Abstract
Collisions between DNA replication and transcription significantly affect genome organization, regulation, and stability. Previous studies have described collisions between replication forks and elongating RNA polymerases. Although replication collisions with the transcription-initiation or -termination complexes are potentially even more important because most genes are not actively transcribed during DNA replication, their existence and mechanisms remained unproven. To address this matter, we have designed a bacterial promoter that binds RNA polymerase and maintains it in the initiating mode by precluding the transition into the elongation mode. By using electrophoretic analysis of replication intermediates, we have found that this steadfast transcription-initiation complex inhibits replication fork progression in an orientation-dependent manner during head-on collisions. Transcription terminators also appeared to attenuate DNA replication, but in the opposite, codirectional orientation. Thus, transcription regulatory signals may serve as “punctuation marks” for DNA replication in vivo.
Keywords: collisions, promoter, terminator
Impairment of DNA replication is believed to be a major factor in genomic instability (1–13). Because transcription and replication share the same template, occasional collisions between the two machineries are inevitable and can interfere with replication fork progression. Collisions between the elongating RNA polymerase and the replication fork have been well documented in vitro (14–17) and in vivo in both Escherichia coli (18–20) and Saccharomyces cerevisiae (6, 21, 22). The consensus from those studies was that head-on collisions with elongating RNA polymerase are much more detrimental for replication fork progression than codirectional collisions. Although it was suggested that replication stalling during the head-on collisions with transcription was caused by topological stress in the DNA separating the two machineries (18, 19, 22, 23), we have recently shown that it is caused by their direct, physical interaction (20). These results, combined with the data on preferred codirectional alignment of transcription units with the direction of replication in prokaryotes (23–27), have led to the suggestion that the main disadvantage of the head-on collisions could be their inhibitory effect on DNA replication.
All of the experimental studies cited in the preceding paragraph evaluated the effects of elongating RNA polymerase on the progression of the replication fork. Is there an interplay between the replication machinery and the transcription-initiation complex? To the best of our knowledge, there have been few studies on this matter. One intriguing example was the detection of a polar replication fork pause site at the tRNA locus of S. cerevisiae, which depended on the functionality of both the promoter and the RNA polymerase III (pol III) (22). It was believed that the replication fork was attenuated during the encounter with the elongating RNA polymerase (22). A later study, however, suggested that the pol III-initiation complex was responsible for the replication slowing in this system (6). In our recent study of transcription–replication collisions in E. coli, the synthetic trc promoter was shown to stall the replication fork in an orientation-dependent manner even in the repressed state (20). Unfortunately, we could not distinguish whether this replication stalling was caused by the lactose repressor, RNA polymerase, or both. Thus, a thorough study of the interplay between the replication fork and the transcription-initiation complex seemed to be warranted.
Initiation of transcription in E. coli is a multistep process (for review, see ref. 28). First, RNA polymerase holoenzyme binds to the promoter, forming the closed complex. The second step is the isomerization of the closed complex into the open complex, accompanied by DNA unwinding. Third, the ternary complex is formed during binding of the first NTP. The fourth step is the synthesis of the first ≈10 RNA bases without the movement of RNA polymerase along DNA, resulting in the formation of the so-called stressed complex. This short RNA may be released from the ternary complex, leading to abortive initiation, or it may become the 5′ end of the RNA transcript. Finally, promoter clearance is accompanied by the dissociation of the σ factor and engagement of the core RNA polymerase in the stable elongation complex.
Although the principles of the initial promoter recognition are well understood, the sequence requirements for the further steps in the initiation process are much less clear. Their importance, however, is illustrated by the results of the computational analysis of bacterial genomic DNA, which reveals many false-positive promoters, whereas the true promoters may not even have the highest computational scores (ref. 29 and references therein).
To study collisions between the replication machinery and transcription-initiation complex carefully, we needed a system in which a transcription-initiation complex without additional proteins would stably exist inside the cell. Such a system could be achieved by shifting the equilibrium from the promoter clearance toward abortive initiation. In this case, the majority of short RNA products would be released while the RNA polymerase would remain in its initiating mode. To ensure efficient binding of the RNA polymerase to the promoter, we chose a strong promoter, the bacteriophage T7 early promoter A1 (30). To shift the equilibrium toward abortive initiation, we decided to modify the initial transcribed sequence (ITS) of the promoter, the importance of which for the strength of the promoters and the transition to the productive elongation mode was demonstrated in refs. 31 and 32. Specifically, the original promoter ITS, positioned between +1 and +20, was converted into a 90% AT-rich element, carrying multiple Ts on the nontemplate strand. Our idea was based on the fact that the weak RNA·DNA hybrid (e.g., A·U) causes backtracking (33). In early elongation, RNA is short and cannot form the secondary structure behind RNA polymerase to prevent backtracking. Therefore, hybrid stability is the primary determinant of the transition to productive elongation (34). Recent work on pol II supports this idea (35).
