Abstract
Parasitic nematodes of the genus Trichinella cause significant food-borne illness and occupy a unique evolutionary position at the base of the phylum Nematoda, unlike the free-living nematode Caenorhabditis elegans. Although the forthcoming genome sequence of Trichinella spiralis can provide invaluable comparative information about nematode biology, a basic framework for understanding the history of the genus Trichinella is needed to maximize its utility. We therefore developed the first robust and comprehensive analysis of the phylogeny and biogeographic history of Trichinella using the variation in three genes (nuclear small-subunit rDNA, and second internal transcribed spacer, mitochondrial large-subunit rDNA, and cytochrome oxidase I DNA) from all 11 recognized taxa. We conclude that (i) although Trichinellidae may have diverged from their closest extant relatives during the Paleozoic, all contemporary species of Trichinella diversified within the last 20 million years through geographic colonization and pervasive host switching among foraging guilds of obligate carnivores; (ii) mammalian carnivores disseminated encapsulated forms from Eurasia to Africa during the late Miocene and Pliocene, and to the Nearctic across the Bering Land Bridge during the Pliocene and Pleistocene, when crown species ultimately diversified; (iii) the greatest risk to human health is posed by those species retaining an ancestral capacity to parasitize a wide range of hosts; and (iv) early hominids may have first acquired Trichinella on the African savannah several million years before swine domestication as their diets shifted from herbivory to facultative carnivory.
Keywords: biogeography, mitochondrial DNA, phylogeny, ribosomal DNA
Exceptional biological diversity among nematodes is exemplified by certain attributes of the parasite Trichinella spiralis. Organization of its mitochondrial genome more closely resembles that of coelomate metazoans than that of its presumed closest relatives, the secernentean nematodes (1). In addition, T. spiralis (Dorylaimia) shares a similar proportion (45%) of its ESTs with the nematode Caenorhabditis elegans (Rhabditina) as it does with the fruit fly Drosophila melanogaster (Arthropoda: Drosophilidae) (2). Thus, many ESTs common to T. spiralis and C. elegans are not necessarily specific to nematodes but may be conserved among diverse taxonomic groups of invertebrates. C. elegans is often thought of as a prototypical nematode because of its acceptance as a model for studying biological processes; however, genomic variation among nematodes is extensive and commensurate with their phylogenetic and ecological diversity. Therefore, the forthcoming genome sequence of T. spiralis will contribute substantially to our understanding of nematode biology and the origins of parasitism.
In 1998, Blaxter et al. (3) used genetic data to delineate the phylum Nematoda into supertaxa consisting of five clades. Trichinella, a parasite of vertebrates, occupies a basal lineage (clade I) consisting of free-living Mononchida, plant parasitic Dorylaimida, and entomophagous Mermithida, with which it shares features of early embryogenesis (4) and small-subunit (SSU) rDNA sequences (3). Other clades show similar levels of diversity in host–parasite associations, suggesting that parasites of vertebrates arose independent of one another during the long evolutionary history for nematodes (3). Thus, understanding the origins and persistence of vertebrate parasitism requires phylogenetic and historic information from a broad spectrum of taxa. Among the Trichocephalida, parasitism may have arisen as early as the Paleozoic in the ancestor of the Trichinellidae and the Trichuridae; however, temporal origins of extant species of Trichinella remain unexplored.
The genus Trichinella comprises a monophyletic lineage in the Trichinellidae, the putative sister to the Trichuridae (Capillariinae, Trichurinae, and Trichosomoidinae). The superfamily Trichinelloidea to which Trichinella belongs is phylogenetically diagnosed by the stichosome, a region of the glandular esophagus, and the bacillary bands, an assembly of structural characters otherwise unknown among the nematodes. Trichinellidae is further diagnosed by a unique life cycle where worms undergo complete development within a single vertebrate host (autohexeroxeny) and by first-stage larvae, which localize within skeletal muscle cells as infectious organisms. Although eating raw or undercooked pork infected with T. spiralis accounts for most human cases of trichinellosis, zoonotic transmission also occurs from species that circulate among wildlife. Eleven distinct taxa are now recognized in mammals, birds, crocodilians, and saurians (5), characterized by either a thick (encapsulated) or a very thin (nonencapsulated) collagen membrane enveloping the parasite-infected muscle cell.
Despite extensive development of biochemical and genetic methods to distinguish taxa (6), evolutionary relationships among genotypes of extant Trichinella remain enigmatic. Delineating the phylogeny and biogeography of Trichinella will substantially increase the comparative value of its genome sequence and help elucidate the determinants of zoonotic risk among species. On a broader scale, this information will provide us an opportunity to investigate ecological and evolutionary forces that promote diversification in complex host–parasite systems.
