Abstract
Although it is well established that Hispanics generally have a mixed Native American, African, and European ancestry, the dynamics of admixture at the foundation of Hispanic populations is heterogeneous and poorly documented. Genetic analyses are potentially very informative for probing the early demographic history of these populations. Here we evaluate the genetic structure and admixture dynamics of a province in northwest Colombia (Antioquia), which prior analyses indicate was founded mostly by Spanish men and native women. We examined surname, Y chromosome, and mtDNA diversity in a geographically structured sample of the region and obtained admixture estimates with highly informative autosomal and X chromosome markers. We found evidence of reduced surname diversity and support for the introduction of several common surnames by single founders, consistent with the isolation of Antioquia after the colonial period. Y chromosome and mtDNA data indicate little population substructure among founder Antioquian municipalities. Interestingly, despite a nearly complete Native American mtDNA background, Antioquia has a markedly predominant European ancestry at the autosomal and X chromosome level, which suggests that, after foundation, continuing admixture with Spanish men (but not with native women) increased the European nuclear ancestry of Antioquia. This scenario is consistent with historical information and with results from population genetics theory.
Keywords: genetic history, migration, latinos, Founder effect, cultural selection
The European colonial expansion to the Americas resulted in massive population movements from the Old World to the New World. During the period of Iberian rule (15th–19th centuries) over 1,000,000 Spanish and Portuguese migrated to the New World (1). In addition, some 5 million Africans were forcefully taken to Iberian America as a result of the Atlantic slave trade (2, 3). The European colonization of the Americas decimated the native population, resulting in what is arguably the most drastic population collapse in human history. By the end of the 16th century a native population of some 50 million was reduced to ≈10% of its original size (4, 5). These dramatic demographic changes ultimately resulted in the establishment of populations with various types and degrees of admixture across the vast region of Iberian control in the Americas. The dynamics of this process are, however, not well known (4, 6). In particular, information for the century of expansion of the colonial frontier (i.e., the “Period of Conquest” in the 16th century) is almost nonexistent. Not surprisingly, this is the period when admixture is likely to have been most extensive (4, 6).
We have been studying the admixture history of a population from a mountainous region of northwest Colombia (Antioquia). Spain began exploring this area early in the 16th century (7), the first named town (Santa Fe de Antioquia) being founded in 1541 in a lowland river valley. Migration from lowland areas to highland areas occurred throughout the 16th and early 17th centuries, culminating in the foundation of the towns of San Lorenzo de Aburrá (currently Medellín) in 1675 and Marinilla in 1690 (in the highland valleys of Aburrá and Oriente, respectively). At the arrival of the Spanish, the native population inhabiting these highland valleys was scarce, pioneer chronicles mentioning a few thousand native individuals early in the 16th century (8). After the colonial period, Antioquia remained relatively isolated from further immigration, and the population expanded mostly by internal growth. During the 19th century, the admixed population that developed in the valleys of Aburra and Oriente formed an important source for the settlement of a large part of Western Colombia (7, 9).
We previously examined the maternal and paternal ancestry of a sample of Antioquian individuals ascertained in the provincial capital but with a widespread ancestry in the region. Y chromosome lineages were found to be ≈94% European, 5% African, and 1% Amerind, whereas mtDNA lineages in this sample are ≈90% Amerind, 8% African, and 2% European (10, 11). The marked difference in the paternal (Y chromosome) and maternal (mtDNA) ancestry indicates a biased pattern of mating at the foundation of Antioquia, admixture involving mostly immigrant men and native women. A similar phenomenon has now been described for several other Latin American populations (11–14). The elevated Native American maternal ancestry detected in Antioquia was, however, unexpected because this population has been considered by historians and genealogists as primarily of Spanish descent (15, 16). Furthermore, available classical marker data (blood groups and proteins) suggest that the genetic background of Antioquia is largely (≈70%) European, with substantially smaller Native American and African contributions (≈15% each) (17). If this admixture estimate is correct, we would expect the mtDNA lineages to be no more than ≈30% Native American. The markedly higher than expected frequency of Native American mtDNA lineages in Antioquia could relate to population substructure in the region and the fact that these studies examined different population samples. However, if true, this discordance could also indicate a deviation from a model of founding admixture followed by isolation.
Here we perform a detailed assessment of admixture estimates obtained from uniparental (Y chromosome and mtDNA) and biparental (autosomal and X chromosome) genetic markers in Antioquia. We examined mtDNA, Y chromosome, and surname diversity in six founder municipalities from the highland valley of Oriente and contrast these data with the general Antioquian sample characterized previously (10, 11). These data indicate little population subdivision in the founder gene pool of Antioquia and provide strong support for the postcolonial isolation of this population. The biparental marker data confirm that the European ancestry of Antioquia is markedly higher than expected based on the frequency of Native American mtDNA lineages. A simple interpretation for this observation is that native women were involved in admixture mostly at the foundation of the population but that gene flow from Spanish men occurred over several generations. This would have resulted in a rapid increase in the European ancestry for autosomal and X-linked polymorphisms, despite Antioquia retaining an almost entirely Native American mtDNA background.