We showed that the amount of RNA transcribed from this mutant promoter drastically decreased both in vitro and in vivo, whereas the RNA polymerase binding to it and the open-complex formation remained effective. This transcription-initiation complex turned out to be inhibitory to replication fork progression in the head-on, but not in the codirectional, orientation. Unexpectedly, we also observed replication stalling at codirectionally positioned transcription terminators. We conclude that transcription-initiation and -termination elements could be “punctuation marks” for DNA replication because they attenuate replication fork progression in an orientation-dependent manner.
Results
AT-Rich ITS Prevents RNA Polymerase Transition to the Productive Elongation Mode Without Affecting the Open-Complex Formation.
We compared the bacteriophage T7 early promoter A1 (promoter A hereafter) with its mutant derivative, where the sequence downstream from the transcription start site was changed such that the resultant +1 to +20 region became 90% AT-rich (promoter B hereafter) (Fig. 1A). Fig. 1B shows that this change drastically increased “promoter futility” in vitro: the amount of the 25-nt RNA transcript from promoter B was negligible compared with original promoter A. The fact that the product on template B has a slightly greater mobility than that on template A could be the result of the differences in their RNA sequences, i.e., high pyrimidine content of the mutant transcript, secondary structures, or accidental RNA polymerase slippage on the mutant template. Our approach does not detect RNA chains that go into reiterative U addition. Thus, we cannot rule out that a fraction of RNA polymerase molecules carry out reiterative synthesis at the mutant promoter. What is important for our purposes, however, is that RNA polymerase does not switch into the productive elongation mode there.
Fig. 1.
AT-rich ITS abolishes promoter activity. (A) Nucleotide sequences of the original bacteriophage T7 early promoter A1 and its mutant derivative, in which the region from +1 to +20 became 90% AT-rich promoter B. The −35, −10, and +1 positions are underlined; the AT-rich stretch is highlighted. (B) In vitro transcription assay on the templates of the promoters A and B. The position of the anticipated 25-nt-long product is marked by an arrow. (C) Northern blot analysis of the transcriptional activity of the promoters A and B in vivo. Only informative portions of the corresponding autoradiographs are shown in Left. (Right) Quantification of the results. HO, head-on orientation of the promoter to the direction of replication in the plasmids; CD, codirectional orientation. Maps of the plasmids pA-HO, pB-HO, pA-CD, and pB-CD are shown in Figs. 3A and 4A. TR, promoter-specific transcript; RNA I, plasmid-derived RNA used for the normalization. (D) Chemical probing of the open-complex formation in situ. Two different primers were used to detect modifications of the template (right half) and nontemplate (left half) strands in promoters A and B (pA-HO and pB-HO plasmids, respectively). Asterisks, primer extension reactions on the in situ chloroacetaldehyde-modified templates; G, A, T, and C, Sanger sequencing reactions on control templates with the same primers. The −10 and +1 positions of the promoters are marked.
We have cloned promoters A and B upstream from transcription terminators T1T2 from the rrnB operon, creating transcriptional cassettes whose defined 400-nt RNA transcripts would serve as readouts of their activities in vivo. The resulting cassettes were cloned into the pTrc99-derived vector (20) in two orientations relative to the direction of replication, head-on (HO) or codirectionally (CD). The Northern blot hybridization analysis shown in Fig. 1C demonstrates that activity of the mutant promoter was significantly (5- to 10-fold) reduced compared with the wild-type promoter in both orientations relative to the direction of replication. Note, however, that for the wild-type promoter, the amount of transcript was twice higher in the head-on than in the codirectional orientation.
To compare the relative efficiencies of RNA polymerase binding and open-complex formation for the two promoters, we performed their chemical probing in situ. We used chloroacetaldehyde, which modifies the base-pairing positions of adenine, cytosine, and, to a lesser extent, guanine. Consequently, this drug is an excellent probe for single-stranded DNA, which efficiently detects promoter unwinding during open-complex formation (36). Cells with various plasmids were treated with chloroacetaldehyde followed by plasmid DNA isolation and detection of modified DNA bases by primer extension. Fig. 1D demonstrates that the two DNA strands in the area of −10 to +2 were modified in both control and mutant promoters. Quantitative analysis of these results showed that the mutant promoter was modified 2.5-fold stronger on the nontemplate strand and 1.5-fold stronger on the template strand compared with the wild-type promoter. Consequently, the drastically diminished activity of promoter B is not the result of inefficient RNA polymerase binding and open-complex formation. We believe, therefore, that transcriptional impairment, caused by the AT-rich ITS, is the result of the inability of the RNA polymerase to undergo the transition to the productive elongation mode. Mutant promoter B was thus chosen to study the interplay between the steady-state transcription-initiation complex and DNA replication in vivo.