Three central questions fueled our curiosity about the evolution of Trichinella. (i) Can prolonged coevolution with distinct tetrapod groups explain why encapsulated species of Trichinella are restricted to eutherian mammals and nonencapsulated species also infect saurian, crocodilian, avian, eutherian, and metatherian hosts (5, 7), or have more recent ecological and biogeographic events influenced current host associations? (ii) Why are most species and genotypes with broad host ranges restricted to well defined geographical localities? (iii) Did humans become infected with Trichinella in the Neolithic before domesticating pigs, or did adaptation after pig domestication cause certain species of Trichinella to threaten human health? To explore these and related questions, we generated the first robust hypothesis for the phylogeny of Trichinella based on multilocus DNA sequence information from all ecologically and genetically recognized species and genotypes. We then interpreted these results with reference to the evolutionary history and biogeography of their hosts.
Results
The general features of the three comparative data sets are summarized in Table 1. Significant variation in the rate of evolution across sites was identified in each model. In no case did enforcing a molecular clock significantly worsen the likelihood of the data.
Table 1.
Characteristics of sequence alignments and their evolutionary models
| Data set* | No. of bases† |
Model type‡ | C:T/Tv | G:A/Tv | Base frequencies |
I | Γ shape | Clock§ |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Var. | Inf. | A | C | G | ΔInL | df > P | ||||||
| SSU | 1,929 | 717 | 417 | TrNefΓ | 2.80 | 2.06 | 0.250 | 0.250 | 0.250 | 0 | 0.408 | 20.3 | 14 > 0.12 |
| 11mit | 1,193 | 276 | 194 | HKY Γ I | 15.60 | 15.60 | 0.299 | 0.170 | 0.174 | 0.49 | 0.492 | 9.84 | 9 > 0.35 |
| 3gene | 1,273 | 204 | 78 | HKY Γ | 3.21 | 3.21 | 0.277 | 0.165 | 0.200 | 0 | 0.156 | 1.86 | 6 > 0.93 |
*Data sets are as follows: SSU, 10 of 11 Trichinella genotypes including outgroup taxa Trichuris muris, Trichuris suis, Adnoncholaimus sp., Pirapulus caudatus, Mormis nigrescens, and Mylonchulus arenicolu; 11mit, mitochondrial LSU rDNA and COI from all 11 Trichinella taxa; 3gene, mitochondrial LSU rDNA, COI, and ITS-2 from all eight encapsulated species and genotypes of Trichinella.
†Total, variable (Var.), and informative (Inf.) number of bases under the parsimony criterion.
‡TrNef, a model consisting of a single transversion rate but two distinct transition rates. HKY models correspond to that of Hasegawa et al. (8) in assuming unequal base frequencies and rates of transition and transversion substitutions. Γ, variation in the substitution across nucleotide sites modeled as a gamma distribution with an estimated shape parameter; I, the estimated proportion of invariant sites; Tv, transversion.
§Difference in log likelihood (ΔInL) trees evaluated with and without the enforcement of a molecular clock. df = (number of taxa) −2. P represents statistical significance in the reduction of likelihood owing to enforcement of a molecular clock.
An initial analysis of the SSU rDNA variation served to evaluate Trichinella phylogeny within the broader context of nematode evolution. In this regard, the SSU rDNA sequences reflect a minimally differentiated, monophyletic assemblage (Fig. 1) where the maximum likelihood (ML) pairwise distance never exceeded 0.0052 among encapsulated species or 0.0135 among all species. These values are substantially less than the ML distance (0.2) separating Trichuris muris from Trichuris suis. If the locus evolved in each lineage at ≈0.4 × 10−9 per site per year (4% over 100 MY), which typifies many metazoan phyla (9), then extant species of Trichinella shared a common ancestor ≈16 million years (MY) ago, and encapsulated taxa first began diversifying within the last 9 MY (Table 2). Only by postulating an implausible 18-fold rate decrease in divergence rate could the extant diversity among Trichinella species date to the origins of distinct tetrapod host groups that occurred some 300 MY B.P. Although species of Trichinella are poorly differentiated from one another at the SSU rDNA, this locus markedly distinguishes them from species of Trichuris. The extant pairwise difference between Trichinella and Trichuris exceeds 16%, and ML estimates of their genetic distance exceed 22%. Assuming a per lineage rate of 4% over 100 MY for SSU rDNA evolution, the most recent common ancestor of Trichinella and Trichuris may have existed 275 MY B.P.
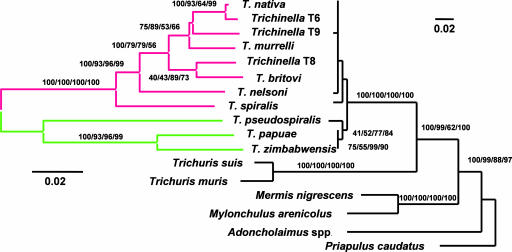
Fig. 1.