Results
mtDNA Haplogroup Diversity.
The frequency of the four major Native American mtDNA haplogroups (haplogroups A–D) and of nonnative haplogroups in Oriente and in the general Antioquian sample examined previously (10) are shown in Table 1. In all municipalities, haplogroup A is the most common, with frequencies ranging from 0.48 to 0.75. Haplogroup B is the second most frequent, representing 0.19–0.47 of those observed in the region. Altogether, haplogroups A and B account for 77–100% of mtDNAs in Oriente municipalities. Haplogroups C and D and non-Native American haplogroups are relatively rare in all samples. In total, native haplogroups have a frequency of 0.97 in Oriente. Haplogroup diversity is highest in Marinilla, the oldest municipality in the region and the only one where all haplogroups are observed. Overall, Oriente shows a lower haplogroup diversity than the general Antioquian sample (0.55 vs. 0.65). Based on the mtDNA haplogroup frequencies, the overall FST in Oriente is 0.015, with no significant differentiation between any of the samples examined nor between Oriente and the general Antioquian sample.
Table 1.
Frequency of Native American mtDNA haplogroups A–D and nonnative haplogroups in Oriente (six municipalities) and in a general Antioquian sample
| Regions | n | mtDNA haplogroup |
H | ||||
|---|---|---|---|---|---|---|---|
| A | B | C | D | Nonnative | |||
| Oriente | |||||||
| Marinilla | 26 | 0.58 | 0.19 | 0.12 | 0.04 | 0.08 | 0.63 |
| Peñol | 42 | 0.48 | 0.45 | 0.05 | 0.00 | 0.02 | 0.58 |
| Santuario | 37 | 0.51 | 0.43 | 0.00 | 0.00 | 0.05 | 0.56 |
| Carmen | 36 | 0.5 | 0.47 | 0.03 | 0.00 | 0.00 | 0.54 |
| Guatapé | 17 | 0.65 | 0.35 | 0.00 | 0.00 | 0.00 | 0.48 |
| Granada | 24 | 0.75 | 0.25 | 0.00 | 0.00 | 0.00 | 0.39 |
| Total | 182 | 0.55 | 0.38 | 0.03 | 0.01 | 0.03 | 0.55 ± 0.02 |
| Antioquia | 80 | 0.45 | 0.38 | 0.06 | 0.01 | 0.10 | 0.65 ± 0.03 |
H indicates haplogroup diversity. The data for Antioquia are from Carvajal-Carmona et al. (10).
Y Chromosome Haplogroup Diversity.
Table 2 shows the frequency of major Y chromosome haplogroups in Oriente municipalities and in the general Antioquian sample of Carvajal-Carmona et al. (10). The most common haplogroups in all population samples are P-M45* and Y*, with frequencies ranging between 0.57 and 0.92 and between 0.08 and 0.40, respectively. These two haplogroups account for 0.83–1.0 of Y chromosomes in Oriente municipalities. Overall, the frequency of Native American (Q-M3) and African (E-M2) haplogroups in Oriente is respectively 0.04 and 0.05. As for mtDNA, Y haplogroup diversity in Oriente municipalities is lower than in the general Antioquian sample (0.47 vs. 0.56), and the highest diversity is observed in Marinilla. The overall FST across municipalities is 0.022 with no significant differentiation in haplogroup frequencies and no significant difference between Oriente and the general Antioquian sample.
Table 2.
Y chromosome haplogroup frequency in Oriente (six municipalities) and in a general Antioquian sample
| Regions | n | Y haplogroup |
H | ||||
|---|---|---|---|---|---|---|---|
| P-M45* | Q-M3 | Y* | DE-YAP | E-M2 | |||
| Oriente | |||||||
| Marinilla | 21 | 0.57 | 0.04 | 0.29 | 0.05 | 0.05 | 0.55 |
| Peñol | 13 | 0.76 | 0.08 | 0.08 | 0.00 | 0.08 | 0.42 |
| Santuario | 13 | 0.92 | 0.00 | 0.08 | 0.00 | 0.00 | 0.15 |
| Carmen | 10 | 0.60 | 0.00 | 0.40 | 0.00 | 0.00 | 0.53 |
| Guatapé | 12 | 0.66 | 0.00 | 0.17 | 0.00 | 0.17 | 0.55 |
| Granada | 23 | 0.78 | 0.00 | 0.22 | 0.00 | 0.00 | 0.36 |
| Total | 92 | 0.70 | 0.04 | 0.20 | 0.01 | 0.05 | 0.47 ± 0.05 |
| Antioquia | 80 | 0.58 | 0.01 | 0.33 | 0.04 | 0.04 | 0.56 ± 0.04 |
Surname Diversity in Oriente.
