Abstract
Voltage-gated ion channels control generation and propagation of action potentials in excitable cells. Significant progress has been made in understanding structure and function of the voltage-gated ion channels, highlighted by the high-resolution open-state structure of the voltage-gated potassium channel, Kv1.2. However, because the structure of the closed state is unknown, the gating mechanism remains controversial. We adapted the rosetta membrane method to model the structures of the Kv1.2 and KvAP channels using homology, de novo, and domain assembly methods and selected the most plausible models using a limited number of experimental constraints. Our model of Kv1.2 in the open state is very similar in overall topology to the x-ray structure of this channel. Modeling of KvAP in the open state suggests that orientation of the voltage-sensing domain relative to the pore-forming domain is considerably different from the orientation in the Kv1.2 open state and that the magnitude of the vertical movement of S4 is significantly greater. Structural modeling of closed state of Kv1.2 suggests gating movement that can be viewed as a sum of two previously suggested mechanisms: translation (2–4 Å) plus rotation (≈180°) of the S4 segment as proposed in the original “sliding helix” or “helical screw” models coupled with a rolling motion of the S1–S3 segments around S4, similar to recent “transporter” models of gating. We propose a unified mechanism of voltage-dependent gating for Kv1.2 and KvAP in which this major conformational change moves the gating charge across the electric field in an analogous way for both channels.
Keywords: membrane protein, rosetta method, voltage-gated ion channel
Voltage-gated potassium (Kv) channels are members of the voltage-gated ion channel superfamily (1, 2), which is important for initiation and propagation of action potentials in excitable cells. They are composed of four identical or homologous subunits, each containing six transmembrane segments: S1–S6. Segments S1–S4 form the voltage-sensing domain (VSD), and segments S5 and S6 connected by the P loop, which is involved in ion selectivity, comprise the pore-forming domain (PD). S4 has four gating-charge-carrying arginines (R1–R4) spaced at intervals of three amino acid residues, which are highly conserved and are thought to play a key role in coupling changes in membrane voltage to opening and closing of the pore (3–5). In the Kv channels ≈13 electronic charges cross the membrane electrical field per channel between the closed and open states (6–8).
High-resolution structures of the bacterial potassium channel KvAP and the mammalian potassium channel Kv1.2 recently have been solved (9–11). Although the KvAP structures showed the VSD in a nonnative orientation with respect to the membrane and the PD, the Kv1.2 structure captured the VSD in a conformation that is thought to represent the open state of the channel. In addition, the structure reveals that the VSD from one subunit interacts closely with the PD of the adjacent subunit in a clockwise direction when the channel structure is viewed from the extracellular side of the membrane (9). However, the closed-state structure of these channels remains unknown, and the mechanism of action of the voltage sensor in translocating gating charge is a subject of controversy. The original “sliding helix” or “helical screw” models of gating posited that S4 moves outward along a clockwise spiral path through the protein structure upon depolarization, making sequential interactions with negatively charged residues (12–14). This movement was detected directly by chemical-labeling studies (15–18). However, extensive studies of labeling of substituted cysteine residues in S4 revealed that only a narrow waist was protected from reaction, leading to the concept that the gating charges of S4 move through a narrow pathway between open internal and external vestibules (19, 20), which we term the gating pore. This movement of the gating charges of S4 through a narrow gating pore is supported by experiments in which mutations of the S4 arginines were shown to create an ion conductance pathway for protons (21, 22) or for cations (23, 24) through the modified gating pore, indicating the presence of a translocation pathway through the protein for S4. However, even with these refinements, S4 movement alone seems unlikely to translocate three charges per subunit because analysis of the motions of substituted fluorescent probes using several distinct strategies indicate that there is only a small (2–4 Å) outward translocation during gating (25–28) and that the gating-charge-carrying arginines must move through a narrowly focused membrane electrical field (22, 29). These findings led to the proposal that the VSD acts like a “transporter” in which accessibility of the S4 gating charges changes from internal to external side due to the relative motions of the S1–S4 segments (27). In contrast to all of these findings, a “paddle” model of gating suggested by the x-ray structure of KvAP proposed that the S4 moves 15–20 Å through the lipid bilayer during the gating process (30, 31). A major goal of our structural modeling work is to develop a gating model that reconciles these seemingly inconsistent views.
Structural models of the voltage-gated ion channels in the closed and open states have been proposed based on multiple modeling strategies (10, 27, 32–37), including a specific gating model based on a transporter mechanism (27). Laine et al. (32) proposed that the VSD of one subunit interacts with the PD of the neighboring subunit in the clockwise direction if viewed from the extracellular side of the membrane, based on a combination of chemical crosslinking and modeling, and this proposal has been confirmed by the Kv1.2 structure (9). Recently, considerable progress has been made in de novo high-resolution structure prediction of water-soluble proteins (38, 39), novel protein-fold design (40), and modeling of protein–protein interactions (41) using the rosetta method (42–44). In addition, the rosetta method was adapted to de novo modeling of multipass α-helical transmembrane proteins, and significant parts of these proteins were predicted with root-mean-square deviation (rmsd) <4.0 Å from the native structure (45).