Midpoint rooted minimum evolution trees reconstructed from all known encapsulated (red) and nonencapsulated (green) species and genotypes of Trichinella based on the variation in mitochondrial LSU and COI DNA (on the left) and SSU rDNA (on the right). Topological support is indicated by Bayesian posterior probabilities and by ML, minimum evolution (using ML distances), and parsimony bootstrap replicate analyses (B/ML/ME/P). Bootstrap support was reconstructed from 100-bp replicates.
Table 2.
Estimates of divergence dates (MY B.P.)
| Genotypes | SSU rDNA |
Mt DNA |
||
|---|---|---|---|---|
| Obs* | Model* | Obs† | Model‡ | |
| All | 16.1 | 16.8 | 7.0 | 11.1 |
| T1 vs. T2 | 6.8 | 9.3 | 3.1 | 2.7 |
| T6 vs. T2 | 0 | 0 | 0.32 | 0.14 |
Assumes that observed (Obs) or ML (Model) pairwise distances accrue at 4% per 100 MY (∗), 2% per MY (†), or 5.2% per MY (‡).
Diverse tree-building approaches yielded consistent and nearly complete phylogenetic resolution among all 11 Trichinella congeners when compared at the two mtDNA genes (Fig. 1), or among all eight encapsulated species when the second internal transcribed spacer (ITS-2) sequences were also included (data not shown). Data suggest that the earliest diversifications among extant encapsulated species gave rise to T. spiralis and then to Trichinella nelsoni. The next diversification event would have separated the remaining encapsulated forms from an ancestor unique to Trichinella T8 and Trichinella britovi. This relationship is inferred by 98% and 77% bootstrap replicates in the distance and parsimony analyses of the mtDNA data, respectively, and by 75% of the ML bootstrap replicates and 78% of the Bayesian posterior distribution when ITS-2 data were included. Whether Trichinella T9 diverged from the common ancestor of Trichinella nativa and Trichinella T6 (weakly preferred by all analyses of mtDNA data alone) or from the ancestor of Trichinella murrelli (as indicated when ITS-2 variation is considered) remained unresolved. Each hypothesis received equivalent statistical support and required equivalent numbers of parsimony-informative changes. Thus, of >2 million rooted, bifurcating trees, only five are demonstrably compatible with the variation in our multilocus data set; on these trees we base our subsequent discussion of Trichinella evolution.
Discussion
The phylogenetic hypothesis generated herein affords us an opportunity to explore the history of host–parasite coevolution and biogeography for species of Trichinella. A vast interval, lasting perhaps 275 MY, separated the divergence between Trichinellidae and Trichuridae from the radiation of extant species of Trichinella (Fig. 1). The 11 recognized taxa are genetically and biologically delineated into two distinct clades, predicated in part on the presence or absence of a well defined, intramuscular collagen cyst. Ancestral lineages of Trichinella that lacked such capsules evidently infected tetrapods (Fig. 2).
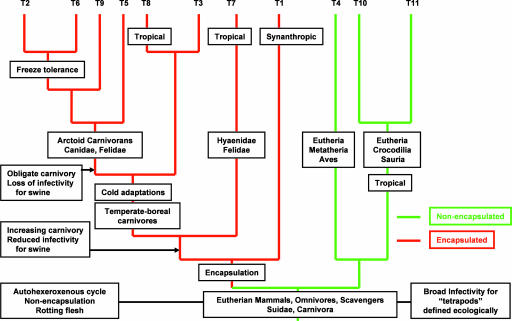
Fig. 2.
Host associations and primary traits for the life history of Trichinella during diversification. Emphasized are shifts in patterns of transmission from omnivorous to carnivorous hosts, adaptations to changes in environmental temperatures, and retention of plesiomorphic traits among members of the nonencapsulated clade (green). Genotype abbreviations are consistent with Table 3.
A deep coevolutionary association had been proposed to explain the unique ability of nonencapsulated species to infect saurians, crocodilians, other nonavian archosaurs (Trichinella papuae and Trichinella zimbabwensis), and birds (Trichinella pseudospiralis), in contrast to the restriction of encapsulated species to synapsid and mammalian hosts (10, 11). However, our data undermine the view that these two parasite groups coevolved exclusively with distinct tetrapod host groups. Instead, ancestral Trichinella likely infected a guild of eutherian omnivores and carnivores including Suidae, Hyaenidae, and Felidae (Fig. 2). Divergence of the primary subclades would have followed in Eurasia during the late Tertiary. Encapsulated forms subsequently diversified through geographic colonization, host switching, and isolation in temperate, boreal, and Arctic habitats across the Holarctic, whereas nonencapsulated forms retained the capacity to infect eutherian mammals and became secondarily associated with some Metatheria, Sauria, Crocodilia, and Aves (Fig. 2). Although resolving the history of host colonization by nonencapsulated species will require additional data, the shallow divergence among T. papuae and T. zimbabwensis suggests that these species were only recently acquired by crocodilians and are not relicts of an ancient coevolutionary association. Thus, extant host distributions became established only since the Miocene.