Replication Fork Progression Is Inhibited During Head-On Collision with the Transcription-Initiation Complex in Vivo.
We evaluated replication fork progression through a transcription cassette by using the two-dimensional electrophoretic analysis of replication intermediates (37). Briefly, replication intermediates of ColE1-derived, unidirectionally replicating plasmids from E. coli cells were cleaved immediately upstream from the origin, and the resultant bubble-shaped intermediates were resolved in a two-dimensional gel, forming the so-called bubble arc (38). This arc is smooth if the replication fork progressed with the same speed throughout the plasmid; if replication was slowed down at a particular position, a distinct bulge appears on the arc because of the accumulation of replication intermediates of the defined size and shape. We have previously applied this approach successfully to detecting transcription–replication collision in E. coli cells (20).
The general map of our plasmids, in which transcription cassettes face the replication origin head-on, is presented in Fig. 2A alongside the schematic drawing of the anticipated bubble arc. The experimental data in Fig. 2B (Left) show that the replication is slowed down throughout the whole area transcribed from the wild-type promoter, in accord with our previous results (20). [Note that the Y arc underneath the bubble arc comes from replicating plasmid dimers (39) and also shows extensive transcription–replication collisions, albeit with a worse resolution. The strength of this Y arc depends on the proportion of plasmid dimerization and varies substantially among various constructs and experiments.] For the mutant promoter B, the results are strikingly different (Fig. 2B Right); replication stalling is mostly evident at a defined spot in the whole plasmid, which roughly corresponds to the position of the promoter. Fig. 2C shows the quantitative analysis of the replication data, obtained from a different experiment. Clearly, most of the replication stalling occurs at the promoter in the pB-HO plasmid, whereas the stalling is evenly spread throughout the transcribed region in promoter A.
Fig. 2.
Replication stalling by the head-on oriented transcription-initiation complex. (A Left) Map of the plasmids pA-HO and pB-HO. Transcription cassettes with promoters A and B, respectively, are oriented head-on to the direction of replication; nucleotide position 1 in the plasmids corresponds to the replication start site; positions of the promoters and T1T2 terminators are shown. (A Right) Schematic representation of the two-dimensional gel electrophoresis of replication intermediates. The bubble arc consists of replication intermediates, starting from the small bubble at the origin of replication (at the bottom) and going up to the biggest, fully replicated bubble (at the top). Replication stalling is detected as a thickening of a particular segment of the arc. P, promoter; T, terminator; arrow, direction of transcription. (B) Two-dimensional gel electrophoresis of replication intermediates. Replication stalling is evident as a long segment that corresponds to the whole transcribed area in the pA-HO plasmid (Left) or as a bulge that corresponds to the promoter in the pB-HO plasmid (Right). (C) Quantitative analysis of the replication arcs around the transcribed units in plasmids pA-HO and pB-HO. The experiment shown here is independent of the one shown in Fig. 4B. (D) Scheme for mapping replication pause sites by in-gel digestion of replication intermediates (for details, see Results). (Left) The vertical lines show the positions of the restriction sites immediately upstream (PstI) and downstream (EcoRI) from the promoter (P). Replication intermediates are shown as bubbles, and the bold bubble corresponds to stalled replication intermediates. (Center and Right) During EcoRI digestion (Center), stalled intermediates become Y-shaped and move to the line, whereas after PstI digestion (Right), they remain bubble-shaped and stay on the arc. (E) Experimental mapping of stalled replication intermediates by in-gel digestion with EcoRI (Left) or PstI (Right). Stalled replication intermediates are shown by arrows. Note the underreplicated stalled intermediates migrating between the arc and the line (for details, see Results).
To confirm that replication stalling indeed occurs at the mutant promoter, we have undertaken fine mapping of the pause site. To this end, we have used a modification of the two-dimensional gel-electrophoretic analysis of the replication intermediates, where an extra in-gel restriction digestion was performed during separation in the first dimension. The schematic representation of this approach is illustrated in Fig. 2D. During in-gel digestion, a fraction of replication intermediates converts into identical Y-shaped molecules that migrate as a horizontal line in the second dimension. When digestion occurs immediately downstream from the promoter (EcoRI), replication intermediates stalled at the promoter must shift from the bubble arc to the horizontal line. In contrast, when the digestion occurs immediately upstream from the promoter (PstI), the same stalled intermediates must remain on the bubble arc (Fig. 2E). The majority of stalled intermediates remain on the bubble arc during the PstI digestion. After EcoRI digestion, one fraction of stalled intermediates moves to the line, and another fraction migrates between the bubble arc and the line. The latter intermediates apparently correspond to DNA molecules in which the lagging strand around the EcoRI site was underreplicated, resulting in incomplete digestion and “butterfly-like” structures (11, 40). The third fraction of the stalled intermediates remained on the arc. Those intermediates reflect the residual activity of the mutant promoter evident from Fig. 1C. Notwithstanding the latter fraction, the fact that a noticeable amount of stalled intermediates moved away from the bubble arc during EcoRI digestion is sufficient to conclude that replication stalling occurred at the promoter during collision with the transcription-initiation complex.
