Abstract
Inactivation of the XRCC4 nonhomologous end-joining factor in the mouse germ line leads to embryonic lethality, in association with apoptosis of newly generated, postmitotic neurons. We now show that conditional inactivation of the XRCC4 in nestin-expressing neuronal progenitor cells, although leading to no obvious phenotype in a WT background, leads to early onset of neuronally differentiated medulloblastomas (MBs) in a p53-deficient background. A substantial proportion of the XRCC4/p53-deficient MBs have high-level N-myc gene amplification, often intrachromosomally in the context of complex translocations or other alterations of chromosome 12, on which N-myc resides, or extrachromosomally within double minutes. In addition, most XRCC4/p53-deficient MBs harbor clonal translocations of chromosome 13, which frequently involve chromosome 6 as a partner. One copy of the patched gene (Ptc), which lies on chromosome 13, was deleted in all tested XRCC4/p53-deficient MBs in the context of translocations or interstitial deletions. In addition, Cyclin D2, a chromosome 6 gene, was amplified in a subset of tumors. Notably, amplification of Myc-family or Cyclin D2 genes and deletion of Ptc also have been observed in human MBs. We therefore conclude that, in neuronal cells of mice, the nonhomologous end-joining pathway plays a critical role in suppressing genomic instability that, in a p53-deficient background, routinely contributes to genesis of MBs with recurrent chromosomal alterations.
Two major pathways of DNA double-strand break (DSB) repair in mammalian cells are homologous recombination and nonhomologous DNA end-joining (NHEJ) (1). NHEJ is used in all somatic cells to repair general DSBs, such as those resulting from normal metabolism or external agents such as ionizing radiation (IR). In addition, NHEJ repairs RAG endonuclease-introduced DSBs that are intermediates in the developing lymphocyte-specific V(D)J recombination process. Currently, there are six well characterized NHEJ factors. The Ku70 and Ku80 proteins form a DNA end-binding complex that recruits and activates the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) that, in turn, activates the Artemis DNA end-processing factor (2). In the major characterized NHEJ pathway, end-ligation employs DNA Ligase 4 (Lig4) and its requisite cofactor, XRCC4 (2). Inactivation of any of the NHEJ factors in mice leads to a lack of mature B and T lymphocytes (“SCID” phenotype) due to the inability to assemble antigen receptor genes. NHEJ deficiency also leads to additional cellular defects due to the impaired ability to repair general DSBs that, for XRCC4 or Lig4 deficiency, include impaired proliferation, premature senescence, IR sensitivity, and genomic instability (2). In mice, XRCC4 or Lig4 deficiency results in embryonic lethality associated with severe apoptosis of newly generated, postmitotic neurons throughout the developing nervous system (3). Both the embryonic lethality and neuronal apoptosis are p53-dependent responses, because they can be rescued by p53 deficiency (4, 5). However, p53 deficiency does not rescue NHEJ defects, because mice deficient for XRCC4 or Lig4 and p53 still have impaired V(D)J recombination and a SCID phenotype (4, 5).
XRCC4/p53 or Lig4/p53 doubly deficient mice routinely succumb to RAG-dependent progenitor B cell (pro-B) lymphomas that harbor complex chromosomal translocations (“complicons”) that lead to c-myc amplification (4–6). The generation of such complicons results from RAG-initiated DSBs at the Ig heavy chain locus on mouse chromosome 12 being fused downstream of the c-myc locus on chromosome 15 to form a dicentric chromosome, which generates amplified c-myc genes via a breakage-fusion-bridge mechanism (6). Such complex translocations and amplifications are commonly found in progressive stages of solid tumors (7). In this context, impaired NHEJ can contribute to the development of tumors outside of the immune system, as evidenced by enhanced development of sarcomas with recurrent translocations in Ink4a/Arf-deficient mice heterozygous for a Lig4-inactivating mutation (8). Moreover, XRCC4/p53-, Lig4/p53-, and Artemis/p53-doubly deficient mice also develop medulloblastomas (MBs) (6, 9, 10), a primitive neuroectodermal tumor that arises predominantly from neoplastic transformation of granule cell precursors in the developing cerebellum (11). The mechanism by which NHEJ contributes to suppression of MBs in a p53-deficient background has not been elucidated, because NHEJ/p53-deficient mice succumb so rapidly to pro-B lymphomas.
Genetic alterations in human MBs include activation of sonic hedgehog (Shh) signaling, which is involved in normal granule cell precursor development, via mutation or deletion of patched (Ptc), as well as inactivation of p53 and amplification of various genes, including c-myc and N-myc (11). There are several mouse MB models based on heterozygosity for Ptc or overexpression of genes, such as c-myc or IGF-2, in neuronal cells (11). Although MBs are rare in p53-deficient mice, p53 deficiency accelerates MB development in Ptc+/− mice (12) and promotes MB development in various mutant mouse backgrounds that affect DNA repair and/or the DSB response (6, 10, 13–17). In the latter context, the fact that XRCC4 and Lig4 deficiency results in p53-dependent apoptosis of newly generated neurons is intriguing considering that the only other cells shown to undergo this phenomenon are progenitor lymphocytes, which undergo p53-dependent apoptosis because of the inability to repair RAG-generated DSBs (4, 5). However, the nature of the presumed DNA DSBs that lead to the widespread apoptosis of newly generated neurons is unknown. By analogy to studies of lymphoid tumors, one means to elucidate potential sites of recurrent genomic instability would be via analysis of tumors. Therefore, to further characterize the role of NHEJ in neuronal development and to identify pathways that lead to MB formation in XRCC4/p53-deficient mice, we have conditionally deleted XRCC4 in nestin-expressing cells of normal and p53-deficient mice.