Encapsulated forms of Trichinella evolved entirely with eutherian hosts (Fig. 2). Among these species, T. spiralis has been globally translocated by an array of domesticated and synanthropic mammals and is now perpetuated largely through a synanthropic cycle involving swine and a sylvatic cycle involving both swine and carnivores (5). Their Eurasian origins are suggested by tree topology, the geography of extant species (Figs. 2 and 3), and elevated genetic diversity among geographically localized isolates from East Asia (G.L.R., unpublished data). This finding implies that encapsulated species may have originated in Eurasia, where the Suidae and numerous carnivorous groups diversified as well (12, 13).
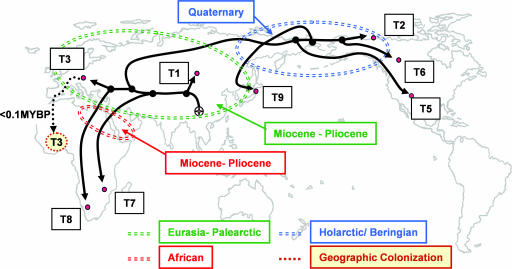
Fig. 3.
Historical biogeography for species of the encapsulated clade of Trichinella. The cladogram for species of Trichinella has been mapped onto a global projection for range expansion. Subsequent to origins in Eurasia, Trichinella became distributed with its carnivorous hosts in Africa, broadly in the Palearctic, and later into the Nearctic. Emphasized is the role for independent events of biotic expansion and geographic colonization into Africa (Miocene/Pliocene/Pleistocene) and through Beringia into the Nearctic (Quaternary) as drivers for speciation. Genotype abbreviations are consistent with Table 3.
It is unlikely that encapsulated lineages colonized Africa before the breakup of Gondwana, because endemic forms are absent from South America, Australia, New Zealand, and much of Australasia. Such biogeographic evidence corroborates our belief that extant species radiated much later than (i) the diversification of basal eutherians (near the termination of the Cretaceous), (ii) the extinction of an archaic carnivorous fauna (Creodonta and Hyaenodonta in the Late Oligocene), or (iii) the divergence between Eutheria and Metatheria (14–16). Instead, encapsulated and nonencapsulated clades probably separated during the mid-Miocene as temperate ecosystems changed, followed by the diversification of extant species within the last 15–20 MY. The hosts of T. spiralis, T. nelsoni, Trichinella T8, and T. britovi all suggest an ancestral association with swine and carnivores (Fig. 2) (5). Interestingly, Suidae also completed their divergence from the Tayassuidae in the Lower Miocene (15–20 MY B.P.) (12, 13).
Transmission of Trichinella has depended on omnivory, carnivory, and scavenging of carrion among Eurasian and African paleoguilds (17). Omnivory continues to contribute to the transmission of the basal encapsulated lineages, T. spiralis and T. nelsoni. In contrast, a transition to facultative and finally to obligate carnivory characterizes T. britovi and the crown species of the encapsulated clade, respectively (Fig. 2). Encapsulated forms radiated through host switching within guild assemblages among Mustelidae, Ursidae, Canidae, Felidae, and Hyaenidae in regional settings ecologically isolated by stadial climate cycles in the late Pliocene and Pleistocene.
Trichinella genotypes now endemic to Africa and those distributed in the amphiberingian region appear to have resulted from episodic periods of biotic expansion, isolation, and host switching consistent with a “taxon pulse” model of biological diversification (18, 19). Three independent expansion events from Eurasia explain the occurrence of T. nelsoni, Trichinella T8, and T. britovi (20) in Africa (Fig. 3). These were arguably facilitated by the formation of land connections to Eurasia and Europe during the middle to upper Miocene, Pliocene, and Pleistocene and by incursions of Eurasian carnivores and omnivores into Africa (12, 14, 21, 22). T. nelsoni and Trichinella T8 have been reported only in sub-Saharan Africa, where they circulate among felids, hyaenids, and to a lesser degree among suids (5, 20). Carnivores, including felids, hyaenids, and viverrids (Feloidea) and canids, ursids, and mustelids (Arctoidea) radiated in Eurasia and subsequently disseminated to Africa from the Lower Miocene to the Quaternary. A more recent expansion of T. britovi into northern and western Africa from the Palearctic, during or since the terminal glaciation of the Pleistocene, is suggested by the genetic uniformity among isolates from these geographical regions (20) (Fig. 3).
