Abstract
Regulation of gene expression by tissue-specific transcription factors involves both turning on and turning off transcription of target genes. Runx3, a runt-domain transcription factor, regulates cell-intrinsic functions by activating and repressing gene expression in sensory neurons, dendritic cells (DC), and T cells. To investigate the mechanism of Runx3-mediated repression in an in vivo context, we generated mice expressing a mutant Runx3 lacking the C-terminal VWRPY, a motif required for Runx3 interaction with the corepressor Groucho/transducin-like Enhancer-of-split (TLE). In contrast with Runx3−/− mice, which displayed ataxia due to the death of dorsal root ganglia TrkC neurons, Runx3VWRPY−/− mice were not ataxic and had intact dorsal root ganglia neurons, indicating that ability of Runx3 to tether Groucho/TLE is not essential for neurogenesis. In the DC compartment, the mutant protein Runx3VWRPY− promoted normally developed skin Langerhans cells but failed to restrain DC spontaneous maturation, indicating that this latter process involves Runx3-mediated repression through recruitment of Groucho/TLE. Moreover, in CD8+ thymocytes, Runx3VWRPY− up-regulated αE/CD103-like WT Runx3, whereas unlike wild type, it failed to repress αE/CD103 in CD8+ splenocytes. Thus, in CD8-lineage T cells, Runx3 regulates αE/CD103 in opposing regulatory modes and recruits Groucho/TLE to facilitate the transition from activation to repression. Runx3VWRPY− also failed to mediate the epigenetic silencing of CD4 gene in CD8+ T cells, but normally regulated other pan-CD8+ T cell genes. These data provide evidence for the requirement of Groucho/TLE for Runx3-mediated epigenetic silencing of CD4 and pertain to the mechanism through which other Runx3-regulated genes are epigenetically silenced.
Keywords: αE/CD103 gene repression, CD4 silencer, CD8 lineage lymphocytes, epigenetic silencing, transcriptional repression
Mammalian Runx3 is one of three transcription factors that comprise the RUNX family (1–4). RUNX proteins regulate lineage-specific gene expression in developmental pathways (2, 3) and also could be involved in autoimmune diseases (5). Loss of Runx3 function is associated with defects in neurogenesis and thymopoiesis, and with the development of colitis, gastritis, and asthma-like features (6–13). Regulation of gene expression by tissue-specific transcription factors involves both turning on and turning off transcription of target genes. Runx3 acts as a bifunctional regulator, which up-regulates but also down-regulates, gene expression (14). How does Runx3 act in vivo both as an activator and a repressor of target genes?
It is believed that a DNA-bound Runx3 elicits repression by tethering corepressors such as the transducin-like Enhancer-of-split (TLE) (15), the mammalian homolog of Drosophila Groucho (Gro) (16), to a subset of target promoters (14, 17, 18). However, the biological significance and in vivo targets of Runx3-mediated transcriptional repression are largely unknown. We (19) and others (20) have shown that both Runx1 and Runx3 interact with the corepressor Gro/TLE through a conserved motif of five amino acids VWRPY, located at the C terminus of RUNX proteins. Gene knockouts of Runx1 and Runx3 demonstrated that during thymopoiesis, they act as transcriptional repressors of CD4 and as growth regulators of CD8-lineage T lymphocytes (11, 12, 21). Rescue experiments by using in vitro-cultured fetal liver cells and knock-in chimera mice have indicated that the VWRPY motif of Runx1 plays a role during early T cell development in regulation of CD4 expression and T cell homeostasis (22, 23). On the other hand, using enforced expression by a retroviral system in organ cultures indicated that the VWRPY motif was not required for either Runx1- or Runx3-mediated repression of CD4 (24). To resolve this discrepancy and study the mechanism of Runx3-mediated repression of negatively regulated target genes in vivo, we generated mice expressing a mutant Runx3 lacking the VWRPY motif (Runx3VWRPY−).