Results
Conditional Deletion of XRCC4 in Nestin-Expressing Neuronal Cells.
We used the Cre-loxP method to conditionally inactivate XRCC4 in mice. For this purpose, we introduced loxP sites on either side of XRCC4 exon 3 in TC1 ES cells to generate the Xc allele and, in addition, we generated ES cells in which exon 3 had been deleted via Cre recombination to generate the X− allele (Fig. 5, which is published as supporting information on the PNAS web site). The X− allele recapitulates that used to generate mice inactivated for XRCC4 in the germ line (3). We also generated X−/− ES cells by high G418 selection and used these cells to confirm expected phenotypic consequences of XRCC4 deficiency (not shown). Subsequently, we used (Xc/+) and (X+/−) ES cells to transmit the mutations into the 129sv/ev mouse germ line. To confirm that the Xc allele recapitulated the phenotype of our original XRCC4 knockout allele (5), we intercrossed the X+/− mice with p53−/− mice to generate X−/−p53−/− mice. As predicted, we observed development of X−/−p53−/− pro-B lymphomas (data not shown) that harbored the same chromosomal translocations observed in the original XRCC4/p53-deficient mice (5).
We used a nestin-Cre transgene to conditionally delete the Xc allele, because this transgene is specifically expressed and deletes loxP-flanked sequences in neuronal progenitor cells (NPCs) (18). Mice homozygous for the Xc allele (Xc/c mice) were bred with X+/− mice that harbored a nestin-Cre transgene (N mice) to generate NXc/− mice. NXc/− mice were born at Mendelian ratios, survived through adulthood (Fig. 1A and C) and lacked overt physiological abnormalities (data not shown). NXc/− mice were not more cancer prone than controls (Table 2, which is published as supporting information on the PNAS web site). By Southern blotting, deletion of the Xc in NXc/− adult mice was nearly complete in the cortex and cerebellum of the adult brain but not in other examined tissues (Fig. 1B). Absence of XRCC4 gene expression was confirmed by RT-PCR and Western blotting (Fig. 6A, which is published as supporting information on the PNAS web site) and by immunohistochemical staining of cryosections of NXc/− brains with anti-XRCC4 antibody (Fig. 6B). Surprisingly, histological analyses of several NXc/− embryos (n = 4) at embryonic days 14.5–15.5 did not reveal obvious neuronal abnormalities in their nervous system (Fig. 6C), even though nestin-Cre is expressed in NPCs (18) and neuronal cell death in XRCC4-deficient mice is observed at the postmitotic neuronal stage (3).
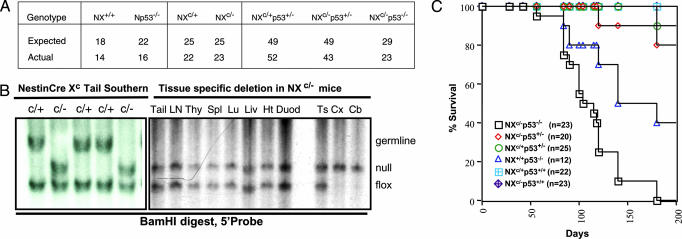
Fig. 1.
Conditional inactivation of XRCC4. (A) Expected and actual numbers of progeny born from nestin Cre, XRCC4 floxed, p53 WT, or p53 deficient crosses in this study. (B Left) Southern blotting of BamHI-digested tail DNA obtained from NXc/+ (c/+) and NXc/− (c/−) progeny of Xc/c mice bred to NX+/− mice. (B Right) Adult NXc/− tissue Southern blot shows specific deletion of Xc allele only in cortex (Cx) and cerebellum (Cb). Ln, lymph node; Thy, thymus; Spl, spleen; Lu, lung; Liv, liver; Ht, heart; Duod, duodenum; Ts, testes. XRCC4 WT (germ line), floxed (flox), and deleted (null) alleles detected by using depicted 5′ probe. (C) Kaplan Meier survival curve of mice described in this study. n, number of cohort mice.
Increased Mortality from Early Onset MBs in p53-Deficient NXc/− Mice.
To determine whether conditional inactivation of XRCC4 in nestin-expressing neuronal cells of p53-deficient mice enhances MB development, we intercrossed NX+/−p53+/− and Xc/cp53+/− mice to generate cohorts of NXc/−p53−/− (n = 23), NXc/−p53+/− (n = 20), NXc/+p53+/− (n = 25), NXc/−(n = 23), and NXc/+(n = 22) mice. Small cohorts of NX+/+p53−/− (n = 12) and NX+/+p53+/+ mice (n = 12) also were monitored. All genotypes were born at Mendelian ratios and appeared normal at birth (Fig. 1A and Table 1). NXc/−p53−/− mice appeared healthy up to ≈10–11 weeks of age but became moribund between 12 and 14 weeks of age; with 20 of 23 dead by 25 weeks (Fig. 1C). Survival of NX+/+p53−/− mice was comparable with p53−/− mice, with 50% dead by 21–22 weeks of age, with thymic lymphoma and solid tumors as the most common lesions (Fig. 1C and Table 1). NXc/−p53+/− mice generally survived beyond 16 months.
Table 1.