Carnivorans, including ursids, canids, and felids, are principally responsible for the dispersal of Holarctic species (5), which likely expanded across Western Europe, through Beringia, and into North America (Fig. 3 and Table 2). Isolation across Beringia appears not to have commenced with the initial opening of Bering Strait 4.8–5.5 MY B.P. (23), but in association with cyclical and episodic glacial–interglacial stages during the Pleistocene. Ursids, mustelids, canids, and felids, all important hosts for Nearctic species of Trichinella, dispersed and became established in the Nearctic during the late Pliocene and Quaternary (24, 25). Increasingly cold and insular conditions in the Arctic during the late Miocene and Pliocene favored the development of cold-adapted forms at a time when Beringia may have represented a filter bridge for host–parasite assemblages limited by arctic and subarctic conditions.
Like other taxa (26, 27), Trichinella expanded through Beringia during the Pliocene and Quaternary and subsequently experienced isolation events that promoted speciation during stadial–interstadial cycles. Isolation during the Quaternary across the Bering Land Bridge, and north and south of the Laurentide and Cordilleran ice, likely drove the divergence of T. murrelli and crown taxa including Trichinella T9, T. nativa, and Trichinella T6 (Fig. 3). Trichinella T9 may represent a peripheral isolate on the islands of Japan derived from a widespread ancestral population that occupied a Holarctic distribution. The divergence of Trichinella T6 from T. nativa may have occurred during a subsequent episode of allopatry in Beringia and periglacial habitats south of the Cordilleran and Laurentide glaciers (28). If so, the contemporary Holarctic range for T. nativa would have arisen from a secondary geographic colonization of the high-latitude Holarctic (−5°C January isotherm) during the Wisconsin or terminal Pleistocene. Indeed, secondary contact zones in western North America have developed for some hosts tracking the retreat of glaciers (29). Populations of T. nativa may have expanded into high arctic habitats of Canada and Greenland during the Holocene, as has been postulated for other Holarctic parasitic nematodes and their mammalian hosts (26, 30, 31). Alternatively, assuming that peripheral isolation played a role in diversifying these crown species, T. nativa may instead represent an ancestral and persistent high-latitude population in the Holarctic.
To date, no endemic species of Trichinella has been identified in the Neotropics even though felids, procyonids, canids, and mustelids colonized South America after formation of the Panamanian Isthmus during the Pliocene/Pleistocene ≈3.0 MY B.P. (32). In South America only T. spiralis has been found, presumably the result of anthropogenic translocation. Among sylvatic Trichinella adapted for survival in harsh, cold, and xeric environments, historical constraints and life history canalization may have precluded Trichinella from expanding into the Neotropics across the Panamanian Isthmus.
Human Trichinellosis
Did the divergence of T. spiralis coincide with the domestication of pigs? If so, its independent history would have commenced no earlier than the Neolithic, and a shallow genetic divergence would be evident between the parasites of wild and domesticated suids. Instead, T. spiralis appears to have commenced its independent evolutionary trajectory several MY before Sus scrofa was first domesticated (33) because none of the enzootic species share an especially close relationship with T. spiralis. Thus, the ecological setting in which hominids first acquired trichinellosis may parallel that of Taenia tapeworms (34) and lice (35). Hunting prey also pursued by felids, canids, and hyaenids may have exposed early humans to a suite of parasites already well established on the savannahs of Africa during the late Tertiary and Quaternary. Accordingly, hominids would have first acquired Trichinella when their diets shifted from herbivory to scavenging and facultative carnivory during the Pliocene or early Pleistocene, (17, 36–38).
Recent host adaptations may have enhanced the risk to human health posed by certain species of Trichinella; however, a broad host range characterizes basal lineages and therefore represents the ancestral condition. The capacity of nonencapsulated forms to infect a broad array of terrestrial vertebrates suggests they pose a significant zoonotic risk as well (11, 38, 39). By contrast, the capacity to infect swine is diminished among more recently radiated species (T. britovi and T. nelsoni) and is nonexistent among the crown species (T. nativa, T. murrelli, Trichinella T6, and Trichinella T9) (5).
Conclusions
Although the Trichinellidae and the Trichuridae apparently diverged as early as the Paleozoic, contemporary species of Trichinella diversified during the age of modern Eutheria. The following observations support this line of reasoning: (i) the genetic diversity among extant species of Trichinella accumulated within the last 15–20 MY; (ii) all extant species infect Eutherian carnivores or rodents; and (iii) infections in saurians, crocodilians, metatherians, and birds appear secondary rather than relictual. If, as we contend, extant diversity originated in Eurasia and disseminated to both Africa and the Nearctic during the late Tertiary, Trichinella should resemble other parasites for which host switching rather than strict cospeciation was a major diversifying force (40). These patterns underscore the role of episodic expansion, isolation, and host switching in promoting Trichinella diversification and highlight the exceptional dissemination potential conferred to T. spiralis by its association with domesticated swine and synanthropic rodents.