We modified the rosetta membrane method (45) to combine homology and de novo modeling with a goal of modeling structures of Kv1.2 and KvAP in open and closed conformations, guided by limited experimental constraints derived from clear structure–function data. We assessed the validity of the models by evaluating their consistency with experimental data not used in their generation. We then examined the validated models to draw insights about the gating mechanism of Kv channels. Our results provide a well defined mechanism for S4 movement through a combination of the motions proposed in the previous sliding helix/helical screw and transporter models and potentially reconcile the apparently contradictory experimental results on bacterial KvAP channels vs. Kv1.2 and other eukaryotic ion channels.
Results and Discussion
Model of Kv1.2 in the Open State.
The x-ray structure of the Kv1.2 channel (9) has missing electron density in segments S1 and S3 and in the loops connecting segments S1, S2, S3, and S4. We used the rosetta membrane method to model these regions as described in Methods. Fig. 1A shows the transmembrane region of a single subunit of Kv1.2. Our model predicts that the R3 arginine in S4 (R300 in Kv1.2) interacts with E1 in S2 (E226 in Kv1.2) (Fig. 1 B and C). R4 (R303 in Kv1.2) is also positioned near E1 and may interact with it in the native protein (Fig. 1 B and C). Several other salt-bridge interactions also form between residues in S1, S2, S3, and S4 (see Fig. 6, which is published as supporting information on the PNAS web site). These salt bridges, which stabilize the open-state structure, represent interactions between amino acid residues that are highly conserved among eukaryotic Kv channels (46–48).
Fig. 1.
Homology model of the transmembrane region of Kv1.2 in the open state. (A) Side view of the ribbon representation of the rosetta membrane model of a single subunit of Kv1.2. Regions of the Kv1.2 structure modeled using the backbone coordinates of the unidentified residues in the x-ray structure of Kv1.2 as a template by the rosetta membrane homology method are shown in red (see Methods). Regions of the Kv1.2 structure modeled using the identified residues in the x-ray structure of Kv1.2 (9) as a template by the rosetta membrane homology method are shown in blue. Regions of the Kv1.2 structure modeled using the rosetta membrane de novo method are shown in green. Transmembrane segments are labeled from S1 to S6, the selectivity filter helix is labeled P, and the position of the S4–S5 linker is indicated by an arrow. N and C termini residues of the transmembrane region are labeled N and C, respectively. (B) Side view of the VSD segments S1–S4 only (colored individually) of the model shown in A. Side chains of gating-charge-carrying arginines in S4 (labeled R1–R4) and E226 in S2 (labeled E1) are shown in stick representation. Blue, red, and cyan atoms in the side chains shown represent nitrogen, oxygen, and carbon atoms, respectively. (C) View of the model shown in B from the extracellular side of the membrane. All structural figures presented in this work were generated using molscript (61) and raster3d (62).
Domain Assembly of Kv1.2 in the Open State.
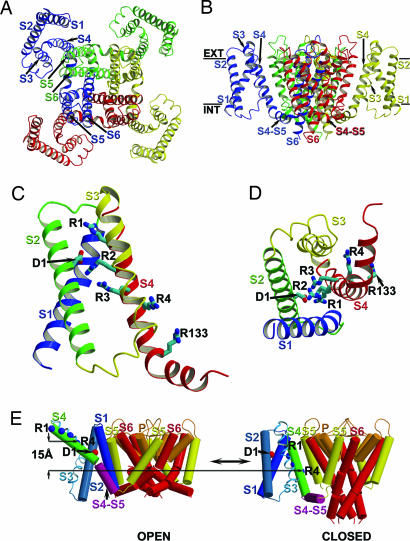
We used the x-ray structures of the separate VSD and PD of Kv1.2 in the open state to test whether the rosetta membrane domain assembly method (see Methods) can predict the interaction of the VSD with the PD using constraints from well established structure–function data for the final selection of models. Amino acid side chains are represented approximately in the rosetta membrane method, and hence the accuracy of the models is limited to 2–4 Å at best (45). Among the 10 largest clusters of models (see Methods), the third cluster agreed with experimental data demonstrating proximity of residues at the extracellular ends of S4 from one subunit and S5 from the adjacent subunit (32, 35–37) (Fig. 2A and B). This model was very similar to the x-ray structure of Kv1.2 in the open state (Fig. 2 C and D), with rms deviation over 260 residues of only 2.0 Å from the native structure (9). Other models in this cluster also were similar to the native structure (see Methods). The success of the domain-assembly method in predicting an open state model that closely matches the native structure encouraged us to use this method to construct a model of Kv1.2 in the closed state.
Fig. 2.