Using mice homozygous for the mutant allele (Runx3VWRPY−/− mice) in comparison with WT and null (Runx3−/−) mice, we derived previously unavailable information on the positive and negative functions of Runx3 in the in vivo context of the animal. Gro/TLE-dependent and -independent functions of Runx3 now are demonstrated in dorsal root ganglia (DRG) TrkC sensory neurons, dendritic cells (DC), and CD8-lineage T lymphocytes, and CD8+ T cell-specific target genes, whose regulation by Runx3 requires the recruitment of Gro/TLE, are identified.
We show that in contrast to Runx3−/− mice that display ataxia and growth retardation (9), Runx3VWRPY−/− mice grew normally and were not ataxic, indicating that the ability of Runx3 to recruit Gro/TLE is not essential for these processes. Runx3 plays an important role in DC development (7). DC are bone marrow-derived cells specialized in uptake, processing, and presentation of antigens to T cells (25) and play an important role in maintenance of self-tolerance (26). Tissue-resident DC are normally maintained at an immature state by immunosuppressive cytokines such as TGF-β, secreted by the surrounding cellular environment (26). In DC compartment, Runx3 functions as a component in the TGF-β signaling pathway and has a dual role: It promotes development of epidermal Langerhans cells (LC), a distinct skin DC population, and restrains maturation of tissue-resident DC (7). Runx3VWRPY− was able to promote LC development but failed to restrain DC maturation, indicating that this latter function involves interactions of Runx3 with Gro/TLE.
During T cell development, the mutant Runx3VWRPY− positively regulated αE/CD103 expression in CD8+ thymocytes, as was previously reported for WT Runx3 (27). However, unlike WT Runx3, the mutant failed to repress αE/CD103 in peripheral CD8+ T cells. These data demonstrate that within the same cell lineage, Runx3 regulates the same gene in opposing regulatory modes, and that the mechanism underlying the transition from activation to repression requires recruitment of Gro/TLE. Moreover, in developing CD8+ T cells, Runx3VWRPY− was unable to repress the CD4 gene, which is otherwise epigenetically silenced (11, 28–30) but regulated normally the expression of other pan-CD8+ T-cell genes. These data provide evidence for Gro/TLE requirement for Runx3-mediated epigenetic silencing of CD4 and pertain to the mechanism through which other Runx3-regulated genes are epigenetically silenced.
Results
Expression of Runx3VWRPY− Protein and Phenotypic Features of Runx3VWRPY−/− Mice.
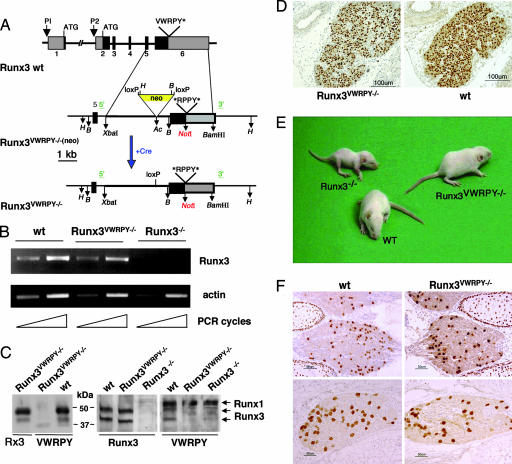
To obtain Runx3VWRPY−/− mice, we first generated a mutant allele Runx3VWRPY−, in which the codons of the five C-terminal amino acids VWRPY, were changed to the stop codon UAG and to codons encoding substituted amino acids designed to create a new NotI site (Fig. 1 A; see also Fig. 5, which is published as supporting information on the PNAS web site). To facilitate selection of positive ES cells, a lox-P-flanked neomycin (neo) cassette was inserted into intron no. 5 (Fig. 1A). Chimeric mice were generated and used to pass on the mutant Runx3VWRPY− through the germ line. To eliminate potential phenotypic effects caused by the neo gene, it was removed by crossing heterozygous Runx3VWRPY− mice onto a PGK-Cre transgenic mice (ref. 32; Fig. 1A). RT-PCR and Western blot analyses were used to demonstrate expression of Runx3VWRPY− mRNΑ and protein (Fig. 1 B and C). The absence of VWRPY motif in Runx3 protein, derived from the mutant allele, was confirmed by using antibodies specific to the VWRPY pentapeptide (Fig. 1C). Immunohistochemistry on sections from Runx3VWRPY−/− mice, homozygous to the VWRPY minus mutant allele, revealed thymus architecture and staining pattern of Runx3VWRPY− that were similar to WT (Fig. 1D).