We conclude, therefore, that the replication pause site is located between the EcoRI and PstI sites, i.e., at the mutant promoter. These data show that a replication fork can stall when encountering a transcription-initiation complex head-on in E. coli cells.
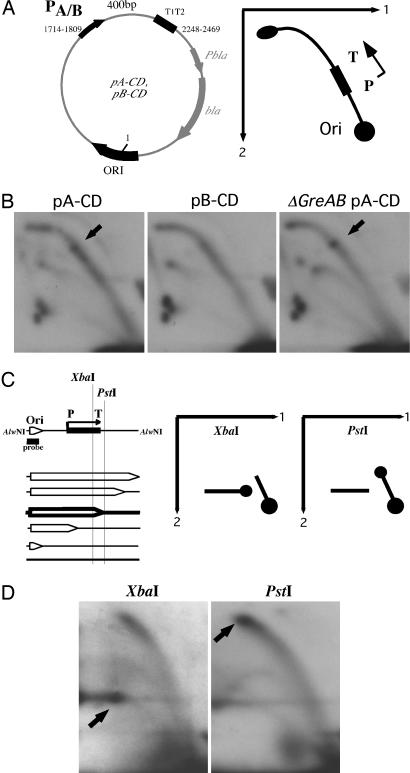
Codirectionally Positioned Transcription Terminators Inhibit the Replication Fork Progression.
To study the effect of the transcription-initiation complex on replication fork progression in the codirectional orientation, we have inverted both the wild-type and mutant transcription cassettes, generating the plasmids shown in Fig. 3A. On an anticipated bubble arc, the transcription cassette would face up, with the promoter positioned proximally and terminators positioned distally to the replication origin.
Fig. 3.
Replication stalling by the codirectionally oriented transcription terminators. (A Left) Map of plasmids pA-CD and pB-CD. Transcription cassettes with promoters A or B, respectively, are oriented codirectionally with the direction of replication; nucleotide position 1 corresponds to the replication start site; positions of the promoters and T1T2 terminators are indicated. (A Right) Schematic representation of the two-dimensional gel electrophoresis of replication intermediates. Unlike Fig. 2, a transcription cassette faces up, i.e., away from the origin. (B) Two-dimensional gel electrophoresis of replication intermediates. Replication stalling is evident in the pA-CD plasmid at the position that corresponds to the position of the transcription terminators (arrow). (C). A scheme of mapping stalled intermediates by in-gel digestion is shown. Replication intermediates were separated in the first dimension followed by digestion with the XbaI and PstI enzymes, flanking the T1T2 terminators (T). (D) Experimental mapping of stalled intermediates. During XbaI digestion (Left), stalled intermediates become Y-shaped and move to the line, whereas during PstI digestion (Right), they remain bubble-shaped and stay on the arc.
Fig. 3B Left and Center shows that, in contrast with the head-on orientation, there is no evident replication stalling by either of the codirectionally oriented promoters A or B. One can see a slight thickening of the replication arc in the whole area transcribed from the wild-type promoter, which reflects a modest replication slow-down during collisions with the codirectionally moving RNA polymerase (17). Unexpectedly, however, there is a more prominent replication stall at the transcription terminators T1T2 in the codirectionally located wild-type cassette. Could this stalling be the result of occasional RNA polymerase backtracking at the terminators? To address this possibility, we looked at the replication stalling at the transcription terminators in the greA,greB knockout strain (which is otherwise isogenic to the wild-type strain used in this study). The GreA and GreB proteins (41) are known to assist RNA polymerase in escaping from backtracked conformation by activating its endonucleolytic activity, which leads to RNA cleavage, formation of the new 3′ end in the active center, and resumption of transcription elongation (42). The replication stalling at terminators was indeed increased in the greA,greB mutant (Fig. 3B Right).
To confirm additionally that the latter replication pausing was caused by the transcription terminators, we have mapped the pause site by following the same logic described in the previous section (Fig. 3C). The experimental data in Fig. 3D show that stalled intermediates moved to the line during in-gel digestion with XbaI but remained on the bubble arc if treated with PstI. Consequently, the replication stall zone is located between these two restriction sites, coinciding with the transcription terminators.