The rosetta membrane domain assembly model of the open state of Kv1.2. (A) View of the Kv1.2 model from the extracellular side of the membrane. All four subunits are colored individually. Segments S1–S6 for the blue-colored subunit and S5–S6 for the green-colored subunit are labeled accordingly. (B) View of the model in A from the side of the membrane. Segments S1–S4, S6, and S4–S5 linker for blue- and yellow-colored subunits are labeled accordingly. Approximate position of the first residue in the S4–S5 linker is indicated by an arrow for blue- and yellow-colored subunits. Extracellular and intracellular edges of the hydrocarbon core of the membrane are marked by solid bars and labeled “EXT” and “INT,” respectively. (C) View from the extracellular side of the membrane of the Kv1.2 structure (shown in blue) and the best rosetta membrane model (shown in orange) of the open state of Kv1.2 superimposed over the PD residues. Only a single VSD is shown attached to the tetramer of the PD for clarity. (D) View of the models shown in C from the side of the membrane.
Model of Kv1.2 in the Closed State.
We applied the rosetta membrane domain assembly method to build a model of Kv1.2 in the closed state as described in Methods. Based on experimental evidence that E1 in S2 (E283 in Shaker; E226 in Kv1.2) is positioned near R1 in S4 (R362 in Shaker; R294 in Kv1.2) in the closed state (23), we constrained the centroids of those residues to be within 8.0 Å from each other (see Methods). Other experimental data were not used during modeling. We examined the 10 largest clusters of models (see Methods). To account for transfer of ≈3 charge units per subunit (6–8), we selected the third largest cluster of models in which at least two of the four potential gating-charge-carrying arginines in S4 were exposed to the intracellular water-accessible environment. Fig. 3A–D shows one of the selected models of Kv1.2 in the closed state. Other models in this cluster were very similar except for the distance between S2 and S3 (see Methods). Thus, the precise position of S3 may not be accurately defined in our best models.
Fig. 3.
The rosetta membrane domain assembly model of the closed state of Kv1.2. (A) View of the Kv1.2 model from the extracellular side of the membrane. The model is colored and labeled as in Fig. 2A. (B) View of the model in A from the side of the membrane. The model is colored and labeled as in Fig. 2B. The S4–S5 linker for blue and red subunits is labeled accordingly. (C) Side view of the VSD only of the model in A. The model is colored and labeled as in Fig. 1B. (D) View of the model in C from the intracellular side of the membrane. (E) Kv1.2 models of the closed and open states shown in cylinder representation. Only a single VSD is shown attached to the tetramer of the PD for clarity. Transmembrane segments S1–S6 and P-loop are colored by a rainbow scheme from blue to red. The S4–S5 linker is in purple. Approximate positions of the Cα atoms of the first and fourth gating charge-carrying arginines in S4 (labeled as R1 and R4 and colored in blue) and E226 in S2 (labeled E1 and colored in red) are shown in sphere representation. All intracellular and extracellular loops, except for the S4–S5 linker, are represented by curved lines for simplicity. Vertical translation of R4 between the closed and open states along the membrane normal vector and relative to the plane of the membrane is indicated by arrows. S3 is represented by ribbon to show clearly the positions of the gating-charge-carrying arginines in S4.
S4 in our closed-state model of Kv1.2 is in close proximity to S5 from the adjacent subunit (Fig. 3 A and B), in agreement with results suggesting this topological arrangement in the closed state (35–37, 49). These segments packed significantly more tightly in our closed-state model compared with the open-state structure, which is realistic because the hydrophobic face of the S4 helix rotated ≈180° from the lipid environment in the open state to the protein environment at the S4–S5 interface in the closed state (Fig. 3 C and D). However, S4 is not completely buried within the protein in the closed state because some of its hydrophobic residues are exposed to lipid between S1 from one subunit and S5 from the adjacent subunit (Fig. 3A).
Gating Movements of the Kv1.2 VSD.
Comparison of the closed-state model and the open-state structure of Kv1.2 suggests a series of linked structural changes during the transition from the closed to the open conformation (Fig. 3E; see also Fig. 7, Tables 1 and 2, and Movies 1–3, which are published as supporting information on the PNAS web site). S4 moves ≈3 Å outward relative to the selectivity filter, consistent with experimental data on eukaryotic Kv channels (16, 17, 22, 26–29, 50, 51), and L293 and R297 in S4 move 1–2 Å relative to the pore axis between the closed and open states, in agreement with distances measured by luminescence resonance energy transfer (LRET) for homologous residues (L361 and R365) in the Shaker Kv channel (28). In addition, S4 rotates clockwise ≈180° about its axis, and the extracellular part of S4 changes its tilt angle from ≈10° to ≈45° relative to the membrane normal vector. The combined outward translocation and clockwise rotation during activation closely resemble the spiral motion of this segment that was first proposed in the sliding helix and helical screw models of gating (12, 13) and subsequently has been supported by extensive experimental results (15–20, 25, 26, 50).