Materials and Methods
All species and genotypes of Trichinella (Table 3) were maintained in Swiss–Webster mice. Eviscerated mouse carcasses were digested with pepsin (1%) and HCl (1%), and the released muscle larvae were collected and washed through multiple exchanges in water. Genomic DNA was isolated by proteinase K and SDS digestion and treated with RNase. At least three full-length clones of PCR-amplified SSU rDNA were sequenced from each species and genotype by using Trichinella-specific primers (GenBank accession no. U60231). PCR-amplified mitochondrial large-subunit (LSU) rDNA and cytochrome oxidase I (COI) genes were directly sequenced. Three multiple sequence alignments were constructed by using clustal-x and visually confirmed. The first data set (18S) aligned the SSU rDNA of Trichinella species to homologues from several outgroup taxa (Table 1). To obtain better resolution of intrageneric relationships, we analyzed a second data set (11-mit) incorporating two mtDNA loci (COI and LSU rDNA) from all 11 species and genotypes of Trichinella. Inasmuch as the ITS-2 sequences of the eight encapsulated Trichinella taxa could not be aligned reliably with those of the nonencapsulated species, only the interrelationships among encapsulated forms were further explored by using a third data set (3-gene) by combining the ITS-2, COI, and mitochondrial LSU rDNA gene sequences. Likelihood ratio tests were used to identify which of 56 nucleotide substitution models best fit each of these three data sets by using model test 3.0.4 (41) (Table 1) and to evaluate the compatibility of each data set with a molecular clock (42).
Table 3.
Trichinella strains used in the analysis
| Parasite | Genotype | ISS code |
|---|---|---|
| T. spiralis | T1 | ISS003 |
| T. nativa | T2 | ISS010 |
| T. britovi | T3 | ISS005 |
| T. murrelli | T5 | ISS035 |
| Trichinella T6* | T6 | ISS040 |
| T. nelsoni | T7 | ISS029 |
| Trichinella T8* | T8 | ISS124 |
| Trichinella T9* | T9 | ISS409 |
| T. pseudospiralis† | T4 | ISS013 |
| T. papuae‡ | T10 | ISS572 |
| T. zimbabwensis‡ | T11 | ISS1029 |
ISS, Istituto Superiore di Sanità. Additional information can be obtained from www.iss.it/site/trichinella/.
*Genetically distinct but taxonomically undefined taxa.
†Nonencapsulated species infectious for birds in addition to mammals.
‡Nonencapsulated species infectious for reptiles in addition to mammals.
Phylogenies were reconstructed from bootstrap replicates of these alignments under the criteria of minimum evolution and ML by using the optimized model as implemented by paup* (43). Additionally, a Bayesian approach was used to simultaneously optimize tree topology and nucleotide substitution parameters, including six independent nucleotide substitution rates, the proportion of invariant sites, the shape parameter of the gamma distribution, and the frequency of each nucleotide, by using mr. bayes 3.0 (44). Of a total 10 million generations, 1 of every 1,000 was sampled after discarding a “burn in” period of 100,000 generations. ML searches performed under topological constraints were also used to evaluate the relative statistical support of competing topologies.
Genetic distances were used to estimate the ages of divergence among species and genotypes of Trichinella. Several approaches were considered for each data set to derive a temporal range. For SSU rDNA, a per-lineage calibration rate of 0.4 × 10−9 per site per year was assumed based on data generally supported for metazoan phyla (9). For mtDNA, data were initially assumed to diverge at a rate of 0.01 × 10−6 per site per year per lineage (i.e., 2% uncorrected pairwise divergence per MY) (45, 46). A second approach used distance and rate estimates derived from ML models, assuming an instantaneous substitution rate of 0.0259 × 10−6 per site per year (47, 48). Alternatively, mtDNA evolution rates (1.1–1.4% uncorrected and 1.4–1.7% ML distance per pair per MY) were derived by assuming that the initial diversification of T. murrelli resulted from isolation south of the Laurentide glaciers after 2.5–3.0 MY B.P.
Host associations and historical biogeography were examined in relation to the phylogenetic hypothesis for Trichinella. Primary hosts, patterns of life history, and geographical distribution (5) were mapped and optimized onto the phylogenetic tree by using mcclade 4.0 (49). The implications of accepting more disparate rate calibrations were also considered during this examination process.
Acknowledgments
We thank K. D. Murrell and R. Fayer for critical reading of the manuscript.
Abbreviations
- SSU
small-subunit
- LSU
large-subunit
- ITS-2
second internal transcribed spacer
- ML
maximum likelihood
- MY
million years
- COI
cytochrome oxidase I.