Discussion
To look at the interplay between transcription-initiation complexes and DNA replication, we have constructed a promoter that supports efficient formation of closed and open complexes with RNA polymerase but prevents promoter clearance. This end was achieved by converting the ITS (+1 to +20 region) of the very strong A1 promoter of the bacteriophage T7 into the 90% AT-rich sequence without altering any other position. This change led to a dramatic (up to 10-fold) decrease in transcriptional activity both in vitro (Fig. 1B) and in vivo (Fig. 1C), without decreasing the efficiency of open-complex formation (Fig. 1D). Interestingly, the activity of the mutant promoter was not decreased further in the greA,greB knockout E. coli strain, and the addition of the GreA and GreB proteins did not improve the efficiency of this promoter in vitro (data not shown). Thus, GreAB proteins fail to facilitate promoter clearance in our case, indicating that either the abortive initiation is responsible for the impairment of our mutant promoter or that the RNA polymerase backtracking is so overwhelming that Gre factors do not make any difference.
The mutated promoter seemed, therefore, ideal for studying collisions between transcription-initiation complexes and the replication fork in vivo. For the replication studies, we have chosen a bacterial plasmid (43) that contains the entire pBR322 replication origin, including pasL and pasH sites required for the efficient switching from pol I- to pol III-mediated DNA replication (44). Our transcription cassettes in various orientations (Figs. 2A and 3A) were positioned ≈1,700 bp downstream from the replication start site and ≈1,300 bp beyond the pasH site, i.e., in the area solidly replicated by the DNA polymerase III holoenzyme. Thus, the data obtained in this plasmid system should be perfectly applicable to E. coli chromosomal replication.
We observed potent inhibition of DNA replication during head-on collision of the fork with the mutant promoter (Fig. 2 A–C). Mapping of this replication pause site by in-gel digestion of replication intermediates immediately upstream and downstream from the promoter confirmed that the replication fork was indeed halted at the promoter (Fig. 2 D and E).
When wild-type promoter A faced replication head-on, we observed a severe replication stalling throughout the whole transcribed area (Fig. 2B Left), in accord with our previous study (20). That study was carried out in the E. coli DH5α strain (20), which carries mutations in some important components of DNA metabolism. Because our current studies performed in the wild-type, MG1655 Rph+ strain gave identical results, head-on transcription–replication collisions could significantly influence nucleic acid metabolism in wild-type E. coli.
Unlike what we saw in our previous study (20), however, we saw a trace of replication stalling during codirectional collisions with elongating RNA (Fig. 3B Left). Although it was diminutive compared with that for the head-on collisions (compare Figs. 2B Left and 3B Left), this study demonstrates replication stalling by codirectional collisions with transcription in vivo. Such collisions between phage replisomes and bacterial RNA polymerases were, so far, detected only in vitro (14, 17). Clearly, however, the head-on collisions with either elongating or initiating RNA polymerase have much more dire consequences for DNA replication. It is tempting to speculate, therefore, that the front edge of the RNA polymerase could be a potent contrahelicase (45).
The amount of full-length transcript generated at the wild-type promoter is approximately twice as high in its head-on orientation as in its codirectional orientation (Fig. 1C). This difference was not the result of a variation in copy number for different plasmids because promoter-specific transcripts were normalized to the amount of RNA I in all cases. RNA I is the origin-specific negative regulator of ColE1-like plasmid replication (for review, see ref. 46), and its abundance directly reflects the plasmid copy number. We believe, therefore, that the difference in the mRNA levels depending on the promoter orientation could be caused by different outcomes of transcription–replication collisions.
Most essential genes in bacteria are oriented codirectionally with replication. It was therefore suggested that the deleterious consequences of head-on transcription–replication collisions could be the result of the formation of truncated transcripts and, consequently, truncated proteins, serving as dominant–negative forms of essential proteins (27). Our transcriptional data are in disagreement with this hypothesis because we see more full-length transcripts produced from head-on, rather than codirectionally oriented, promoters.
While studying codirectional collisions, we unexpectedly observed replication stalling in the plasmid region that corresponded to the transcription terminators T1T2 (Fig. 3 A and B). Mapping of this replication pause site has indeed confirmed that it coincides with the terminators (Fig. 3 C and D). Why would the replication fork pause at this regulatory element? A possible explanation could be that a fraction of RNA polymerase molecules may not dissociate from the template at a terminator, but remain bound in some form of a trapped or backtracked complex (47). If this explanation is correct, one would expect a stronger terminator-caused replication stalling in the greA,greB mutant (Fig. 3B Right). This explanation is also supported by the footprinting, attributed to RNA polymerase, observed at some terminators (48). Further experiments are needed to address the existence and structure of such complexes both in vitro and in vivo.
Altogether, our data show the replication fork stalling during its head-on encounter with the transcription-initiation complex or its codirectional encounter with the transcription-terminator complex. Notably in both instances, the replication fork is stalled after passing the coding region. It is plausible, therefore, that transcription-initiation and -termination elements could serve as polar punctuation marks for DNA replication, i.e., attenuate the replication fork progression as it traverses the coding areas (Fig. 4). This attenuation could provide extra room for the repair or gene conversion machineries to clear the coding areas off the newly acquired mutations, thus helping to maintain the integrity of the coding regions of sparsely transcribed bacterial genes.
Fig. 4.