In the transition from the closed to open state, the S4–S5 linker moves ≈10 Å tangentially around the PD, ≈4 Å away from the pore axis, and ≈4 Å outward. Residues in the S4–S5 linker remain in contact with the same set of residues in the intracellular end of the S6 from the same subunit in both the closed and open states. This continuous contact may allow the S4–S5 linker to pull the associated S6 segment to bend and open the pore as the S4 voltage sensor moves outward and rotates. These results agree with experimental observations demonstrating interactions between the S4–S5 linker and the intracellular end of S6 in several channels (52–55) and a proposal by Long et al. (56) that this interaction plays an important role in voltage-dependent gating by coupling conformational changes in the VSD with closing and opening of the pore.
The S1, S2, and S3 segments move around S4 in the clockwise direction (Fig. 3E), so that the intracellular end of S3 moves closer to the S4–S5 linker. Rotation of S4 around its axis and simultaneous rolling of S2 around S4 allow sequential interactions of the four gating-charge-carrying residues in S4 (R294, R297, R300, and R303 in Kv1.2) with the conserved negatively charged residue in S2 (E226 in Kv1.2) as proposed previously (46–48). Importantly, S4 rotates relative to S3 during the conformational change in contrast to the movement of these two helices as a rigid helical hairpin as predicted in the paddle model of gating (30). Substitutions of the gating-charge-carrying arginines in voltage-gated K+ and Na+ channels with histidine or glutamine creates state-dependent proton- or ion-conducting pores within the VSD (22–24). These data support mechanisms of gating in which the arginine side chains move through a narrow gating pore formed between extracellular and intracellular water-filled vestibules during conformational changes in the VSD between the closed and open states. Thus, the coordination of the small outward movement and clockwise rotation of S4 with the rolling motion of the S1–S3 around S4 results in the movement of three gating-charge-carrying arginine residues from internal to external exposure in the transition to the open state (see Fig. 3E and Movies 1–3 and also Figs. 8 and 9, which are published as supporting information on the PNAS web site). This rolling motion of S1–S3 is similar in concept to the transporter models of gating-charge movement that were proposed based on fluorescent imaging experiments that demonstrated small transmembrane movement of S4 (27).
Model of KvAP in an Open/Inactivated State.
The x-ray structures of the full-length KvAP channel reported to date have their VSD structure disrupted such that important interactions with the PD are apparently lost because of absence of native lipid bilayer environment (10, 11). The x-ray structure of the separate VSD of KvAP (10) is similar in overall helical topology to the structure of the VSD of Kv1.2 and also has the first three gating-charge-carrying arginines facing the extracellular side of the membrane. It therefore may represent an open or open/inactivated conformation of the VSD. We used this VSD structure and the open-state structure of the PD of KvAP (10) to model the transmembrane region of the KvAP (see Methods). Our largest cluster of models generated without constraints from experimental data resembled the Kv1.2 structure in that the VSD interacts with the PD of the adjacent subunit in clockwise direction, as viewed from the extracellular side (see Methods). Many models in this cluster were in good agreement with EPR spectroscopy accessibility data for the open/inactivated state of the KvAP (57), and we used the EPR data to search for the most favorable model among the 10 largest clusters (see Methods). The most favorable rosetta membrane model of KvAP in the open state (Fig. 4) has significant differences from the Kv1.2 structure (9) and from the previous KvAP model (11) (see Fig. 10, which is published as supporting information on the PNAS web site). The most striking difference in predicted topology is the position of S1 in close contact with the pore-forming S5 and S6 in KvAP model (Fig. 4 A and B), compared with its location on the outside edge of the VSD-PD complex in the Kv1.2 structure (9). In addition, S4 is positioned more on the periphery of the KvAP model and does not interact directly with the PD (Fig. 4 A and B), in contrast to the location of S4 near the PD in the Kv1.2 structure (9). Our model fits the predictions of lipid accessibility from EPR data for 97% of the amino acid residues studied by Cuello et al. (57) in the VSD, but S4 in our model of the open/inactivated state of KvAP appears to be significantly further outward than its expected interfacial position based on the EPR accessibility data (see Supporting Text and Fig. 11, which are published as supporting information on the PNAS web site).
Fig. 4.
The rosetta membrane domain assembly model of an open/inactivated state of KvAP. (A) View of the KvAP model from the extracellular side of the membrane. The model is colored and labeled as in Fig. 2A. (B) View of the model in A from the side of the membrane. The model is colored and labeled as in Fig. 2B. Approximate position of the first residue in the S4–S5 linker is indicated by arrow for blue and yellow subunits. (C) Side view of the VSD only (colored individually) of the model in A. Side chains of gating-charge-carrying arginines in S4 (labeled R1–R4), R133 in S4, and D62 in S2 (labeled D1) are shown in stick representation. Atoms in the side chains shown are colored as in Fig. 1B. (D) View of the model in C from the extracellular side of the membrane.
Model of KvAP in the Closed State.