Footnotes
References
- 1.Lavrov D. V., Brown W. M. Genetics. 2001;157:621–637. doi: 10.1093/genetics/157.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parkinson J., Mitreva M., Whitton C., Thomson M., Daub J., Martin J., Schmid R., Hall N., Barrell B., Waterston R. H., et al. Nat. Genet. 2004;36:1259–1267. doi: 10.1038/ng1472. [DOI] [PubMed] [Google Scholar]
- 3.Blaxter M. L., De Ley P., Garey J. R., Liu L. X., Scheldeman P., Vierstraete A., Vanfleteren J. R., Mackey L. Y., Dorris M., Frisse L. M., et al. Nature. 1998;392:71–75. doi: 10.1038/32160. [DOI] [PubMed] [Google Scholar]
- 4.Voronov D. A., Panchin Y. V., Spiridonov S. E. Nature. 1998;395:28. doi: 10.1038/25637. [DOI] [PubMed] [Google Scholar]
- 5.Pozio E., Zarlenga D. S. Int. J. Parasitol. 2005;35:1191–1204. doi: 10.1016/j.ijpara.2005.07.012. [DOI] [PubMed] [Google Scholar]
- 6.Zarlenga D. S., La Rosa G. Vet. Parasitol. 2000;93:279–292. doi: 10.1016/s0304-4017(00)00346-0. [DOI] [PubMed] [Google Scholar]
- 7.Gauthier J. A., Kluge A. G., Rowe T. In: Phylogeny and Classification of the Tetrapods, Volume 1: Amphibians, Reptiles, and Birds. Benton M. J., editor. Oxford: Clarendon; 1987. pp. 103–155. [Google Scholar]
- 8.Hasegawa M., Kishino H., Yano T. J. Mol. Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 9.Aris-Brosou S., Yang Z. Syst. Biol. 2002;51:703–714. doi: 10.1080/10635150290102375. [DOI] [PubMed] [Google Scholar]
- 10.Pozio E., Foggin C. M., Marucci G., La Rosa G., Sacchi L., Corona S., Rossi P., Mukaratirwa S. Int. J. Parasitol. 2002;32:1787–1799. doi: 10.1016/s0020-7519(02)00139-x. [DOI] [PubMed] [Google Scholar]
- 11.Pozio E., Marucci G., Casulli A., Sacchi L., Mukaratirwa S., Foggin C. M., La Rosa G. Parasitology. 2004;128:333–342. doi: 10.1017/s0031182003004542. [DOI] [PubMed] [Google Scholar]
- 12.Cooke H. B. S., Wilkinson A. F. In: Evolution of African Mammals. Maglio V. J., Cooke H. B. S., editors. Cambridge, MA: Harvard Univ. Press; 1978. pp. 435–482. [Google Scholar]
- 13.Bowen G. J., Clyde W. C., Koch P. L., Ting S., Alroy J., Tsubamoto T., Wang Y., Wang Y. Science. 2002;295:2062–2065. doi: 10.1126/science.1068700. [DOI] [PubMed] [Google Scholar]
- 14.Savage R. J. G. In: Evolution of African Mammals. Maglio V. J., Cooke H. B. S., editors. Cambridge, MA: Harvard Univ. Press; 1978. pp. 249–267. [Google Scholar]
- 15.Luo Z. X., Ji Q., Wible J. R., Yuan C. X. Science. 2003;302:1934–1940. doi: 10.1126/science.1090718. [DOI] [PubMed] [Google Scholar]
- 16.Cifelli R. L., Davis B. M. Science. 2003;302:1899–1900. doi: 10.1126/science.1092272. [DOI] [PubMed] [Google Scholar]
- 17.Lewis M. E. J. Hum. Evol. 1997;32:257–288. doi: 10.1006/jhev.1996.0103. [DOI] [PubMed] [Google Scholar]
- 18.Erwin T. L. In: Vicariance Biogeography: A Critique. Nelson G., Rosen D. E., editors. New York: Columbia Univ. Press; 1981. pp. 159–196. [Google Scholar]
- 19.Halas D., Zamparo D., Brooks D. R. J. Biogeogr. 2005;32:249–260. [Google Scholar]
- 20.Pozio E., Pagani P., Marucci G., Zarlenga D. S., Hoberg E. P., De Meneghi D., La Rosa G., Rossi L. Int. J. Parasitol. 2005;35:955–960. doi: 10.1016/j.ijpara.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 21.Ferrao A. L., Brooks D. R. J. Biogeogr. 2005;32:1291–1299. [Google Scholar]
- 22.Briggs J. C. Global Biogeography. Amsterdam: Elsevier; 1995. [Google Scholar]
- 23.Sher A. Nature. 1999;397:103–104. [Google Scholar]
- 24.Kurtén B., Anderson E. Pleistocene Mammals of North America. New York: Columbia Univ. Press; 1980. [Google Scholar]
- 25.Rausch R. L., Babero B. B., Rausch V. R., Schiller E. L. J. Parasitol. 1956;42:259–271. [PubMed] [Google Scholar]