Table 3 shows the frequency of the 15 most common surnames in Oriente based on census information for 1786 and 2004. In aggregate these 15 surnames account for 59% and 49% of all surnames in these two periods. By comparison, in the French Canadian population, the lowest diversity has been reported in Ile de la Madeleine, where the most common 15 surnames have a combined frequency of 42% (18). Of the 15 most common surnames in Oriente, 11 are the same in 1786 and 2004. During this period, the population increased 107-fold (177,903 vs. 1,668), whereas the number of surnames increased 13-fold (1,432 vs. 114). Founder surnames (registered at the foundation of Marinilla in 1690) currently have an aggregate frequency of 60% in the region (results not shown).
Table 3.
Frequency (%) of the 15 most common surnames in Oriente (for 1786 and 2004) and the total frequency (%) of the 15 most common surnames in French Canadian regions
| Oriente, Colombia |
French Canada |
||||
|---|---|---|---|---|---|
| 1786 |
2004 |
||||
| Surname | Frequency | Surname | Frequency | Region | Total Frequency |
| Giraldo | 9 | Giraldo | 8 | Ile de la Madeleine | 42 |
| Gomez | 7 | Gomez | 6 | Charlevoix | 37 |
| Duque | 6 | Ramirez | 5 | Sanguenay | 27 |
| Salazar | 5 | Zuluaga | 5 | Saint Laurent | 24 |
| Quintero | 5 | Quintero | 4 | Beauce | 19 |
| Ramirez | 4 | Garcia | 3 | Cote Nord | 14 |
| Zuluaga | 4 | Duque | 3 | Gaspesie | 13 |
| Agudelo | 3 | Castaño | 3 | Quebec | 10 |
| Garcia | 3 | Aristizabal | 3 | Bois Franc | 9 |
| Marin | 3 | Salazar | 2 | Mauricie | 9 |
| Aristizabal | 3 | Lopez | 2 | Abitibi | 8 |
| Idarraga | 3 | Alzate | 2 | Canton Est | 8 |
| Ocampo | 2 | Montoya | 2 | Laurentides | 8 |
| Castaño | 2 | Cardona | 1 | Montreal | 2 |
| Valencia | 2 | Valencia | 1 | ||
| Total frequency | 59 | Total frequency | 49 | ||
Surnames included in both periods for Oriente are in bold. Data for French Canada are from ref. 20.
Y Microsatellite Haplotype Diversity of Founder Surnames.
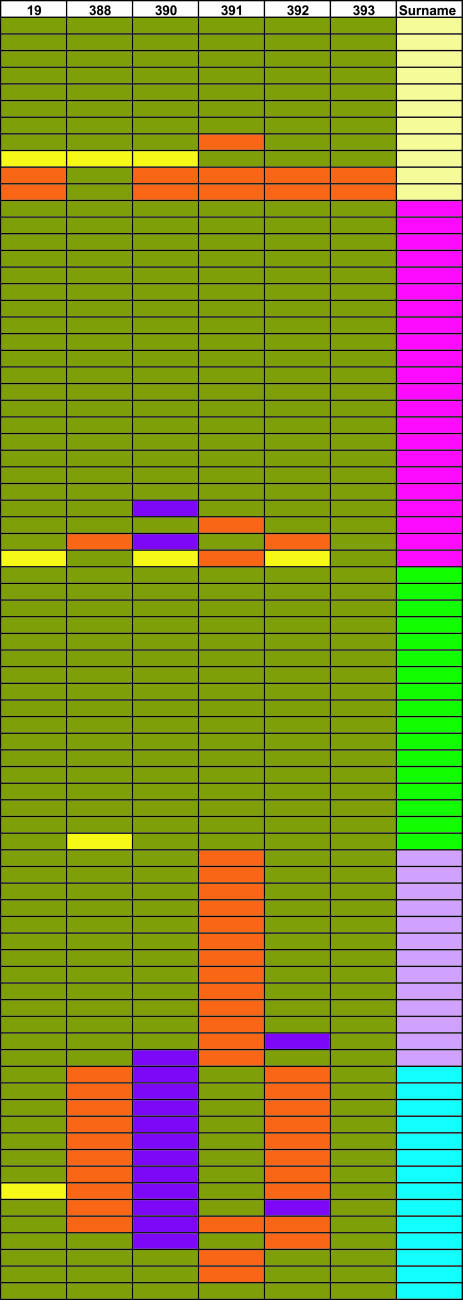
Genealogical information indicates that five of the most common surnames present in Oriente in 2004 (Table 3) were introduced in the region each by a single Spanish immigrant in the mid-17th century (15). Six-locus microsatellite haplotypes of a sample of individuals with each of these five surnames in Oriente are shown in Fig. 1. For two surnames, all individuals typed carry the same haplotype or a haplotype with a one-step difference. For the three other surnames, the modal haplotype plus one-step neighbors represent between 73% and 91% of all chromosomes (Table 4). Assuming that one-step neighbors of the surname modal haplotype result from mutation at a microsatellite locus (occurring after the introduction of each surname) allows an estimate of mutation rate to be obtained. Taking a generation time of 25 years and a mean date for the introduction of the five surnames examined in 1,650 results in an average mutation rate for the six microsatellites tested of 1.86 × 10−3 per generation. Other than one-step neighbors other haplotypes observed differ from the surname modal haplotype by three or more differences (Fig. 1) and most likely represent instances of nonpaternity. Averaging over the five surnames examined and with the same assumptions used above results in a mean nonpaternity rate of 0.74 × 10−2 per generation.