A model of transcription regulatory elements, serving as punctuation marks for DNA replication. P, promoter; T, terminator. For details, see Discussion.
Methods
Construction of Promoters.
Both templates were generated by PCR-directed mutagenesis from T7A1 promoter template (49) with DNA polymerase (Phusion; Fermentas, Hanover, MD) and DNA oligonucleotides (IDT, Coralville, IA): 5′-AAAACTGCAGTCCAGATCCCGAAAATTTATCAA-3′ (sense primer for both templates) 5′-CTGTTGAATTCGGTTGGCGGAAAGAATAAATTAAAAAGATGGCTGTAAGTATCCTATAGG-3′ (antisense primer for template B) 5′-CTGTTGAATTCGGTTGGATGTTCGCCGTGGCCCTCTCGAT-3′ (antisense primer for template A).
In Vitro Transcription Assay.
RNA polymerase (2 pmol) was mixed with 10 pmol of DNA in 20 μl of TB50 (10 mM Tris·HCl, pH 7.2/50 mM KCl/10 mM MgCl2) for 5 min at 37°C followed by the addition of the primer ApUpC (10 μM), GTP + ATP + CTP (100 μM each for template A), or ATP + UTP + CTP (100 μM each for template B) and 10 μl of TB50-equilibrated NeutrAvidin–agarose beads (Pierce) for 7 min. Next, the beads were washed three times with 1 ml of TB300 and twice with TB50. After washing, 1.5 μl of [α-32P]CTP (3,000 Ci/mmol; 1 Ci = 37 GBq) was added along with 25 μM UTP (for template A) or 25 μM GTP (for template B) for 5 min at room temperature to obtain a 25-mer elongation complex in each case, followed by washing three times with TB100. The reactions were quenched by the addition of an equal volume of stop solution (8 M urea/20 mM EDTA/1× TBE/0.25% bromophenol blue/0.25% xylene cyanol). The products were separated by electrophoresis in a 12% polyacrylamide gel containing 8 M urea.
Plasmids.
The pA-HO and pB-HO plasmids were obtained by cloning the promoters A and B, respectively, between the PstI and EcoRI sites of the pTrc-HO/T1T2-400 plasmid (20), replacing the original trc promoter. The pA-CD and pB-CD plasmids were made by inversion of the whole transcription cassette between the PstI and HindIII sites of the pA-HO and pB-HO plasmids, respectively.
Electrophoretic Analysis of Replication Intermediates.
Isolation of replication intermediates from bacteria, their separation by two-dimensional neutral/neutral gel electrophoresis, and mapping of the replication stop zones were performed as described in ref. 40. Quantification of the results was done with a Storm PhosphorImager and imagequant software (Amersham Biosciences).
Chemical Probing of DNA in Vivo.
Chemical probing of intracellular plasmid DNA was carried out as in ref. 50. The primer extension reactions, using AACTGCCGGAAATCGTCGTG (for nontemplate strand) and CAGGGTTATTGTCTCATGAG (for template strand) primers, were carried out with a Sequenase Version 2.0 DNA sequencing kit (USB Corp.) according to the manufacturer’s instructions, except that the extension mix was added to the final concentration of 45 mM for each dNTP. In parallel, untreated plasmid DNA was used for the Sanger sequencing with the same primers and the same kit. Identical amounts of chloroacetaldehyde-treated plasmid DNA were compared in all cases. The efficiency of modification was quantified with a Storm PhosphorImager and imagequant software.
RNA Analysis.
RNA was isolated from 1 ml of bacterial cultures grown in LB with 100 μg/ml ampicillin to OD600 = 0.5 according to the Carol Gross laboratory protocol for total RNA isolation from E. coli (http://bugarrays.stanford.edu/protocols), and 1/10th of the total RNA was analyzed by Northern blotting in ULTRAhyb oligonucleotide hybridization buffer (Ambion, Austin, TX). Each RNA sample was run on the gel twice and hybridized separately to both the A·B promoter-specific transcript probe (TR) CTCCAGAGCGATGAAAACGTTTCAGTTTGCTCATGGAAAACGG and the RNA I-specific probe ACCACCGCTACCAGCGGTGGTTTGTTTGCCGGATCAAGAG. The data were quantified with a Storm PhosphorImager and imagequant software. For every RNA sample, the value of the TR product was normalized to the amount of the RNA I product. RNA analysis was repeated three times for each plasmid, starting from the fresh bacterial culture every time.
Acknowledgments
We thank Vadim Nikiforov and Konstantin Severinov for helpful discussions and the anonymous reviewers for invaluable comments and editorial suggestions. This work was supported by National Institutes of Health Grants GM60987 (to S.M.M.), GM58750 (to E.N.), and GM72814 (to E.N.).