Fig. 1.
Runx3VWRPY−/− mice expressing the mutant protein display a normal outward phenotype. (A) Disruption of the Gro/TLE binding site (VWRPY) in Runx3. A scheme is shown of Runx3 genomic locus, targeting vector, and mutant allele. The targeting vector spans the XbaI-BamHI fragment of intron 5 and exon 6. Relevant restriction sites are shown (H, HindIII; B, BglII; Ac, AccI). The 3′ and 5′ external probes used for the Southern blot analysis, shown in Fig. 5, are marked in green. The floxed neo gene later was removed, giving rise to Runx3VWRPY−. (B) Semiquantitative RT-PCR analysis of RNA isolated from thymocytes of WT, Runx3−/−, and Runx3VWRPY−/− mice. Representative results of three independent experiments by using RNA obtained from three different mice are shown. Similar results were obtained by using RNA isolated from spleens of these mice. (C) Western blot analysis of proteins extracted from HEK-293 cells (Left) transfected with vectors expressing either WT Runx3 or Runx3VWRPY− cDNAs (Center) (CMV-Runx3 and CMV- Runx3VWRPY− in Fig. 6) and from the spleen (Right) of WT, Runx3−/− and Runx3VWRPY−/− mice. Blots were reacted with antibodies against either Runx3 (31) or VWRPY as designated. Arrows indicate the 48- and 46-kDa Runx3 proteins and the 50-kDa Runx1 protein, which reacts with anti-VWRPY Ab. (D) Immunostaining with anti-Runx3 antibodies of thymus sections from Runx3VWRPY−/− and WT mice showing similar patterns of Runx3 staining. (Magnification: ×10.) (E) Picture depicts 1-month-old WT, Runx3−/−, and Runx3VWRPY−/− mice. Runx3−/− mice are smaller and have a clearly recognizable phenotype, characterized by severe ataxia and posture abnormalities, whereas WT and Runx3VWRPY−/− mice have indistinguishable phenotypes. (F) Immunostaining of DRG sections from WT and Runx3VWRPY−/− embryonic day 14.5 embryos with anti-Runx3 (Upper) and embryonic day 17.5 embryos with anti-parvalbumin (Lower) showing similar patterns of immunostaining (Magnification: ×20.)
Previous studies showed that newborn Runx3−/− mice had sensory ataxia, due to the death of TrkC+ neurons in DRG, and that mice displayed a reduced growth rate (8, 9). In contrast, Runx3VWRPY−/− mice had an apparent phenotype indistinguishable from WT mice; they were not ataxic and displayed normal growth (Fig. 1E). Further analysis showed that contrary to Runx3−/− mice, which lack expression of Runx3 (9), expression of Runx3VWRPY− protein in DRG of Runx3VWRPY−/− mice was similar to that in WT mice (Fig. 1F), indicating that TrkC+ neurons in Runx3VWRPY−/− mice are viable. More convincingly, immunostaining of the calcium-binding protein parvalbumin, a marker of TrkC+ neurons (9, 33), gave a similar pattern in DRG from WT and Runx3VWRPY−/− mice (Fig. 1F). We conclude that Runx3VWRPY− is capable of supporting neurogenesis of TrkC+ neurons when expressed from its endogenous locus and that Runx3 capacity to recruit Gro/TLE is not essential for this process.
Enhanced Spontaneous Maturation of Runx3VWRPY−/− DC.