Fig. 1.
Y microsatellite haplotypes for five common surnames in Oriente. Each row represents a different individual. The rightmost column indicates surnames. The other six columns represent Y-microsatellite loci (the prefix DYS has been omitted). Alleles and surnames have been color-coded with each color representing a different allele or surname.
Table 4.
Y chromosome microsatellite haplotype frequency and diversity (H) for five common surnames in Oriente
| Surname color in Fig. 1 | n | Modal haplotype | Modal + 1 step | Other | H |
|---|---|---|---|---|---|
| Turquoise | 14 | 0.500 | 0.786 | 0.214 | 0.297 |
| Yellow | 11 | 0.636 | 0.727 | 0.273 | 0.370 |
| Magenta | 22 | 0.818 | 0.909 | 0.091 | 0.131 |
| Green | 17 | 0.941 | 1.000 | 0.000 | 0.020 |
| Lavender | 13 | 0.846 | 1.000 | 0.000 | 0.051 |
Genetic Background and Admixture Dynamics in Antioquia.
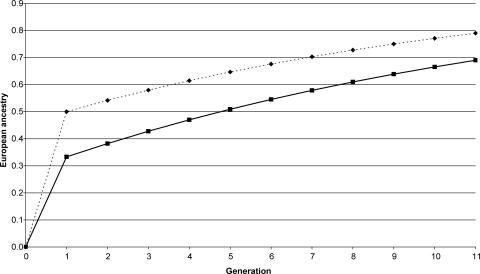
Table 5 shows estimates of admixture for Antioquia obtained with different marker systems. Autosomal data indicate that the ancestry of the population is ≈79% European, 15% Amerind, and 6% African. X-linked markers also show a predominant European ancestry (69%), with smaller Amerind (25%) and African (6%) contributions. Fig. 2 gives the expected increase in the European ancestry of Antioquia over time for autosomal and X-linked markers based on the migration model described in Materials and Methods. The scenario explored assumes that migration was restricted to the colonial period, comprising approximately the first 250 years after foundation (≈10 generations, for a generation time of 25 years). Taking qt+1 = 0.79, the current proportion of European ancestry based on autosomal data (Table 5), results in estimates for gene flow parameter m and mm of 0.083 and 0.166, respectively (from Eqs. 3 and 4 in Materials and Methods). For X-linked markers, qt+1 = 0.69 (Table 5) and the corresponding values for m and mm are 0.074 and 0.218, respectively.
Table 5.
Admixture proportions in Antioquia
| Genetic system | N | European | African | Amerind |
|---|---|---|---|---|
| Y chromosome | 80 | 94 ± 6 | 5 ± 5 | 1 ± 3 |
| mtDNA | 113 | 2 ± 1 | 8 ± 3 | 90 ± 3 |
| Autosomes | 80 | 79 ± 6 | 6 ± 2 | 16 ± 6 |
| X chromosome | 96 | 69 ± 10 | 6 ± 9 | 25 ± 6 |
Fig. 2.
The expected change in European ancestry over time when the rate of gene flow per generation m = 0.083 and 0.074 for autosomal (dotted line) and X-linked (solid line) loci, respectively, obtained using recursion Eq. 2 in Materials and Methods.
Discussion
Our analyses provide strong evidence of the postcolonial isolation of Antioquia. By comparison to French Canada (a relatively isolated population of similar age to Antioquia) Oriente shows lower surname diversity than Ile de la Madeleine, the most isolated French Canadian region. The fairly stable frequency of common surnames in Oriente between 1786 and 2004 is consistent with the notion that the time of greatest immigration to Antioquia was before the 18th century and that the subsequent population expansion was driven mostly by internal growth. The reduced heterogeneity of the male founders in Oriente based on historical information is supported by the Y chromosome microsatellite haplotype data: Five of the most common surnames (currently accounting for >25% of surnames in Oriente) were very likely introduced by a single founder each (15). Consistent with the introduction of each surname by a single founder, the estimated mean microsatellite mutation rate and the mean illegitimacy rate in Oriente are within the range of those reported in independent studies (19–23).