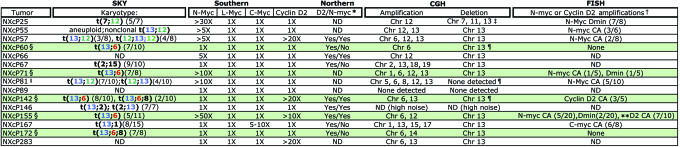
Summary of analyses of NXc/−p53−/− medulloblastoma
Tumors containing 13;6 translocations are highlighted in green. Number of metaphases containing recurrent translocation are shown unbolded in parentheses. ND, not determined.
*Cyclin D2 (D2) and N-myc total RNA levels expressed as compared with P4 brain determined by Northern blot analyses (Fig. 9B). Yes, denotes expression; No, no expression detected. Overexpression of CyclinD2 is detected in the majority of the medulloblatomas.
†CA, complex chromosomal amplications; Dmin, double minutes.
‡Specific heterozygous loss of Ptc detected in NXcP25 by CGH (data not shown).
§By SKY and single-chromosome paints, these tumors have 13;6 translocations, two copies of chromosome 6 (e.g., NXcP60 and NXcP142, Fig. 3A) and one copy of chromosome 13. NXcP172, with t(13;6;8) has one copy of chromosome 13 in all metaphases analyzed and metaphases containing one or two copies of chromosome 6.
¶Loss of patched (Ptc) on chromosome 13 detected by FISH (see Fig. 4).
‖Additional t(13;12;13) complex translocations were also observed in NXcP81, shown in Fig. 3A.
**NXcP155, 1 metaphase exhibited both N-myc CA and Dmins (1/20).
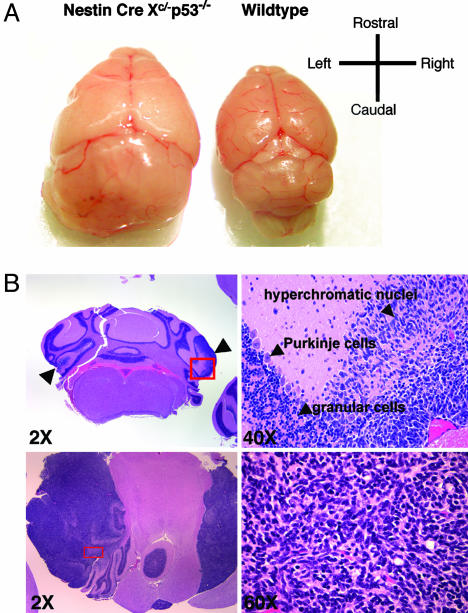
Analyses of several NXc/−p53−/− mice at 4 and 8.5 weeks revealed multifocal tumor masses expanding from the granular layer of the cerebellum (Fig. 2B Upper Left), which consisted of densely packed cells containing hyperchromatic, round to spindle-shaped nuclei (Fig. 2B Right) with high mitotic indices. Tumor cells occasionally formed neuroblastic rosettes characteristic of classical variant of MBs (ref. 19; data not shown). At 12–14 weeks, the skulls of NXc/−p53−/− mice showed rapid morphological enlargement (data not shown). Eighteen NXc/−p53−/− mice analyzed histologically had aggressive MBs (Fig. 2A), located in the cerebellar vermis (data not shown) or hemisphere (Fig. 2B Lower Left; NXcP67). NXcP67 further exhibited metastases in the spinal cord and two tumors (NXcP25 and NXcP81) contained regions of large anaplastic cells with pleomorphic nuclei, characteristics of large cell MBs (ref. 19; data not shown). The majority of NXc/−p53−/− MBs expressed glial fibrillary acidic protein neural marker and the NeuN mature neuronal marker (Fig. 7, which is published as supporting information on the PNAS web site), features characteristic of advanced stage/progressive human MBs (19, 20).
Fig. 2.
Medullobastoma development in NXc/−p53−/− mice. (A) Brain dissected from an NXc/−p53−/− mouse (NXcP25; Left) harboring MB and brain dissected from an age matched WT mouse (Right). (B Upper Left) ×2 magnification of cross section of brain from an 8.5-week-old NXc/−p53−/− mouse stained with hematoxylin/eosin. Multifocal tumor masses in the molecular layer are indicated by arrowheads. (B Upper Right) ×40 magnification of the boxed region, in red, is shown. Arrowheads indicate tumor cells with hyperchromatic nuclei, Purkinge cells, granular cells. (B Lower Left) ×2 magnification of cross section of NXcP67 brain harboring hemispheric (on the left) and meninges (on the right) tumor expansion. (B Lower Right) ×60 magnification of the boxed region, in red, is shown.
Recurrent Chromosomal Translocations in NXc/−p53−/− MBs.
