn
transposon
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF180939, AY014398–AY014401, AF324210, AF324211, AF324482–AF324484, and AF324934–AF324946).
Vaisvila, R., Morgan, R. & Raleigh, E., Ninth International Congress of Bacteriology and Applied Microbiology, Aug. 15–20, 1999, Sidney, Australia, abstr. BP09.27.
References
- 1.Ochman H, Lawrence J G, Groisman E A. Nature (London) 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 2.Davies J E. Science. 1994;264:375–382. doi: 10.1126/science.8153624. [DOI] [PubMed] [Google Scholar]
- 3.Sundström L, Radström P, Swedberg G, Sköld O. Mol Gen Genet. 1988;213:191–201. doi: 10.1007/BF00339581. [DOI] [PubMed] [Google Scholar]
- 4.Stokes H W, Hall R M. Mol Microbiol. 1989;3:1669–1683. doi: 10.1111/j.1365-2958.1989.tb00153.x. [DOI] [PubMed] [Google Scholar]
- 5.Ouellette M, Bissonnette L, Roy P H. Proc Natl Acad Sci USA. 1987;84:7378–7382. doi: 10.1073/pnas.84.21.7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinez E, de la Cruz F. EMBO J. 1990;9:1275–1281. doi: 10.1002/j.1460-2075.1990.tb08236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall R M, Collis C M. Mol Microbiol. 1995;15:593–600. doi: 10.1111/j.1365-2958.1995.tb02368.x. [DOI] [PubMed] [Google Scholar]
- 8.Rowe-Magnus D A, Mazel D. Curr Opin Microbiol. 1999;2:483–488. doi: 10.1016/s1369-5274(99)00004-1. [DOI] [PubMed] [Google Scholar]
- 9.Hall R M, Stokes H W. Genetica. 1993;90:115–132. doi: 10.1007/BF01435034. [DOI] [PubMed] [Google Scholar]
- 10.Sundstrom L. APMIS Suppl. 1998;84:37–42. doi: 10.1111/j.1600-0463.1998.tb05646.x. [DOI] [PubMed] [Google Scholar]
- 11.Stokes H W, O'Gorman D B, Recchia G D, Parsekhian M, Hall R M. Mol Microbiol. 1997;26:731–745. doi: 10.1046/j.1365-2958.1997.6091980.x. [DOI] [PubMed] [Google Scholar]
- 12.Recchia G D, Hall R M. Microbiology. 1995;141:3015–3027. doi: 10.1099/13500872-141-12-3015. [DOI] [PubMed] [Google Scholar]
- 13.Levesque C, Brassard S, Lapointe J, Roy P H. Gene. 1994;142:49–54. doi: 10.1016/0378-1119(94)90353-0. [DOI] [PubMed] [Google Scholar]
- 14.Collis C M, Kim M J, Stokes H W, Hall R M. Mol Microbiol. 1998;29:477–490. doi: 10.1046/j.1365-2958.1998.00936.x. [DOI] [PubMed] [Google Scholar]
- 15.Mazel D, Davies J. Cell Mol Life Sci. 1999;56:742–754. doi: 10.1007/s000180050021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall R M, Collis C M. Drug Resist Updates. 1998;1:109–119. doi: 10.1016/s1368-7646(98)80026-5. [DOI] [PubMed] [Google Scholar]
- 17.Poirel L, Le Thomas I, Naas T, Karim A, Nordmann P. Antimicrob Agents Chemother. 2000;44:622–632. doi: 10.1128/aac.44.3.622-632.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mazel D, Dychinco B, Webb V A, Davies J. Science. 1998;280:605–608. doi: 10.1126/science.280.5363.605. [DOI] [PubMed] [Google Scholar]
- 19.Trucksis M, Michalski J, Deng Y K, Kaper J B. Proc Natl Acad Sci USA. 1998;95:14464–14469. doi: 10.1073/pnas.95.24.14464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heidelberg J F, Eisen J A, Nelson W C, Clayton R A, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Umayam L, et al. Nature (London) 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rowe-Magnus D A, Guerout A-M, Mazel D. Res Microbiol. 1999;150:641–651. doi: 10.1016/s0923-2508(99)00127-8. [DOI] [PubMed] [Google Scholar]
- 22.Barker A, Clark C A, Manning P A. J Bacteriol. 1994;176:5450–5458. doi: 10.1128/jb.176.17.5450-5458.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Felsenstein, J. (1995) phylip (Dept. of Genetics, Univ. of Washington, Seattle).
- 25.Page, R. D. M. (1996) treeview (Division of Environmental and Evolutionary Biology, Institute of Biomedical and Life Sciences, Univ. of Glasgow, Glasgow, U.K.).
- 26.Sirko A, Zatyka M, Sadowy E, Hulanicka D. J Bacteriol. 1995;177:4134–4136. doi: 10.1128/jb.177.14.4134-4136.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feller G, Thiry M, Arpigny J-L, Mergeay M, Gerday C. FEMS Microbiol Lett. 1990;66:239–244. [Google Scholar]
- 28.Chambers S P, Prior S E, Barstow D A, Minton N P. Gene. 1988;68:139–149. doi: 10.1016/0378-1119(88)90606-3. [DOI] [PubMed] [Google Scholar]
- 29.Mazel D, Dychinco B, Webb V, Davies J. Antimicrob Agents Chemother. 2000;44:1568–1574. doi: 10.1128/aac.44.6.1568-1574.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gravel A, Messier N, Roy P H. J Bacteriol. 1998;180:5437–5442. doi: 10.1128/jb.180.20.5437-5442.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jain R, Rivera M C, Lake J A. Proc Natl Acad Sci USA. 1999;96:3801–3806. doi: 10.1073/pnas.96.7.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ochman H, Wilson A C. J Mol Evol. 1987;26:74–86. doi: 10.1007/BF02111283. [DOI] [PubMed] [Google Scholar]
- 33.Van Cott, E. M. & Wilson, G. G. (New England Biolabs) (1991) U.S. Patent 4983542.
- 34.Martin C, Timm J, Rauzier J, Gomez-Lus R, Davies J, Gicquel B. Nature (London) 1990;345:739–743. doi: 10.1038/345739a0. [DOI] [PubMed] [Google Scholar]
- 35.Jones M E, Peters E, Weersink A M, Fluit A, Verhoef J. Lancet. 1997;349:1742–1743. doi: 10.1016/S0140-6736(05)62954-6. [DOI] [PubMed] [Google Scholar]
- 36.Recchia G D, Hall R M. Trends Microbiol. 1997;5:389–394. doi: 10.1016/S0966-842X(97)01123-2. [DOI] [PubMed] [Google Scholar]
- 37.Demain A L. Biochem Soc Symp. 1983;48:117–132. [PubMed] [Google Scholar]
- 38.Ogawa A, Takeda T. Microbiol Immunol. 1993;37:607–616. doi: 10.1111/j.1348-0421.1993.tb01683.x. [DOI] [PubMed] [Google Scholar]
- 39.van Dongen W M A M, van Vlerken M M A, DeGraaf F K. Mol Gen (Life Sci Adv) 1987;6:85–91. [Google Scholar]
- 40.Cheetham B F, Katz M E. Mol Microbiol. 1995;18:201–208. doi: 10.1111/j.1365-2958.1995.mmi_18020201.x. [DOI] [PubMed] [Google Scholar]
- 41.Hendrix R W, Smith M C, Burns R N, Ford M E, Hatful G F. Proc Natl Acad Sci USA. 1999;96:2192–2197. doi: 10.1073/pnas.96.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.