The analyses performed here show little evidence of genetic stratification among founder Antioquiam municipalities and confirm the existence of an important discordance between the biparental and uniparental estimates of admixture (Table 5). Tables 1 and 2 indicate that the largely predominant Spanish male and Native American female ancestry initially observed in a broad sample from Antioquia (10) is a common pattern in the province, with little variation across founder municipalities. In addition, the admixture estimates (Table 5) confirm a largely predominant European ancestry in Antioquia, as suggested by classical markers (17). The level of European ancestry estimated from DNA data are in fact somewhat higher than that calculated from classical markers (79% vs. 70%, respectively) (17). Considering that the African ancestry of Antioquia is essentially uniform across marker systems (≈5–8%) (Table 5) and ignoring the marginal contribution of Amerind men (≈1%) and European women (≈2%), a scenario of founding admixture by European men and native women followed by isolation should result in European/Amerind ancestry ratios of ≈1 for autosomal markers. Table 5 indicates that this ratio is, in fact, substantially higher: 4.94 (79%/16%). A simple explanation of this observation is that following the initial admixture at the foundation of Antioquia, continuing gene flow from Europeans but not from Native Americans increased the European ancestry of the population. The fact that this increase in European ancestry did not affect mtDNA suggests that gene flow after initial foundation was mediated mostly through males. This scenario is supported by the X chromosome data. Because women contribute two times more X chromosomes than men, admixture estimates obtained from X chromosome markers are highly sensitive to migration of women and less sensitive to drift than mtDNA. Under the assumption of male-only migration, the European/Amerind ancestry ratio based on X chromosome markers is expected to be one-half the autosomal estimate. Remarkably, the estimate obtained here is only slightly higher than expected: 2.76 (69%/25%) vs. 2.47. This concordance reinforces the notion that the demographic impact of immigrant Spanish women in Antioquia was very small relative to that of Spanish men.
The simple quantitative model explored here results in estimates of gene flow after the initial admixture event that are consistent for autosomal and X chromosome markers. The migration rates calculated are necessarily approximations but the mean values obtained are not inconsistent with historical information. It seems likely that the highest rates of European gene flow occurred in the first few generations after foundation because Spanish emigration to America was highest during the mid-16th to mid-17th centuries (1). At the time, the local population was also in the early stages of growth and would have been impacted more strongly by immigrant gene flow than in later generations. The first population register carried out in the Aburrá Valley (in 1675) recorded ≈3,000 individuals, of which 18% were categorized as white, 56% mestizo, 9% native, and 17% African (8). It is noticeable that the recognized admixed population was already by far the largest group. Also, although “white” is a particularly unreliable category, it included a substantial fraction of recent Spanish male immigrants, many of which were the founders of surnames that are presently common in the region (8, 15).
Historical information also suggests that the influx of Spanish males during the 16th and 17th centuries was unlikely to be accompanied by a parallel incorporation into the population of similar numbers of native females. This discordance could primarily relate to the initially low Native American population density in Antioquia and the rapid demographic collapse of the native population. Another likely contributing factor is that Spanish immigrants arriving after the period of initial exploration and settlement preferentially had offspring with mestizo (admixed) women rather than with natives. It is known that the offspring of the first conquistadors and native women often inherited the social status of Europeans. However, as colonial society became consolidated it increasingly discouraged new Spanish immigrants from marrying natives and favored individuals of greater Spanish ancestry. Social discrimination based on ancestry culminated in the establishment of a caste system during the 18th century that reserved many privileges (such as access to higher education or government office) only to individuals purportedly having no Native American ancestry. These circumstances are likely to have made even more difficult a continuing incorporation of Native American women into the Antioquian population. In this historical context, an interesting possibility is that a process of “cultural” selection could also have contributed to the increase in the European ancestry of Antioquia. Using detailed demographic data, it has been shown in French Canadians that a correlation exists across generations in the number of offspring a family has, a process that can result in rapid changes in allele frequency (24, 25). The transmission of social status associated with particular male lineages has recently been invoked to explain the high frequency of specific Y chromosome haplotypes in Asia (26, 27). Because, in populations such as Antioquia, Spanish ancestry has historically been associated with higher social status and probably with greater reproductive success, it is possible that cultural selection also impacted the current genetic makeup of these populations.
There is currently considerable interest in the potential of admixed populations for mapping human disease genes and Hispanics seem well suited for this approach (28). However, some caution seems warranted because specifics of the dynamics of gene flow will affect how best to implement this approach. For instance, continuous gene flow implies higher levels of linkage disequilibrium than admixture followed by isolation, resulting in a variable density of markers required for mapping in populations with different admixture dynamics (29). The history of admixture varied among Hispanic populations depending, among other things, on the size and number of migratory waves to specific regions and the native population density in the areas being occupied. As shown here, populations such as Antioquia that developed in regions with a low initial native population density and with a history of isolation after the colonial period are likely to have undergone mostly an early admixture. Regions with high initial native population densities (e.g., Mexico, Central America, and the central Andes) or that received large waves of immigrants recently (e.g., parts of southern South America) are still undergoing important admixture. It seems likely that each type of population will present advantages and disadvantages for admixture mapping. In any event, optimal application of this approach in Hispanics will require that the strategy used is adjusted to the specific admixture history of the population from where patients are being ascertained.