To further develop the comparison between the Kv1.2 and KvAP models, we also modeled the closed state of KvAP. We used the rosetta homology/de novo method to construct a KvAP model in the closed state, using the Kv1.2 closed-state model as a template (see Methods). This approach assumes that the closed-state structures of KvAP and Kv1.2 are similar, even though there are significant differences between the open-state structures. The center of the largest cluster of models of KvAP in the closed state is shown in Fig. 5. Comparison of the closed- and open/inactivated state models of KvAP suggest substantial structural changes during transition from the closed to open conformation (Fig. 5E and Table 1; see also Movies 4–6, which are published as supporting information on the PNAS web site). S4 in KvAP moves ≈14 Å outward relative to the selectivity filter, rotates clockwise ≈180° about its axis, and moves away from S5 from the adjacent subunit. The S4–S5 linker moves away from S6 in the same subunit, eventually breaking hydrophobic interactions with the intracellular end of S6 (Fig. 5E). S1, S2, and S3 roll around S4 in the clockwise direction, as viewed from the extracellular side. Rotation of S4 around its own axis and simultaneous rolling of S2 around S4 allows for sequential interaction of the four gating-charge-carrying residues in S4 (R117, R120, R123, and R126 in the KvAP) with the negatively charged residue in S2 (D62 in the KvAP). Overall, these predicted gating movements resemble those postulated for Kv1.2 (Fig. 3E), but the movement of S4 is much larger in KvAP and results in a more outward position in the open state.
Fig. 5.
The rosetta membrane homology model of KvAP in closed state. (A) View of the KvAP model from the extracellular side of the membrane. The model is colored and labeled as in Fig. 2A. (B) View of the model in A from the side of the membrane. The model is colored and labeled as in Fig. 2B. The S4–S5 linker for blue and red subunits is labeled accordingly. (C) Side view of the VSD only of the model in A. The model is colored and labeled as in Fig. 4C. (D) View of the model in C from the intracellular side of the membrane. (E) KvAP models of the closed and open/inactivated (labeled as “open”) states shown in cylinder representation and as described in Fig. 3E. Approximate positions of the Cα atoms of the first and fourth gating-charge-carrying arginines in S4 (labeled R1 and R4), R133 in S4, and D62 in S2 (labeled D1) are shown in sphere representation.
Our modeling suggests that the difference in the magnitude of the S4 vertical movement observed between KvAP and Kv1.2 originates mainly from the differences in the open-state structures of these channels. However, we cannot exclude the possibility that S4 may also be positioned even further inward in the closed state structure of KvAP than in Kv1.2, because our KvAP closed-state model suggests a more outward position of S4 than the biotin–avidin accessibility data (see Supporting Text and also Fig. 12, which is published as supporting information on the PNAS web site). In the x-ray structure of the VSD of KvAP, the conserved negatively charged residue in S2 (D62) interacts with R133, which is located seven residues downstream from the R4 arginine, the last gating charge-carrying arginine in S4 of KvAP (10, 11) (Fig. 4 C and D). In contrast, in the x-ray structure of Kv1.2, the homologous negatively charged residue in S2 (E226) interacts with R300, which is R3 arginine in S4 (9) (Fig. 1 B and C). This structural difference places the S4 residues of KvAP ≈10 Å farther toward the extracellular surface in the open/inactivated state (Figs. 3E and 5E). In addition, the S4–S5 linker is oriented at an angle of ≈45° relative to the plane of the membrane and partially lies within the hydrocarbon core of the membrane in the KvAP model (Fig. 5E). In contrast, the S4–S5 linker is positioned parallel to the plane of the membrane at the interface between the polar and hydrocarbon core membrane environments in the Kv1.2 structure (9) (Fig. 3E).
Unified Gating Mechanism for Kv1.2 and KvAP.
Our modeling suggests that the eukaryotic and bacterial Kv channels have significantly different magnitude of S4 vertical movement, an idea that was recently discussed by Tombola and coworkers (58) based on a survey of the literature. The large movement of S4 in KvAP is consistent with structure–function studies on KvAP that used a biotin–avidin accessibility approach (10, 31) but is inconsistent with many experiments on eukaryotic Kv channels (16, 17, 22, 26–29, 50, 51). Our models provide a natural resolution of this apparent discrepancy by showing that the most favored conformations of the VSDs of Kv1.2 and KvAP in the open state are quite different. These two distantly related Kv channels have only ≈20% sequence identity in the transmembrane region, leaving many possibilities for differences in amino acid sequence that may cause differences in structure (see Fig. 13, which is published as supporting information on the PNAS web site). Nevertheless, our models suggest that KvAP may pass through an intermediate state whose structure is similar to the Kv1.2 open state structure during the transition from closed to open/inactivated states (see Movies 7–9, which are published as supporting information on the PNAS web site). In this case, the major conformational change that moves the gating-charge-carrying arginines in S4 through the gating pore would be analogous in Kv1.2 and KvAP, but an additional transition would be required to reach the KvAP open/inactivated state that was characterized structurally, in which R133 interacts with the key negatively charged residue in S2. It is conceivable that Kv1.2 or other eukaryotic ion channels that have a positively charged residue analogous to R133 may potentially reach a second open state or an inactivated state that is similar in structure to the KvAP open/inactivated state. Thus, we propose a unified mechanism of gating in which the major conformational change that moves most of the gating charge across the electric field in the transition from the closed to open state of all voltage-gated ion channels may be similar to Kv1.2 (see Movies 1–3), but KvAP and perhaps some other members of the ion channel superfamily may undergo a second conformational change in which the S4 segment moves considerably farther outward to reach a second open or inactivated state.