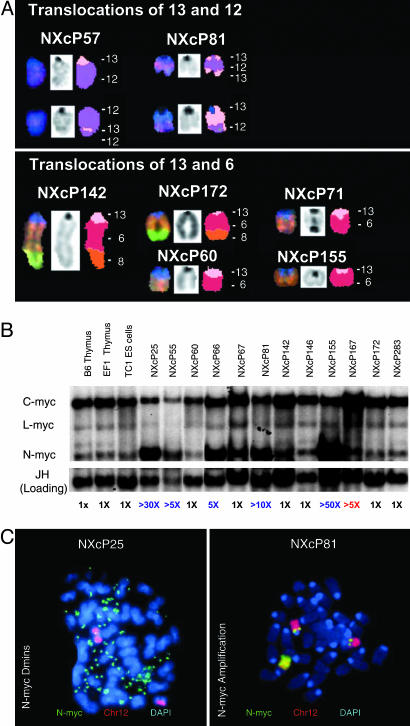
Spectral karyotype analyses revealed all analyzed MBs harbored clonal translocations involving a few specific chromosomes. We defined translocations as “clonal” when we found them in 50% or more of analyzed metaphases (Table 1). Strikingly, 9 of 11 analyzed NXc/−p53−/− MBs harbored clonal translocations involving chromosome 13 (Fig. 3A and Table 1). The most common partner in the chromosome 13 translocations was chromosome 6, which occurred in five of nine independent translocations. Other chromosome 13 translocations involved chromosome 12 (two of nine) and chromosomes 1 and 2 (one each) (Table 1 and Fig. 3A). Of the nine chromosome 13 translocations, three appeared reciprocal, including t(13; 12) and t(12; 13) in NXcP81 and t(13; 2) and t(2; 13) in NXcP146 (Table 1); NXcP57 contained t(13; 12), t(12; 13) and complex t(12; 13; 12) translocations (Fig. 3A and Table 1). Other chromosome 13 translocations either were complex [e.g., t(13; 6; 8) in both NXcP142 and NXcP172] or nonreciprocal [e.g., t(13; 6) in NXcP60, NXcP71, and NXcP155]. All chromosome 13 translocations retained the centromeric portion of chromosome 13 and lost the telomeric portion (Figs. 3A and 4Left Insets and Table 1).
Fig. 3.
Recurrent translocations and amplifications in NXc/−p53−/− MBs. (A) Recurrent translocations involving chromosome 13 determined by SKY analyses of NXc/−p53−/− tumor metaphases. (A Upper) Clonal translocations involving chromosome 12 observed in 2 of 11 tumors. (A Lower) Clonal translocations involving chromosome 6. Translocations of the centromeric region of chromosome 13 onto the distal portion of the q arm of chromosome 6 were the most frequently observed in 5 of 11 tumors. Additional chromosome 13 and unique translocations are shown in Table 1. chr13, pink; chr12, purple; chr6, red; chr8, orange. (B) Southern blotting reveals the majority of NXc/−p53−/− tumors harbor amplified N-myc. Twelve NXc/−p53−/− tumors and control DNA were digested with EcoRI and probed with N-myc, c-myc, L-myc probes and then Ig heavy chain JH probe as loading control. Quantification of N-myc amplifications is shown in blue; c-myc amplification, in red, is labeled below. (C) Amplification of N-myc manifested as double minutes (Dmins) (NXcP25; Left) or as chromosomal amplifications (NXcP81; Right) detected by FISH with an N-myc BAC (in green) and chromosome 12 paint (in red). Unamplified N-myc is located at both chromosome 12 centromeric regions. In NXcP81, the amplified N-myc is located in another chromosomal location.
Fig. 4.
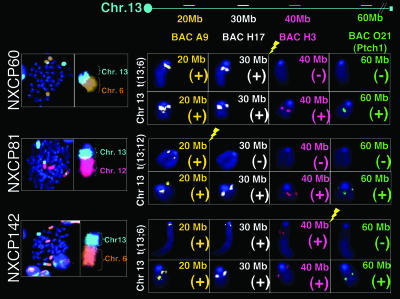
Recurrent deletions involving chromosome 13. FISH analyses of NXc/−p53−/− tumor metaphases were performed by using chromosome 13 BACs. The ideogram at top of figure indicates the position of chromosome 13 BAC probes. All tumors examined had one untranslocated, apparent normal chromosome 13 and one t(13; 6), a complex t(13; 6; 8), or a t(13; 12) marker chromosome. DAPI staining (dark blue) and chromosomes 13 (bright blue), 6 (orange), and 12 (pink) paints are shown. Yellow lightning symbol denotes the chromosome 13 breakpoints. Chromosome 13 translocations consistently result in a single copy loss of the patched genomic locus. Loss of at least a single copy of patched was verified in all tumors analyzed by CGH (Table 1 and Fig. 8, which is published as supporting information on the PNAS web site). Mbp is abbreviated as Mb in figure.
All MBs with chromosome 13 translocations retained a single chromosome 13 that appeared normal. In contrast, MBs with translocations of chromosome 6 often retained two apparently normal copies of chromosome 6, in addition to the translocated portion of chromosome 6 (e.g., NXcP60 and NXcP142; Fig. 4 Left Insets). In addition to the two MBs with reciprocal translocations between chromosomes 12 and 13, another (NXcP25) had a nonreciprocal translocation t(7; 12) (Table 1). The one tumor that did not have a translocation involving either chromosome 13, 6, or 12 (or a combination), had a clonal t(2; 15) (NXcP67; Table 1). We conclude that inactivation of XRCC4 in p53-deficient nestin-expressing neuronal cells leads to the generation of MBs with strikingly recurrent chromosomal translocations.
Recurrent Gene Amplifications in NXc/−p53−/− MBs.