- 26.Hoberg E. P. J. Parasitol. 2005;91:358–369. doi: 10.1645/GE-3466. [DOI] [PubMed] [Google Scholar]
- 27.Cook J. A., Hoberg E. P., Koehler A., Henttonen H., Wickström L., Haukisalmi V., Galbreath K., Chernyavsky F., Dokuchaev N., Lahzuhtkin A., et al. Mammal Study. 2006;30:S35–S44. [Google Scholar]
- 28.La Rosa G., Marucci G., Zarlenga D. S., Casulli A., Zarnke R. L., Pozio E. Int. J. Parasitol. 2003;33:209–216. doi: 10.1016/s0020-7519(02)00258-8. [DOI] [PubMed] [Google Scholar]
- 29.Stone K. D., Flynn R. W., Cook J. A. Mol. Ecol. 2002;11:2048–2063. doi: 10.1046/j.1365-294x.2002.01596.x. [DOI] [PubMed] [Google Scholar]
- 30.Hoberg E. P., Monsen K. J., Kutz S., Blouin M. S. J. Parasitol. 1999;85:910–934. [PubMed] [Google Scholar]
- 31.Hoberg E. P., Kutz S. J., Galbreath K. E., Cook J. J. Parasitol. 2003;89:S84–S95. [Google Scholar]
- 32.Marshall L. G., Webb S. D., Sepkoski J. J., Raup D. M. Science. 1982;215:1351–1357. doi: 10.1126/science.215.4538.1351. [DOI] [PubMed] [Google Scholar]
- 33.Larson G., Dobney K., Albarella U., Fang M., Matisoo-Smith E., Robins J., Lowden S., Finlayson H., Brand T., Willerslev E., et al. Science. 2005;307:1618–1621. doi: 10.1126/science.1106927. [DOI] [PubMed] [Google Scholar]
- 34.Hoberg E. P., Alkire N. L., deq Ueiroz A., Jones A. Proc. R. Soc. London Ser. B. 2001;268:781–787. doi: 10.1098/rspb.2000.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reed D. L., Smith V. S., Hammond S. L., Rogers A. R., Clayton D. H. PLoS Biol. 2004;2:e340. doi: 10.1371/journal.pbio.0020340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Heinzelin J., Clark J. D., White T., Hart W., Renne P., WoldeGabriel G., Beyene Y., Vrba E. Science. 1999;284:625–629. doi: 10.1126/science.284.5414.625. [DOI] [PubMed] [Google Scholar]
- 37.Sponheimer M., Lee-Thorp J. A. Science. 1999;283:368–370. doi: 10.1126/science.283.5400.368. [DOI] [PubMed] [Google Scholar]
- 38.Owen I. L., Gomez Morales M. A., Pezzetti P., Pozio E. Trans. R. Soc. Trop. Med. Hyg. 2005;99:618–624. doi: 10.1016/j.trstmh.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 39.Pozio E., Owen I. L., La Rosa G., Sacchi L., Rossi P., Corona S. Int. J. Parasitol. 1999;29:1825–1839. doi: 10.1016/s0020-7519(99)00135-6. [DOI] [PubMed] [Google Scholar]
- 40.Hoberg E. P. In: Marine Parasitology. Rohde K., editor. Sydney: Commonwealth Scientific and Industrial Research Organization; 2005. pp. 329–339. [Google Scholar]
- 41.Posada D., Crandall K. A. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 42.Huelsenbeck J. P., Rannala B. Science. 1997;276:227–232. doi: 10.1126/science.276.5310.227. [DOI] [PubMed] [Google Scholar]
- 43.Swofford D. L. paup*: Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, MA: Sinauer; 2001. Version 4.0. [Google Scholar]
- 44.Huelsenbeck J. P., Ronquist F. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 45.Brown W., Jr., George M., Jr., Wilson A. C. Proc. Natl. Acad. Sci. USA. 1979;76:1967–1971. doi: 10.1073/pnas.76.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brown W., Prager E. M., Wang A., Wilson A. C. J. Mol. Evol. 1982;18:225–239. doi: 10.1007/BF01734101. [DOI] [PubMed] [Google Scholar]
- 47.Slowinski J. B., Arbogast B. S. Syst. Biol. 1999;48:396–399. doi: 10.1080/106351599260364. [DOI] [PubMed] [Google Scholar]
- 48.Arbogast B., Edwards S. V., Wakeley J., Beerli P., Slowinski J. B. Annu. Rev. Ecol. Syst. 2002;33:707–740. [Google Scholar]
- 49.Maddison D. R., Maddison W. P. macclade. Sunderlund, MA: Sinauer; 2000. Version 4.0. [Google Scholar]