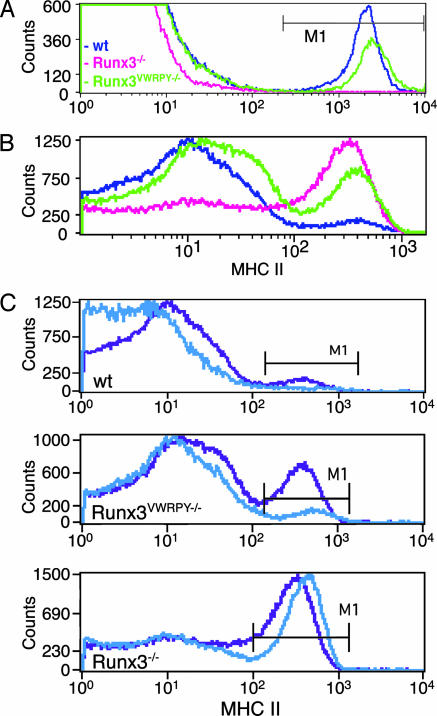
In addition to its function in fate determination of DRG sensory neurons, Runx3 plays a role in development and maturation of skin LC and tissue DC, where it functions as a component in TGF-β signaling pathway (7). TGF-β plays a dual role in the LC/DC compartment: it promotes development of epidermal LC and inhibits maturation of DC. When Runx3 function was lost, Runx3−/− mice lacked epidermal LC, and their DC did not respond to TGF-β-induced maturation inhibition and spontaneously matured (7). Unlike Runx3−/−, skin epidermis of Runx3VWRPY−/− mice contained abundant LC similar to WT littermate mice (Fig. 2A). But in contrast to WT DC, which exhibited a low level of spontaneous maturation, a substantial proportion (≈50%) of Runx3VWRPY−/− DC spontaneously matured, resembling the Runx3−/− phenotype (Fig. 2B). Even exogenously added TGF-β did not completely inhibit the spontaneous maturation of Runx3VWRPY−/− DC (Fig. 2C). Thus, Runx3VWRPY− activity was sufficient to promote TGF-β-dependent development of skin LC but was insufficient to promote TGF-β-induced maturation inhibition of DC. These data show that during LC/DC development, Runx3 mediates both positive and negative cues of TGF-β, some of which require engagement of Gro/TLE.
Fig. 2.
Runx3VWRPY− fully supports the development of LC but is less efficient than WT in restraining the spontaneous maturation of DC. (A) Epidermal LC highly express MHC II (7). Cells from epidermal sheaths of WT, Runx3VWRPY−/−, and Runx3−/− mice were isolated, stained with anti-MHC II mAb, and analyzed by FACS. LC (MHC IIhigh) were present in WT and Runx3VWRPY−/− mice (7.0% and 5.1%, respectively) but not in Runx3−/− mice (0.2%). (B) Runx3VWRPY−/− DC display enhanced spontaneous maturation. Bone marrow-derived DC grown for 8 days were stained with anti-CD11c and anti-MHC II mAb and analyzed by FACS. MHC II levels in CD11c+-gated cells are shown. The average frequencies and SE of mature DC in four independent experiments were as follows: 15.6 ± 2.5% for WT, 47.4 ± 7.3% for Runx3VWRPY−/− and 64.8 ±5.5% for Runx3−/−. (C) Bone marrow-derived DC grown for 8 days in the presence (blue) or absence (purple) of TGF-β (10 ng/ml) were stained with anti-CD11c and anti-MHC II mAb and analyzed by FACS. Shown are MHC II levels in CD11c+-gated cells of WT, Runx3VWRPY−/−, and Runx3−/− mice.
Runx3-Mediated Silencing of the CD4 Gene Requires Recruitment of Gro/TLE.
During thymopoiesis,CD4+CD8+ double-positive thymocytes differentiate into mature single-positive (SP) CD4+ or SP CD8+ cells (34). In SP CD8+ cells, the expression of CD4 is transcriptionally repressed (30, 35). A region of 430 bp, known as the CD4 silencer, is required for this process (29, 30, 36–38). This silencer encompasses two functional Runx-binding sites, which are essential for the irreversible epigenetic silencing of CD4 in mature SP CD8+ cells (11, 28–30). Runx3 is highly expressed in SP CD8+ and, when lost, transcriptional silencing of CD4 is impaired, leading to accumulation of an abnormal population of mature CD8+ T cells in which expression of CD4 is not repressed (11, 12, 21).