Conclusions
Structural modeling of the Kv1.2 and KvAP channels using rosetta suggests that the open state conformations of the voltage sensors of these channels are significantly different, whereas the voltage sensors may be similar in conformation in their closed states. This finding potentially explains the difference in magnitude of the S4 translational movement observed experimentally for these channels. We propose a mechanism of conformational changes of the VSD during channel gating that involves translation and rotation of S4 as proposed in the original sliding helix or helical screw models coupled with a rolling motion of S1–S3 around S4 similar to recent transporter models of gating. Extension of our rosetta models by homology modeling raises the possibility that the major conformational change in the VSD may be similar in these two distantly related Kv channels, thereby unifying the structural basis of their gating. The combination of structure-prediction methods with limited experimental data should become an increasingly powerful approach to complex problems in structural biology as structure prediction methodology matures. We close with the thought that our models, although in better agreement with a wide range of structure–function data than previous ones, require detailed testing by mutagenesis, functional analysis, and further structural work for validation. These models make many specific predictions that will catalyze further research and will serve as rigorous tests of their validity.
Methods
rosetta Membrane Method.
We modified the homology (59) and domain assembly (A. M. Wollacott, A. Zanghellini, and D.B., unpublished data) modes of rosetta (42) to use the membrane environment-specific score function developed for modeling of α-helical transmembrane proteins (45) for modeling of the Kv channels. Multiple sequence alignment information based score derived from the ConSeq server (http://conseq.bioinfo.tau.ac.il) (60) for the Kv1.2 and KvAP sequences was added to the rosetta membrane score function and improved sampling of native-like structures (V.Y.-Y. and D.B., unpublished data). Further details of the modeling procedures and illustrations of additional sequence alignments, starting models, and final clusters are presented in Figs. 14–23, which are published as supporting information on the PNAS web site. Coordinates of all structural models presented in this work are available from V.Y.-Y. upon request (e-mail: yarovoy@u.washington.edu).
Supplementary Material
Acknowledgments
We thank Benoit Roux for critical comments on the manuscript and for Shaker Kv channel models; Roderick Mackinnon for the KvAP channel model; Eduardo Perozo for EPR spectroscopy accessibility data; Todd Scheuer, Jack Schonbrun, and Alexandre Zanghellini for helpful discussion; Keith Laidig for excellent administration of computational resources; and Laura Gonzalez for making movies. This work was supported by National Institute of Mental Health Career Development Research Grant K01 MH67625 (to V.Y.-Y.), a Howard Hughes Medical Institute grant (to D.B.), and National Institutes of Health Grant R01 NS15751 (to W.A.C.).
Abbreviations
- VSD
voltage-sensing domain
- PD
pore-forming domain
- Kv
voltage-gated potassium
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Hille B. Ion Channels of Excitable Membranes. Sunderland, MA: Sinauer Associates; 2001. [Google Scholar]
- 2.Yu F. H., Catterall W. A. Sci. STKE. 2004;2004:re15. doi: 10.1126/stke.2532004re15. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong C. M., Bezanilla F. J. Gen. Physiol. 1974;63:533–552. doi: 10.1085/jgp.63.5.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bezanilla F. Physiol. Rev. 2000;80:555–592. doi: 10.1152/physrev.2000.80.2.555. [DOI] [PubMed] [Google Scholar]
- 5.Sigworth F. J. Q. Rev. Biophys. 1994;27:1–40. doi: 10.1017/s0033583500002894. [DOI] [PubMed] [Google Scholar]
- 6.Schoppa N. E., McCormack K., Tanouye M. A., Sigworth F. J. Science. 1992;255:1712–1715. doi: 10.1126/science.1553560. [DOI] [PubMed] [Google Scholar]