Abbreviations
- CD
codirectional orientation of the promoter to the direction of replication in the plasmid
- HO
head-on orientation
- ITS
initial transcribed sequence
- pA
promoter A in a plasmid
- pB
promoter B in a plasmid
- pol I, II, and III
RNA polymerase I, II, and III, respectively
- RNA I
origin-specific negative regulator of ColE1-like plasmid replication
- TR
A/B promoter-specific transcript probe.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Higgins N. P., Kato K., Strauss B. J. Mol. Biol. 1976;101:417–425. doi: 10.1016/0022-2836(76)90156-x. [DOI] [PubMed] [Google Scholar]
- 2.Cha R. S., Kleckner N. Science. 2002;297:602–606. doi: 10.1126/science.1071398. [DOI] [PubMed] [Google Scholar]
- 3.Prado F., Aguilera A. EMBO J. 2005;24:1267–1276. doi: 10.1038/sj.emboj.7600602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Myung K., Datta A., Kolodner R. D. Cell. 2001;104:397–408. doi: 10.1016/s0092-8674(01)00227-6. [DOI] [PubMed] [Google Scholar]
- 5.Lambert S., Watson A., Sheedy D. M., Martin B., Carr A. M. Cell. 2005;121:689–702. doi: 10.1016/j.cell.2005.03.022. [DOI] [PubMed] [Google Scholar]
- 6.Ivessa A. S., Lenzmeier B. A., Bessler J. B., Goudsouzian L. K., Schnakenberg S. L., Zakian V. A. Mol. Cell. 2003;12:1525–1536. doi: 10.1016/s1097-2765(03)00456-8. [DOI] [PubMed] [Google Scholar]
- 7.Michel B., Grompone G., Floráes M. J., Bidnenko V. Proc. Natl. Acad. Sci. USA. 2004;101:12783–12788. doi: 10.1073/pnas.0401586101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox M. M., Goodman M. F., Kreuzer K. N., Sherratt D. J., Sandler S. J., Marians K. J. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 9.Courcelle J., Hanawalt P. C. Annu. Rev. Genet. 2003;37:611–646. doi: 10.1146/annurev.genet.37.110801.142616. [DOI] [PubMed] [Google Scholar]
- 10.Cleary J. D., Nichol K., Wang Y. H., Pearson C. E. Nat. Genet. 2002;31:37–46. doi: 10.1038/ng870. [DOI] [PubMed] [Google Scholar]
- 11.Samadashwily G. M., Raca G., Mirkin S. M. Nat. Genet. 1997;17:298–304. doi: 10.1038/ng1197-298. [DOI] [PubMed] [Google Scholar]
- 12.Mirkin S. M., Smirnova E. V. Nat. Genet. 2002;31:5–6. doi: 10.1038/ng0502-5. [DOI] [PubMed] [Google Scholar]
- 13.Lahiri M., Gustafson T. L., Majors E. R., Freudenreich C. H. Mol. Cell. 2004;15:287–293. doi: 10.1016/j.molcel.2004.06.034. [DOI] [PubMed] [Google Scholar]
- 14.Liu B., Wong M. L., Tinker R. L., Geiduschek E. P., Alberts B. M. Nature. 1993;366:33–39. doi: 10.1038/366033a0. [DOI] [PubMed] [Google Scholar]
- 15.Liu B., Alberts B. M. Science. 1995;267:1131–1137. doi: 10.1126/science.7855590. [DOI] [PubMed] [Google Scholar]
- 16.Elias-Arnanz M., Salas M. EMBO J. 1999;18:5675–5682. doi: 10.1093/emboj/18.20.5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elias-Arnanz M., Salas M. EMBO J. 1997;16:5775–5783. doi: 10.1093/emboj/16.18.5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.French S. Science. 1992;258:1362–1365. doi: 10.1126/science.1455232. [DOI] [PubMed] [Google Scholar]
- 19.Olavarrieta L., Hernandez P., Krimer D. B., Schvartzman J. B. J. Mol. Biol. 2002;322:1–6. doi: 10.1016/s0022-2836(02)00740-4. [DOI] [PubMed] [Google Scholar]
- 20.Mirkin E. V., Mirkin S. M. Mol. Cell. Biol. 2005;25:888–895. doi: 10.1128/MCB.25.3.888-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takeuchi Y., Horiuchi T., Kobayashi T. Genes Dev. 2003;17:1497–1506. doi: 10.1101/gad.1085403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deshpande A. M., Newlon C. S. Science. 1996;272:1030–1033. doi: 10.1126/science.272.5264.1030. [DOI] [PubMed] [Google Scholar]
- 23.Brewer B. J. Cell. 1988;53:679–686. doi: 10.1016/0092-8674(88)90086-4. [DOI] [PubMed] [Google Scholar]