Certain human neuronal tumors, including MBs and neuroblastomas, often amplify the c-myc or N-myc genes (19, 21, 22). Therefore, we assayed NXc/−p53−/− MBs for Myc-family gene amplification by Southern blotting. These analyses revealed that N-myc (which is located near the centromere of chromosome 12) was amplified, often to very high levels, in 7 of 15 NXc/−p53−/− MB tumors, and c-myc was amplified in 1 of 15 (Fig. 3B and Table 1). Another Myc-family gene, L-myc, was not amplified in any tested MB (Fig. 3B and Table 1). The amplified N-myc genes, as assayed by FISH, usually occurred within chromosomes, sometimes in the context of complex translocations, but in some cases, within double minute chromosomes (Fig. 3C and Table 1). In the tumors analyzed, N-myc amplification generally correlated with greatly increased N-myc expression (Fig. 8B). Chromosomally amplified N-myc genes in metaphases from NXcP71 were observed occasionally within dicentric chromosomes (Table 1 and data not shown), suggesting that amplification may occur via a breakage-fusion-bridge cycle mechanism as observed for N-myc amplification in pro-B lymphomas in mice doubly deficient for Artemis and p53 (10).
To further assay for potential genomic amplifications or deletions, we performed array comparative genomic hybridization (CGH) on genomic DNA from the various MBs (Table 1 and Fig. 9, which is published as supporting information on the PNAS web site). These analyses confirmed amplification of regions of chromosomes 12 and 15 spanning the N-myc and c-myc genes in particular MBs and detected additional amplifications and deletions of sequences on chromosome 6 and chromosome 13 in MBs harboring t(13; 6) translocations (Fig. 9 and Table 1). To determine the region of chromosome 6 amplification, we performed FISH analyses with a series of bacterial artificial chromosome (BAC) probes spaced at 20-Mbp intervals over a chromosome 6 region that contains CyclinD2, a downstream target of the Shh pathway implicated in MB pathogenesis (11, 23) and found that Cyclin D2 was chromosomally amplified in two MBs harboring 13;6 translocations (NXcP142 and NXcP155; Table 1). Southern blotting confirmed Cyclin D2 amplification in these two tumors and in two additional tumors (NXcP57 and NXcP283; Table 1 and Fig. 8A). Cyclin D2 was expressed at high levels in several NXc/−p53−/− MBs (Fig. 8 B and C), with the highest levels in the two MBs with Cyclin D2 amplification (Table 1 and Fig. 8). Although deregulated activation of Wnt signaling can lead to increased Cyclin D2 (or N-myc) expression, β-catenin expression and localization appeared unperturbed in the NXc/−p53−/− MBs (data not shown).
Deletion of Patched in Most NXc/−p53−/− MBs.
By array CGH, we detected single-copy interstitial deletion or complete loss of the distal portion of chromosome 13 in the majority of the NXc/−p53−/− MBs (Table 1 and Fig. 9), which directly correlated with SKY analyses that showed the loss of this chromosomal region in the context of nonreciprocal translocations (Fig. 3A and Table 1). At least two factors, not mutually exclusive, might contribute to the high frequency of chromosome 13 translocations and deletions in NXc/−p53−/− MBs. There may be strong selection for the loss of chromosome 13 regions, and/or there might be a region of chromosome 13 that accumulates DSBs at increased frequency. To test these possibilities, we hybridized metaphases from several different MBs with chromosome 13 BAC probes spanning regions at 20 Mbp to 60 Mbp from the centromere of chromosome 13 (Fig. 3). The BAC at the 60-Mbp region also contained the Ptc tumor suppressor gene. These hybridizations revealed that chromosome 13 breakpoints were separated by tens of millions of bps in different MBs. The NXcP142 breakpoint lay between 40 and 60 Mbp, the NXcP60 breakpoint was located between 30 and 40 Mbp, and the NXcP81 breakpoint was between 20 and 30 Mbp from the centromere (Fig. 4). Despite the variability of chromosome 13 breakpoints, all tested found only one detectable copy of the Ptc gene (Fig. 4). In this context, based on CGH analyses, NXcP25, which does not contain a chromosome 13 translocation, must have an interstitial deletion of the Ptc locus (Table 1 and data not shown). These findings are consistent with the notion that chromosome 13 genetic alterations may be associated, at least in part, with a strong selective pressure for loss of a copy of Ptc in NXc/−p53−/− MBs.
Discussion
We have used conditional gene deletion to assay the effect of XRCC4 inactivation in nestin-expressing neural cells. Germ-line inactivation of XRCC4 leads to embryonic lethality and widespread apoptosis of newly generated neurons in a wave that temporally and spatially follows the onset and cessation of their differentiation from NPCs throughout the CNS (3). Although nestin-expression occurs in NPCs, NXc/− mice survive into adulthood, and we did not observe massive death of newly generated neurons. There could be several explanations for the differing effects of XRCC4 deletion in the germ line versus conditional deletion via Cre under the nestin promoter. One is that neuronal death results from noncell autonomous defects as observed in Rb deficiency (24). Another derives from our previous suggestion that lesions leading to increased neuronal apoptosis may happen at the NPC stage. Thus, incomplete deletion at the NPC stage or the persistence of XRCC4 protein for sometime after deletion may allow enough repair to permit neuronal survival. In this context, defects in the Ku NHEJ factor, which leads to less neuronal death than XRCC4 or Lig4 deficiency, allows mice to develop essentially normal nervous systems (25). We also note that we have not ruled out more subtle defects that may be found with more in-depth analyses.