- 7.Aggarwal S. K., MacKinnon R. Neuron. 1996;16:1169–1177. doi: 10.1016/s0896-6273(00)80143-9. [DOI] [PubMed] [Google Scholar]
- 8.Seoh S. A., Sigg D., Papazian D. M., Bezanilla F. Neuron. 1996;16:1159–1167. doi: 10.1016/s0896-6273(00)80142-7. [DOI] [PubMed] [Google Scholar]
- 9.Long S. B., Campbell E. B., Mackinnon R. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 10.Jiang Y., Lee A., Chen J., Ruta V., Cadene M., Chait B. T., MacKinnon R. Nature. 2003;423:33–41. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]
- 11.Lee S. Y., Lee A., Chen J., MacKinnon R. Proc. Natl. Acad. Sci. USA. 2005;102:15441–15446. doi: 10.1073/pnas.0507651102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Catterall W. A. Trends Neurosci. 1986;9:7–10. doi: 10.1016/0166-2236(93)90193-p. [DOI] [PubMed] [Google Scholar]
- 13.Guy H. R., Seetharamulu P. Proc. Natl. Acad. Sci. USA. 1986;83:508–512. doi: 10.1073/pnas.83.2.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Catterall W. A. Annu. Rev. Biochem. 1986;55:953–985. doi: 10.1146/annurev.bi.55.070186.004513. [DOI] [PubMed] [Google Scholar]
- 15.Yang N., Horn R. Neuron. 1995;15:213–218. doi: 10.1016/0896-6273(95)90078-0. [DOI] [PubMed] [Google Scholar]
- 16.Yang N., George A. L., Jr., Horn R. Neuron. 1996;16:113–122. doi: 10.1016/s0896-6273(00)80028-8. [DOI] [PubMed] [Google Scholar]
- 17.Larsson H. P., Baker O. S., Dhillon D. S., Isacoff E. Y. Neuron. 1996;16:387–397. doi: 10.1016/s0896-6273(00)80056-2. [DOI] [PubMed] [Google Scholar]
- 18.Yusaf S. P., Wray D., Sivaprasadarao A. Pflügers Arch. 1996;433:91–97. doi: 10.1007/s004240050253. [DOI] [PubMed] [Google Scholar]
- 19.Yang N., George A. L., Jr, Horn R. Biophys. J. 1997;73:2260–2268. doi: 10.1016/S0006-3495(97)78258-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguyen T. P., Horn R. J. Gen. Physiol. 2002;120:419–436. doi: 10.1085/jgp.20028636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Starace D. M., Bezanilla F. J. Gen. Physiol. 2001;117:469–490. doi: 10.1085/jgp.117.5.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Starace D. M., Bezanilla F. Nature. 2004;427:548–553. doi: 10.1038/nature02270. [DOI] [PubMed] [Google Scholar]
- 23.Tombola F., Pathak M. M., Isacoff E. Y. Neuron. 2005;45:379–388. doi: 10.1016/j.neuron.2004.12.047. [DOI] [PubMed] [Google Scholar]
- 24.Sokolov S., Scheuer T., Catterall W. A. Neuron. 2005;47:183–189. doi: 10.1016/j.neuron.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 25.Glauner K. S., Mannuzzu L. M., Gandhi C. S., Isacoff E. Y. Nature. 1999;402:813–817. doi: 10.1038/45561. [DOI] [PubMed] [Google Scholar]
- 26.Cha A., Snyder G. E., Selvin P. R., Bezanilla F. Nature. 1999;402:809–813. doi: 10.1038/45552. [DOI] [PubMed] [Google Scholar]
- 27.Chanda B., Asamoah O. K., Blunck R., Roux B., Bezanilla F. Nature. 2005;436:852–856. doi: 10.1038/nature03888. [DOI] [PubMed] [Google Scholar]
- 28.Posson D. J., Ge P., Miller C., Bezanilla F., Selvin P. R. Nature. 2005;436:848–851. doi: 10.1038/nature03819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahern C. A., Horn R. Neuron. 2005;48:25–29. doi: 10.1016/j.neuron.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 30.Jiang Y., Ruta V., Chen J., Lee A., MacKinnon R. Nature. 2003;423:42–48. doi: 10.1038/nature01581. [DOI] [PubMed] [Google Scholar]
- 31.Ruta V., Chen J., MacKinnon R. Cell. 2005;123:463–475. doi: 10.1016/j.cell.2005.08.041. [DOI] [PubMed] [Google Scholar]
- 32.Laine M., Lin M. C., Bannister J. P., Silverman W. R., Mock A. F., Roux B., Papazian D. M. Neuron. 2003;39:467–481. doi: 10.1016/s0896-6273(03)00468-9. [DOI] [PubMed] [Google Scholar]
- 33.Durell S. R., Shrivastava I. H., Guy H. R. Biophys. J. 2004;87:2116–2130. doi: 10.1529/biophysj.104.040618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shrivastava I. H., Durell S. R., Guy H. R. Biophys. J. 2004;87:2255–2270. doi: 10.1529/biophysj.104.040592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elliott D. J., Neale E. J., Aziz Q., Dunham J. P., Munsey T. S., Hunter M., Sivaprasadarao A. EMBO J. 2004;23:4717–4726. doi: 10.1038/sj.emboj.7600484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gandhi C. S., Clark E., Loots E., Pralle A., Isacoff E. Y. Neuron. 2003;40:515–525. doi: 10.1016/s0896-6273(03)00646-9. [DOI] [PubMed] [Google Scholar]