Materials and Methods
Study Population.
For the genetic structure analyses, samples were obtained in the Antioquian municipalities of Marinilla, (El) Peñol, (El) Santuario, Cocorná, Carmen (de Viboral), Granada, and Guatapé. These municipalities are all located in the highland valley of Oriente. Census information for Oriente in the years 1786 and 2004 was reviewed to obtain total population size and surname frequency in each period. The samples examined for autosomal and X chromosome admixture estimation were essentially the same as in our previous studies of mtDNA and Y chromosome polymorphisms (10, 11). A genealogical interview that recorded the places of birth of ancestors up to the great grandparents was carried out to confirm local ancestry of study participants and exclude closely related individuals. A blood or cheek swab sample was collected from each individual, and genomic DNA was extracted by routine laboratory procedures. All participants provided informed consent for this investigation (approved by the Bioethics Committee of Universidad de Antioquia).
Polymorphisms Typed.
We obtained data for four biallelic and six microsatellite markers on the nonrecombining region of the Y chromosome. The biallelic markers examined (and conditions for typing) were DYS271/M2 (30), DYS287 (31), 92R7/M45 (32), and DYS199/M3 (33). These markers allow the identification of major lineages with large frequency differences between Europeans, sub-Saharan Africans, and Native Americans (34, 35). In particular, DYS199/M3 and DYS271/M2 define haplogroups Q-M3 and E-M2 (36), which are restricted to Native Americans and sub-Saharan Africans, respectively. The microsatellite markers typed (as reported by ref. 37) were DYS19, DYS388, DYS390, DYS391, DYS392, and DYS393. The four major Native American mtDNA founder haplogroups (A–D) (38) were identified by typing three polymorphic restriction sites and the 9-bp COII/tRNALys intergenic deletion. The enzymes used and the location of the restriction sites examined were HaeIII (663 bp), HincII (3,259 bp), and AluI (5,176 bp). Experimental conditions for mtDNA typing were as described in ref. 39.
Eight autosomal (AT3, DRD2 TaqID, PV92, LPL, ICAM1, SB19.3, ApoA1, and FY-null) and five X-linked (MID1326, MID1327, MID1705, MID193, and MID220) markers were typed as described in refs. 40–42 and the Marshfield Clinic Research Foundation (Marshfield, WI). Markers were selected based on their informativity for admixture estimation as assessed by large allele frequency differentials between Europeans, Africans, and Amerindians (see Table 6, which is published as supporting information on the PNAS web site). We also typed (as reported) 35 SNPs around exon 44 of the DMD gene on chromosome X in Antioquian males (43). These SNPs allow the definition of 10 major haplogroups in world populations, as detailed in ref. 43.
Data Analysis.
Allele and haplotype frequency, unbiased gene diversity (H), FST (and significance of FST) were obtained from genotype data files using the program arlequin (44). The mean mutation rate at the Y microsatellite loci examined was calculated based on the average squared distance (45) using the approach described in ref. 46. Admixture proportions were estimated by the least-squares approximation of Long (47) as implemented in the admix program kindly provided by J. C. Long (University of Michigan, Ann Arbor).
Male migration rates after foundation were calculated from the autosomal and X chromosome admixture estimates assuming that the founding admixture involved solely Spanish men and Amerind women and that gene flow occurred only from Europe into America. The proportion of European ancestry can be considered equivalent to the frequency in the admixed population of an allele (e.g., A) fixed in Europeans and absent in Native Americans.
Based on traditional gene flow theory (48),
where m is the migration rate per generation, q1 and qt+1 are the frequency of allele A in the admixed population at foundation and in generation t + 1, respectively, and qm is the frequency of A in Europeans (here qm = 1 and is constant over generations).
Substituting qm = 1 in Eq. 1,
and
where c = (1 − qt+1)/(1 − q1).
Because the founding population is the offspring of matings between Amerindian women and European men, q1 is 0.5 for an autosomal locus and 0.333 for an X-linked locus (because twice as many X chromosomes come from the Amerindian mothers as the European fathers).
For an autosomal locus
where mm and mf are the rates of gene flow per generation for men and women, respectively. Because for the model examined here mf = 0, mm = 2m. Analogously, the contribution to gene flow of males and females for an X-linked locus is
and if mf = 0, then mm = 3m.