Specifically eliminating XRCC4 in the NPCs of p53-deficient mice abrogates generation of aggressive pro-B lymphomas observed when XRCC4 and p53 are inactivated together in the germ line, allowing generation of a mouse model for MB. The routine, early onset of MBs in the NXc/−p53−/− background confirms that the lesions necessary to initiate these tumors occur in the context of conditional XRCC4 inactivation via nestin-Cre, even though such inactivation does not cause severe apoptosis in the developing nervous system. The question naturally arises as to why we observe meduloblastomas and not other neural tumors in NXc/−p53−/− mice. One possibility relates to the fact that NPCs in the cerebellum continue dividing and differentiating postnatally (11). However, MBs also were the only neuronal tumor observed in mice in which XRCC4 and p53 were deleted in the germ line, leading to the suggestion that the cerebellum is particularly sensitive to NHEJ deficiency, perhaps because the granule cell precursor population is the most abundant in the nervous system (9). Regardless, our finding that NXc/−p53−/− MBs routinely harbor recurrent genomic alterations clearly supports a role for NHEJ in the maintenance of genomic stability during neurogenesis.
We observed frequent N-myc amplification in NXc/−p53−/− MBs, with some amplifications occurring chromosomally in the absence of obvious translocations, others chromosomally in the context of complicons, and yet others extrachromosomally within double minutes. In NHEJ/p53-deficient pro-B lymphomas, all c-myc or N-myc amplifications occurred within complicons. Such apparent differences in the cytogenetic location of the amplified Myc genes in NHEJ/p53-deficient pro-B and MB tumors, if confirmed by additional analyses, might reflect various factors, including the nature of initiating lesions (e.g., G1 phase RAG DSBs in pro-B cells) or the generally higher degree of amplification in MBs. In any case, N-myc amplification frequently occurs in human neural tumors, including MBs, retinoblastomas, and neuroblastomas (21, 22), again sometimes coupled with translocation to a different chromosome or within double minutes (21). There are several factors, not mutually exclusive, that could contribute to this outcome. One is the high expression of N-myc in the developing nervous system (21, 26). Another would be related to the ability of N-myc to promote proliferation and inhibit differentiation of NPCs into neurons (27). In this regard, N-myc is essential for cerebellar organogenesis and to drive proliferation of granule cell precursors within the EGL in response to Shh signaling (28). Yet another possibility is that N-myc is in a chromosomal region prone to DSBs (21). The frequent amplification of N-myc in Artemis/53-deficient pro-B tumors versus c-myc in other NHEJ/p53-deficient pro-B tumors is consistent with the latter possibility (10).
Unrepaired DSBs due to impaired NHEJ likely leads to the recurrent clonal translocations found in the NXc/−p53−/− NHEJ-deficient background. In NHEJ/p53-deficient lymphoid malignancies, translocation breakpoints were localized to narrow chromosomal regions on chromsome 12 because they were initiated by RAG activity at the Ig heavy chain locus (6). In contrast, breakpoints of recurrent chromosome 13 translocations in NXc/p53−/− MB were distributed over a wide chromosomal region, but all resulted in the loss of the distal chromosome 13 material and the Ptc gene. Because loss of Ptc has been reported for both human MBs and MBs arising in several mouse models (11), the frequent occurrence of chromosome 13 translocations leading to deletion of Ptc in NXc/−p53−/− MB likely reflects strong positive selection for the loss of this gene. The frequent occurrence of chromosome 6 as a partner for chromosome 13 translocations also was striking. Again, the chromosome 6 translocations may alter a gene that confers a selective advantage. In that regard, Cyclin D2, which resides on chromosome 6, is sometimes, but not always, amplified in NXc/−p53−/− MBs with chromosome 6 translocations. Further elucidation of chromosome 6 breakpoints will be necessary to determine whether particular regions of chromosome 6 are involved in the translocations, which might be indicative of selection for alteration of specific genes or, alternatively, of fragile sites or other types of recurrent lesions that could generate recombinagenic DSBs. Further breakpoint analyses also may reveal their potential relationship, if any, to factors that lead to widespread neuronal apoptosis when NHEJ is absent in the germ line.
Methods
Conditional Inactivation of XRCC4 in Mice.
The conditional XRCC4 (Xc) allele was generated by targeting a construct in which exon3 of XRCC4 is flanked by loxP sites into TC1 (129sv/ev) ES cells (Fig. 5). Xc/+ and X+/− ES cells, generated by transient Cre-mediated deletion of the NeoR marker, were transmitted into the germ line. X−/− ES cells were generated by high G418 selection and deletion of NeoR marker as described in ref. 29. X+/− mice also were bred into a p53−/− background (30) to generate X−/−p53−/− mice. Xc/+ and X+/− mice were bred to nestin-Cre transgenic (27) and p53−/− mice (30) and genotyped by Southern blotting with XRCC4 (3), Cre (27), and p53 (30) probes.
Histology and Immunostaining.
Mouse tissues were fixed in Bouins/10% formalin, embedded in paraffin, sectioned, stained with hematoxylin/eosin, or immunostained after antigen retrieval by following standard methods. Immunohistological analyses were performed by using antibodies to glial fibrillary acidic protein (Chemicon), Synaptophysin (Vector Laboratories), NeuN (Chemicon), MIB1/Ki67 (Vector/Novocastra, Burlingame, CA) and β-catenin (Cell Signaling Technology, Beverly, MA). XRCC4 (Santa Cruz Biotechnology) and β3-tubulin (Tuj1, Babco, Richmond, CA) staining of OCT frozen brain sections, detected with secondary antibodies (FITC or Cy5 conjugates) (Jackson ImmunoResearch), and counterstained with DAPI (Sigma).