- 37.Broomand A., Mannikko R., Larsson H. P., Elinder F. J. Gen. Physiol. 2003;122:741–748. doi: 10.1085/jgp.200308927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moult J., Fidelis K., Zemla A., Hubbard T. Proteins. 2003;53(Suppl. 6):334–339. doi: 10.1002/prot.10556. [DOI] [PubMed] [Google Scholar]
- 39.Bradley P., Misura K. M., Baker D. Science. 2005;309:1868–1871. doi: 10.1126/science.1113801. [DOI] [PubMed] [Google Scholar]
- 40.Kuhlman B., Dantas G., Ireton G. C., Varani G., Stoddard B. L., Baker D. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- 41.Schueler-Furman O., Wang C., Bradley P., Misura K., Baker D. Science. 2005;310:638–642. doi: 10.1126/science.1112160. [DOI] [PubMed] [Google Scholar]
- 42.Rohl C. A., Strauss C. E., Misura K. M., Baker D. Methods Enzymol. 2004;383:66–93. doi: 10.1016/S0076-6879(04)83004-0. [DOI] [PubMed] [Google Scholar]
- 43.Gray J. J., Moughon S., Wang C., Schueler-Furman O., Kuhlman B., Rohl C. A., Baker D. J. Mol. Biol. 2003;331:281–299. doi: 10.1016/s0022-2836(03)00670-3. [DOI] [PubMed] [Google Scholar]
- 44.Kuhlman B., O’Neill J. W., Kim D. E., Zhang K. Y., Baker D. J. Mol. Biol. 2002;315:471–477. doi: 10.1006/jmbi.2001.5229. [DOI] [PubMed] [Google Scholar]
- 45.Yarov-Yarovoy V., Schonbrun J., Baker D. Proteins. 2006;62:1010–1025. doi: 10.1002/prot.20817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tiwari-Woodruff S. K., Lin M. A., Schulteis C. T., Papazian D. M. J. Gen. Physiol. 2000;115:123–138. doi: 10.1085/jgp.115.2.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tiwari-Woodruff S. K., Schulteis C. T., Mock A. F., Papazian D. M. Biophys. J. 1997;72:1489–1500. doi: 10.1016/S0006-3495(97)78797-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Silverman W. R., Roux B., Papazian D. M. Proc. Natl. Acad. Sci. USA. 2003;100:2935–2940. doi: 10.1073/pnas.0636603100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lai H. C., Grabe M., Jan Y. N., Jan L. Y. Neuron. 2005;47:395–406. doi: 10.1016/j.neuron.2005.06.019. [DOI] [PubMed] [Google Scholar]
- 50.Ahern C. A., Horn R. J. Gen. Physiol. 2004;123:205–216. doi: 10.1085/jgp.200308993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Phillips L. R., Milescu M., Li-Smerin Y., Mindell J. A., Kim J. I., Swartz K. J. Nature. 2005;436:857–860. doi: 10.1038/nature03873. [DOI] [PubMed] [Google Scholar]
- 52.Lu Z., Klem A. M., Ramu Y. Nature. 2001;413:809–813. doi: 10.1038/35101535. [DOI] [PubMed] [Google Scholar]
- 53.Lu Z., Klem A. M., Ramu Y. J. Gen. Physiol. 2002;120:663–676. doi: 10.1085/jgp.20028696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tristani-Firouzi M., Chen J., Sanguinetti M. C. J. Biol. Chem. 2002;277:18994–19000. doi: 10.1074/jbc.M200410200. [DOI] [PubMed] [Google Scholar]
- 55.Decher N., Chen J., Sanguinetti M. C. J. Biol. Chem. 2004;279:13859–13865. doi: 10.1074/jbc.M313704200. [DOI] [PubMed] [Google Scholar]
- 56.Long S. B., Campbell E. B., Mackinnon R. Science. 2005;309:903–908. doi: 10.1126/science.1116270. [DOI] [PubMed] [Google Scholar]
- 57.Cuello L. G., Cortes D. M., Perozo E. Science. 2004;306:491–495. doi: 10.1126/science.1101373. [DOI] [PubMed] [Google Scholar]
- 58.Tombola F., Pathak M. M., Isacoff E. Y. Neuron. 2005;48:719–725. doi: 10.1016/j.neuron.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 59.Rohl C. A., Strauss C. E., Chivian D., Baker D. Proteins. 2004;55:656–677. doi: 10.1002/prot.10629. [DOI] [PubMed] [Google Scholar]
- 60.Berezin C., Glaser F., Rosenberg J., Paz I., Pupko T., Fariselli P., Casadio R., Ben-Tal N. Bioinformatics. 2004;20:1322–1324. doi: 10.1093/bioinformatics/bth070. [DOI] [PubMed] [Google Scholar]
- 61.Kraulis P. J. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- 62.Merritt E. A., Bacon D. J. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.