Supplementary Material
Acknowledgments
We thank Cátira Bortolini and Francisco Salzano for suggestions on the manuscript and Timothée Flutre for comments on the admixture model and help producing Fig. 2. This work was partly funded by National Institutes of Health Grant R01NS043538, The Wellcome Trust Grant 086052, and a National Alliance for Research on Schizophrenia and Depression (NARSAD) Young Investigator Award (to A.R.L.) D.L. is supported by Canadian Institutes of Health Research Grant MOP-67150.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Burkholder M. A., Johnson L. L. Colonial Latin America. Oxford: Oxford Univ. Press; 2003. [Google Scholar]
- 2.Curtin P. D. The Atlantic Slave Trade: A Census. Madison: Univ. Wisconsin Press; 1969. [Google Scholar]
- 3.Thomas H. The Slave Trade. New York: Simon and Schuster; 1997. [Google Scholar]
- 4.Sanchez-Albornoz N. The Population of Latin America: A History. Berkeley: Univ. California Press; 1974. [Google Scholar]
- 5.Salzano F. M., Bortolini M. C. The Evolution and Genetics of Latin American Populations. Cambridge, U.K.: Cambridge Univ. Press; 2002. [Google Scholar]
- 6.Morner M. Race Mixture in the History of Latin America. Boston: Little Brown; 1967. [Google Scholar]
- 7.Safford F., Palacios M. Colombia: Fragmented Land, Divided Society (Latin American Histories) Oxford: Oxford Univ. Press; 2001. [Google Scholar]
- 8.Alvarez V. M. In: Historia de Medelin. Melo J. O., editor. Medellin, Colombia: Suramericana; 1996. [Google Scholar]
- 9.Parsons J. J. Antioqueno Colonization in Western Colombia. Berkeley: Univ. California Press; 1968. [Google Scholar]
- 10.Carvajal-Carmona L. G., Soto I. D., Pineda N., Ortiz-Barrientos D., Duque C., Ospina-Duque J., McCarthy M., Montoya P., Alvarez V. M., Bedoya G., et al. Am. J. Hum. Genet. 2000;67:1287–1295. doi: 10.1016/s0002-9297(07)62956-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carvajal-Carmona L. G., Ophoff R., Service S., Hartiala J., Molina J., Leon P., Ospina J., Bedoya G., Freimer N., Ruiz-Linares A. Hum. Genet. 2003;112:534–541. doi: 10.1007/s00439-002-0899-8. [DOI] [PubMed] [Google Scholar]
- 12.Green L. D., Derr J. N., Knight A. Am. J. Hum. Genet. 2000;66:989–998. doi: 10.1086/302801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alves-Silva J., da Silva S. M., Guimaraes P. E., Ferreira A. C., Bandelt H. J., Pena S. D., Prado V. F. Am. J. Hum. Genet. 2000;67:444–461. doi: 10.1086/303004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carvalho-Silva D. R., Santos F. R., Rocha J., Pena S. D. Am. J. Hum. Genet. 2001;68:281–286. doi: 10.1086/316931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arango-Mejia G. Genealogias de Antioquia y Caldas. Medellin, Colombia: Litoarte; 1993. [Google Scholar]
- 16.Gutierrez de Pineda V. Familia y Cultura en Colombia. Medellin, Colombia: Univ. Antioquia; 1993. [Google Scholar]
- 17.Sandoval C., de la Hoz A., Yunis E. Rev. Fac. Med. Univ. Nac. Bogota. 1993;41:3–14. [Google Scholar]
- 18.Bourchard G., De Braekeleer M. Histoire d'un Génôme: Population et Génétique dans l'Est du Québec. Quebec, Canada: Presses Univ. Québec; 1991. [Google Scholar]
- 19.Kayser M., Roewer L., Hedman M., Henke L., Henke J., Brauer S., Kruger C., Krawczak M., Nagy M., Dobosz T., et al. Am. J. Hum. Genet. 2000;66:1580–1588. doi: 10.1086/302905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sykes B., Irven C. Am. J. Hum. Genet. 2000;66:1417–1419. doi: 10.1086/302850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macintyre S., Sooman A. Lancet. 1991;338:869–871. doi: 10.1016/0140-6736(91)91513-t. [DOI] [PubMed] [Google Scholar]
- 22.Dupuy B. M., Stenersen M., Egeland T., Olaisen B. Hum. Mutat. 2004;23:117–124. doi: 10.1002/humu.10294. [DOI] [PubMed] [Google Scholar]
- 23.Zhivotovsky L. A., Underhill P. A., Cinnioglu C., Kayser M., Morar B., Kivisild T., Scozzari R., Cruciani F., Destro-Bisol G., Spedini G., et al. Am. J. Hum. Genet. 2004;74:50–61. doi: 10.1086/380911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Austerlitz F., Heyer E. Proc. Natl. Acad. Sci. USA. 1998;95:15140–15144. doi: 10.1073/pnas.95.25.15140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heyer E., Sibert A., Austerlitz F. Trends Genet. 2005;21:234–239. doi: 10.1016/j.tig.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 26.Oakey R., Tyler-Smith C. Genomics. 1990;7:325–330. doi: 10.1016/0888-7543(90)90165-q. [DOI] [PubMed] [Google Scholar]