- 24.Blattner F. R., Plunkett G., III, Bloch C. A., Perna N. T., Burland V., Riley M., Collado-Vides J., Glasner J. D., Rode C. K., Mayhew G. F., et al. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 25.McLean M. J., Wolfe K. H., Devine K. M. J. Mol. Evol. 1998;47:691–696. doi: 10.1007/pl00006428. [DOI] [PubMed] [Google Scholar]
- 26.Nomura M., Morgan E. A. Annu. Rev. Genet. 1977;11:297–347. doi: 10.1146/annurev.ge.11.120177.001501. [DOI] [PubMed] [Google Scholar]
- 27.Rocha E. P., Danchin A. Nat. Genet. 2003;34:377–378. doi: 10.1038/ng1209. [DOI] [PubMed] [Google Scholar]
- 28.Wagner R. Transcription Regulation in Prokaryotes. New York: Oxford Univ. Press; 2000. pp. 58–83. [Google Scholar]
- 29.Huerta A. M., Collado-Vides J. J. Mol. Biol. 2003;333:261–278. doi: 10.1016/j.jmb.2003.07.017. [DOI] [PubMed] [Google Scholar]
- 30.Deuschle U., Kammerer W., Gentz R., Bujard H. EMBO J. 1986;5:2987–2994. doi: 10.1002/j.1460-2075.1986.tb04596.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kammerer W., Deuschle U., Gentz R., Bujard H. EMBO J. 1986;5:2995–3000. doi: 10.1002/j.1460-2075.1986.tb04597.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu L. M., Vo N. V., Kane C. M., Chamberlin M. J. Biochemistry. 2003;42:3777–3786. doi: 10.1021/bi026954e. [DOI] [PubMed] [Google Scholar]
- 33.Nudler E., Mustaev A., Lukhtanov E., Goldfarb A. Cell. 1997;89:33–41. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- 34.Greive S. J., von Hippel P. H. Nat. Rev. Mol. Cell Biol. 2005;6:221–232. doi: 10.1038/nrm1588. [DOI] [PubMed] [Google Scholar]
- 35.Weaver J. R., Kugel J. F., Goodrich J. A. J. Biol. Chem. 2005;280:39860–39869. doi: 10.1074/jbc.M509376200. [DOI] [PubMed] [Google Scholar]
- 36.Dayn A., Malkhosyan S., Mirkin S. M. Nucleic Acids Res. 1992;20:5991–5997. doi: 10.1093/nar/20.22.5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brewer B. J., Fangman W. L. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- 38.Martin-Parras L., Hernandez P., Martinez-Robles M., Schvartzman J. B. J. Mol. Biol. 1991;220:843–855. doi: 10.1016/0022-2836(91)90357-c. [DOI] [PubMed] [Google Scholar]
- 39.Friedman K. L., Brewer B. J. Methods Enzymol. 1995;262:613–627. doi: 10.1016/0076-6879(95)62048-6. [DOI] [PubMed] [Google Scholar]
- 40.Krasilnikova M. M., Samadashwily G. M., Krasilnikov A. S., Mirkin S. M. EMBO J. 1998;17:5095–5102. doi: 10.1093/emboj/17.17.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Borukhov S., Sagitov V., Goldfarb A. Cell. 1993;72:459–466. doi: 10.1016/0092-8674(93)90121-6. [DOI] [PubMed] [Google Scholar]
- 42.Hsu L. M., Vo N. V., Chamberlin M. J. Proc. Natl. Acad. Sci. USA. 1995;92:11588–11592. doi: 10.1073/pnas.92.25.11588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aman E., Ochs B., Abel K.-J. Gene. 1988;69:301–315. doi: 10.1016/0378-1119(88)90440-4. [DOI] [PubMed] [Google Scholar]
- 44.Marians K. J., Soeller W., Zipursky S. L. J. Biol. Chem. 1982;257:5656–5662. [PubMed] [Google Scholar]
- 45.Mohanty B. K., Bairwa N. K., Bastia D. Proc. Natl. Acad. Sci. USA. 2006;103:897–902. doi: 10.1073/pnas.0506540103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kornberg A., Baker T. DNA Replication. New York: Freeman; 1992. pp. 641–647. [Google Scholar]
- 47.Gusarov I., Nudler E. Mol. Cell. 1999;3:495–504. doi: 10.1016/s1097-2765(00)80477-3. [DOI] [PubMed] [Google Scholar]
- 48.Epshtein V., Toulmâe F., Rahmouni A. R., Borukhov S., Nudler E. EMBO J. 2003;22:4719–4727. doi: 10.1093/emboj/cdg452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nudler E., Avetissova E., Markovtsov V., Goldfarb A. Science. 1996;273:211–217. doi: 10.1126/science.273.5272.211. [DOI] [PubMed] [Google Scholar]
- 50.Krasilnikov A. S., Podtelezhnikov A., Vologodskii A., Mirkin S. M. J. Mol. Biol. 1999;292:1149–1160. doi: 10.1006/jmbi.1999.3117. [DOI] [PubMed] [Google Scholar]