Cell Culture, Western Blot, and Molecular Analyses.
Tumor cells were separated into neurospheres by trituration and cultured in serum-free defined media (31) or DMEM media containing 15% FCS and 2× leukocyte inhibitory factor. Western blot analyses were performed by using antibodies to XRCC4 (Santa Cruz Biotechnology), actin (Santa Cruz Biotechnology), Cyclin D2 (Santa Cruz Biotechnology), and tubulin (Sigma). Lymphoid cells were analyzed by flow cytometry as described in ref. 10. Probes for Southern and Northern blots: N-myc, c-myc, Ig heavy chain JH, and LR8 were described in ref. 10. The L-myc genomic probe and GAPDH cDNA were obtained by PCR based on published sequences. Cyclin D2 cDNA probe was a gift from Peter Sicinski's laboratory (Harvard Medical School, Boston). CGH analyses were performed with HindIII- and XbaI-digested biotinylated tumor and reference genomic DNAs on Affymetrix GeneChip MOE 430_2 arrays as described for SNP genotyping on the Affymetrix 100K SNP array pair. Two custom chip description files (cdf) were written: One file defined each probe set as group of four or more probes, which all reside in the same, 4,000 bp or less, XbaI fragment, the other cdf made the same specification for HindIII fragments, and computational analyses were performed by using dchip software (available upon request).
Cytogenetic Analysis of MBs.
Tumors were incubated in serum-free media (31) medium with 100 nM colcemid (Karyo-Max, Invitrogen) for 3–5 h, dissociated, and fixed as described in ref. 32. SKY and FISH were performed by following standard protocols (6). N-myc and c-myc probes were described in ref. 10; Cyclin D2, patched, and additional chromosome 6 and 13 BAC probes were obtained from BACPAC Resources Center (Oakland, CA).
Supplementary Material
Acknowledgments
We thank Sonia Franco, Catherine Quigley, David Lombard, Peter Sicinski, Yuko Fujiwara, Nicole Stokes, Tiffany Borjeson, Alyssa Riley, Laurie Davidson, Laurence Daheron, Cosmos Giallourakis (all at Harvard Medical School); Jean-Baptiste Telliez (Wyeth Research, Cambridge, MA); and Thomas Perlot (Harvard Medical School) for helpful comments, reagents, and assistance and Ron DePinho and Irving Weissman for critical evaluation of the manuscript. This work was supported by National Institutes of Health (NIH) grants (to F.W.A.), NIH Postdoctoral Training Grant T32 CA70083 (to C.T.Y.), American Association of Cancer Research Fellowship PF-05-151-01-MGO (to D.K.), and NIH Grant 1P01-CA10990 (to E.F.). F.W.A. is an investigator of the Howard Hughes Medical Institute. R.M. is a laboratory principal investigator of G.M.
Abbreviations
- BAC
bacterial artificial chromosome
- CGH
comparative genomic hybridization
- DSB
DNA double-strand break
- MB
medulloblastoma
- NHEJ
nonhomologous DNA end-joining
- NPC
neuronal progenitor cell
- pro-B
progenitor B cell
- SKY
spectral karyotyping
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Bassing C. H., Alt F. W. DNA Repair (Amst.) 2004;3:781–796. doi: 10.1016/j.dnarep.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 2.Rooney S., Chaudhuri J., Alt F. W. Immunol. Rev. 2004;200:115–131. doi: 10.1111/j.0105-2896.2004.00165.x. [DOI] [PubMed] [Google Scholar]
- 3.Gao Y., Sun Y., Frank K. M., Dikkes P., Fujiwara Y., Seidl K. J., Sekiguchi J. M., Rathbun G. A., Swat W., Wang J., et al. Cell. 1998;95:891–902. doi: 10.1016/s0092-8674(00)81714-6. [DOI] [PubMed] [Google Scholar]
- 4.Frank K. M., Sharpless N. E., Gao Y., Sekiguchi J. M., Ferguson D. O., Zhu C., Manis J. P., Horner J., DePinho R. A., Alt F. W. Mol. Cell. 2000;5:993–1002. doi: 10.1016/s1097-2765(00)80264-6. [DOI] [PubMed] [Google Scholar]
- 5.Gao Y., Ferguson D. O., Xie W., Manis J. P., Sekiguchi J., Frank K. M., Chaudhuri J., Horner J., DePinho R. A., Alt F. W. Nature. 2000;404:897–900. doi: 10.1038/35009138. [DOI] [PubMed] [Google Scholar]
- 6.Zhu C., Mills K. D., Ferguson D. O., Lee C., Manis J., Fleming J., Gao Y., Morton C. C., Alt F. W. Cell. 2002;109:811–821. doi: 10.1016/s0092-8674(02)00770-5. [DOI] [PubMed] [Google Scholar]
- 7.Padilla-Nash H. M., Heselmeyer-Haddad K., Wangsa D., Zhang H., Ghadimi B. M., Macville M., Augustus M., Schrock E., Hilgenfeld E., Ried T. Genes Chromosomes Cancer. 2001;30:349–363. doi: 10.1002/gcc.1101. [DOI] [PubMed] [Google Scholar]