- 27.Xue Y., Zerjal T., Bao W., Zhu S., Lim S. H., Shu Q., Xu J., Du R., Fu S., Li P., et al. Am. J. Hum. Genet. 2005;77:1112–1116. doi: 10.1086/498583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith M. W., O'Brien S. J. Nat. Rev. Genet. 2005;6:623–632. doi: 10.1038/nrg1657. [DOI] [PubMed] [Google Scholar]
- 29.Pfaff C. L., Parra E. J., Bonilla C., Hiester K., McKeigue P. M., Kamboh M. I., Hutchinson R. G., Ferrell R. E., Boerwinkle E., Shriver M. D. Am. J. Hum. Genet. 2001;68:198–207. doi: 10.1086/316935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hammer M. F., Horai S. Am. J. Hum. Genet. 1995;56:951–962. [PMC free article] [PubMed] [Google Scholar]
- 31.Seielstad M. T., Hebert J. M., Lin A. A., Underhill P. A., Ibrahim M., Vollrath D., Cavalli-Sforza L. L. Hum. Mol. Genet. 1994;3:2159–2161. doi: 10.1093/hmg/3.12.2159. [DOI] [PubMed] [Google Scholar]
- 32.Hurles M. E., Veitia R., Arroyo E., Armenteros M., Bertranpetit J., Perez-Lezaun A., Bosch E., Shlumukova M., Cambon-Thomsen A., McElreavey K., et al. Am. J. Hum. Genet. 1999;65:1437–1448. doi: 10.1086/302617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Underhill P. A., Jin L., Zemans R., Oefner P. J., Cavalli-Sforza L. L. Proc. Natl. Acad. Sci. USA. 1996;93:196–200. doi: 10.1073/pnas.93.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Underhill P. A., Shen P., Lin A. A., Jin L., Passarino G., Yang W. H., Kauffman E., Bonne-Tamir B., Bertranpetit J., Francalacci P., et al. Nat. Genet. 2000;26:358–361. doi: 10.1038/81685. [DOI] [PubMed] [Google Scholar]
- 35.Underhill P. A., Passarino G., Lin A. A., Shen P., Mirazon L. M., Foley R. A., Oefner P. J., Cavalli-Sforza L. L. Ann. Hum. Genet. 2001;65:43–62. doi: 10.1046/j.1469-1809.2001.6510043.x. [DOI] [PubMed] [Google Scholar]
- 36.Y Chromosome Consortium. Genome Res. 2002;12:339–348. doi: 10.1101/gr.217602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas M. G., Bradman N., Flinn H. M. Hum. Genet. 1999;105:577–581. doi: 10.1007/s004399900181. [DOI] [PubMed] [Google Scholar]
- 38.Torroni A., Schurr T. G., Cabell M. F., Brown M. D., Neel J. V., Larsen M., Smith D. G., Vullo C. M., Wallace D. C. Am. J. Hum. Genet. 1993;53:563–590. [PMC free article] [PubMed] [Google Scholar]
- 39.Bailliet G., Rothhammer F., Carnese F. R., Bravi C. M., Bianchi N. O. Am. J. Hum. Genet. 1994;54:27–33. [PMC free article] [PubMed] [Google Scholar]
- 40.Parra E. J., Marcini A., Akey J., Martinson J., Batzer M. A., Cooper R., Forrester T., Allison D. B., Deka R., Ferrell R. E., et al. Am. J. Hum. Genet. 1998;63:1839–1851. doi: 10.1086/302148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoggart C. J., Parra E. J., Shriver M. D., Bonilla C., Kittles R. A., Clayton D. G., McKeigue P. M. Am. J. Hum. Genet. 2003;72:1492–1504. doi: 10.1086/375613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Collins-Schramm H. E., Phillips C. M., Operario D. J., Lee J. S., Weber J. L., Hanson R. L., Knowler W. C., Cooper R., Li H., Seldin M. F. Am. J. Hum. Genet. 2002;70:737–750. doi: 10.1086/339368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zietkiewicz E., Yotova V., Gehl D., Wambach T., Arrieta I., Batzer M., Cole D. E., Hechtman P., Kaplan F., Modiano D., et al. Am. J. Hum. Genet. 2003;73:994–1015. doi: 10.1086/378777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneider S., Roessli D., Excoffier L. arlequin, A Software for Population Genetic Data Analysis. Genetics and Biometry Lab., University of Geneva; 2000. Version 2000. [Google Scholar]
- 45.Goldstein D. B., Ruiz Linares A., Cavalli-Sforza L. L., Feldman M. W. Genetics. 1995;139:463–471. doi: 10.1093/genetics/139.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bortolini M. C., Salzano F. M., Thomas M. G., Stuart S., Nasanen S. P., Bau C. H., Hutz M. H., Layrisse Z., Petzl-Erler M. L., Tsuneto L. T., et al. Am. J. Hum. Genet. 2003;73:524–539. doi: 10.1086/377588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Long J. C. Genetics. 1991;127:417–428. doi: 10.1093/genetics/127.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hedrick P. W. Genetics of Populations. Boston: Jones and Bartlett; 2005. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.