- 8.Sharpless N. E., Ferguson D. O., O'Hagan R. C., Castrillon D. H., Lee C., Farazi P. A., Alson S., Fleming J., Morton C. C., Frank K., et al. Mol. Cell. 2001;8:1187–1196. doi: 10.1016/s1097-2765(01)00425-7. [DOI] [PubMed] [Google Scholar]
- 9.Lee Y., McKinnon P. J. Cancer Res. 2002;62:6395–6399. [PubMed] [Google Scholar]
- 10.Rooney S., Sekiguchi J., Whitlow S., Eckersdorff M., Manis J. P., Lee C., Ferguson D. O., Alt F. W. Proc. Natl. Acad. Sci. USA. 2004;101:2410–2415. doi: 10.1073/pnas.0308757101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marino S. Trends Mol. Med. 2005;11:17–22. doi: 10.1016/j.molmed.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 12.Wetmore C., Eberhart D. E., Curran T. Cancer Res. 2001;61:513–516. [PubMed] [Google Scholar]
- 13.Lee Y., Miller H. L., Jensen P., Hernan R., Connelly M., Wetmore C., Zindy F., Roussel M. F., Curran T., Gilbertson R. J., McKinnon P. J. Cancer Res. 2003;63:5428–5437. [PubMed] [Google Scholar]
- 14.Tong W. M., Ohgaki H., Huang H., Granier C., Kleihues P., Wang Z. Q. Am. J. Pathol. 2003;162:343–352. doi: 10.1016/S0002-9440(10)63825-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bassing C. H., Suh H., Ferguson D. O., Chua K. F., Manis J., Eckersdorff M., Gleason M., Bronson R., Lee C., Alt F. W. Cell. 2003;114:359–370. doi: 10.1016/s0092-8674(03)00566-x. [DOI] [PubMed] [Google Scholar]
- 16.Celeste A., Difilippantonio S., Difilippantonio M. J., Fernandez-Capetillo O., Pilch D. R., Sedelnikova O. A., Eckhaus M., Ried T., Bonner W. M., Nussenzweig A. Cell. 2003;114:371–383. doi: 10.1016/s0092-8674(03)00567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uziel T., Zindy F., Xie S., Lee Y., Forget A., Magdaleno S., Rehg J. E., Calabrese C., Solecki D., Eberhart C. G., et al. Genes Dev. 2005;19:2656–2667. doi: 10.1101/gad.1368605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graus-Porta D., Blaess S., Senften M., Littlewood-Evans A., Damsky C., Huang Z., Orban P., Klein R., Schittny J. C., Muller U. Neuron. 2001;31:367–379. doi: 10.1016/s0896-6273(01)00374-9. [DOI] [PubMed] [Google Scholar]
- 19.Sarkar C., Deb P., Sharma M. C. Childs Nerv. Syst. 2005;21:272–293. doi: 10.1007/s00381-004-1066-4. [DOI] [PubMed] [Google Scholar]
- 20.Donner L. R. Acta Neuropathol. 2005;109:543–551. doi: 10.1007/s00401-005-0986-8. [DOI] [PubMed] [Google Scholar]
- 21.Kohl N. E., Kanda N., Schreck R. R., Bruns G., Latt S. A., Gilbert F., Alt F. W. Cell. 1983;35:359–367. doi: 10.1016/0092-8674(83)90169-1. [DOI] [PubMed] [Google Scholar]
- 22.Schwab M. Semin. Cancer Biol. 1999;9:319–325. doi: 10.1006/scbi.1999.0126. [DOI] [PubMed] [Google Scholar]
- 23.Park P. C., Taylor M. D., Mainprize T. G., Becker L. E., Ho M., Dura W. T., Squire J., Rutka J. T. J. Neurosurg. 2003;99:534–541. doi: 10.3171/jns.2003.99.3.0534. [DOI] [PubMed] [Google Scholar]
- 24.MacPherson D, Sage J., Crowley D., Trumpp A., Bronson R. T., Jacks T. Mol. Cell. Biol. 2003;23:1044–1053. doi: 10.1128/MCB.23.3.1044-1053.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu Y., Sekiguchi J., Gao Y., Dikkes P., Frank K., Ferguson D., Hasty P., Chun J., Alt F. W. Proc. Natl. Acad. Sci. USA. 2000;97:2668–2673. doi: 10.1073/pnas.97.6.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohl N. E., Gee C. E., Alt F. W. Science. 1984;226:1335–1337. doi: 10.1126/science.6505694. [DOI] [PubMed] [Google Scholar]
- 27.Knoepfler P. S., Cheng P. F., Eisenman R. N. Genes Dev. 2002;16:2699–2712. doi: 10.1101/gad.1021202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kenney A. M., Cole M. D., Rowitch D. H. Development (Cambridge, U.K.) 2003;130:15–28. doi: 10.1242/dev.00182. [DOI] [PubMed] [Google Scholar]
- 29.Mortensen R. M., Conner D. A., Chao S., Geisterfer-Lowrance A. A., Seidman J. G. Mol. Cell. Biol. 1992;12:2391–2395. doi: 10.1128/mcb.12.5.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Donehower L. A., Harvey M., Slagle B. L., McArthur M. J., Montgomery C. A., Jr, Butel J. S., Bradley A. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 31.Svendsen C. N., Caldwell M. A., Ostenfeld T. Brain Pathol. 1999;9:499–513. doi: 10.1111/j.1750-3639.1999.tb00538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rehen S. K., McConnell M. J., Kaushal D., Kingsbury M. A., Yang A. H., Chun J. Proc. Natl. Acad. Sci. USA. 2001;98:13361–13366. doi: 10.1073/pnas.231487398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.