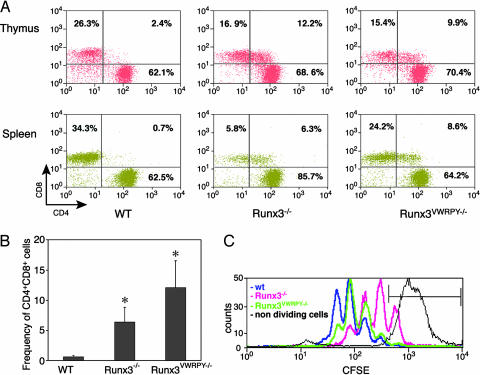
We assessed the distribution of CD4/CD8 among mature (TCRhighHSA−/low) T cells in thymus and spleen of Runx3VWRPY−/− mice in comparison with that of WT and Runx3−/− mice. A profound increase in the proportion of mature CD8+ T cells that also expressed CD4 was observed in both thymus and spleen of Runx3VWRPY−/− mice compared with WT mice (Fig. 3A). The distribution of mature CD4+/CD8+ thymocytes in Runx3VWRPY−/− mice was similar to that in Runx3−/− mice and markedly different from that in WT mice (Fig. 3A Upper; refs. 11 and 12). Interestingly, among splenocytes, the frequency of CD4+CD8+ in Runx3VWRPY−/− mice was even higher than that in Runx3−/− mice (Fig. 3 A Lower). The average (n = 5) frequency of CD4+CD8+ splenic T cells in Runx3VWRPY−/− and Runx3−/− mice was 12.0 ± 4.4% and 6.4 ± 2.4%, respectably, compared with 0.67 ± 0.2% in WT mice (Fig. 3B). These data unequivocally show that silencing of CD4 by Runx3 requires the recruitment of the corepressor Gro/TLE. It also indicates that CD4 expression was significantly less repressed in CD8+ splenocytes of Runx3VWRPY−/− mice as compared with splenocytes of Runx3−/− mice.
Fig. 3.
Recruitment of Gro/TLE is required for Runx3-mediated CD4 silencing but not for Runx3-dependent CD8+ T cell proliferation. (A) Mature thymic and splenic CD8+ T cells of Runx3VWRPY−/− mice also expressed CD4. Thymocytes (Upper) and splenocytes (Lower) from WT, Runx3VWRPY−/−, and Runx3−/− mice were stained with anti-TCR-β, anti-HSA, anti-CD8α, and anti-CD4 mAb and analyzed by FACS. Mature cells were gated as TCRhighHSA−/low. The frequencies of SP CD8, SP CD4, and DP CD4+CD8+ populations are indicated. A representative experiment of five independent analyses is shown. Note the profound reduction of splenic CD8+ T cells in Runx3−/− but not in Runx3VWRPY−/− or WT mice. (B) Frequency of mature splenic CD8+ cells that also expressed CD4 is significantly (∗) increased in Runx3VWRPY−/− (n = 5; P < 0.0001) and Runx3−/− mice (n = 5; P < 0.0003) compared with WT. Frequency of splenic CD4+CD8+ in Runx3VWRPY−/− mice was significantly higher (n = 5; P < 0.015) compared with Runx3−/− mice. (C) Runx3VWRPY−/− splenic CD8+ T cells display normal proliferation. 5,6-Carboxyfluorscein diacetate, succinimidyl ester (CFSE) cell division assay of CD8+ splenocytes isolated from WT (blue), Runx3VWRPY−/− (green), and Runx3−/− (pink) mice. The black line represents a control experiment of cells incubated without anti-CD3 and IL2 and, thus, were nondividing. The overall cell division rate of WT or Runx3VWRPY−/− was similar, whereas that of Runx3−/− cells was much lower. Cells from WT or Runx3VWRPY−/− mice underwent four population divisions compared with only three divisions of Runx3−/− cells. After 72 h in culture, >20% of Runx3−/− cells remained undivided compared with ≈1% and 2% of WT and Runx3VWRPY−/−, respectively.
Using transfection assays, we also assessed the ability of Runx3 to negatively regulate the CD4 silencer in conjunction with Gro/TLE. Reporter constructs, in which human elongation factor promoter, with or without CD4 silencer, regulate luciferase (luc) transcription, were cotransfected into two cell lines (COS-7 and HEK-293) with constructs expressing TLE1 (CMV-TLE1) and constructs expressing either WT Runx3 (CMV-Runx3) or mutant Runx3 (CMV-Runx3VWRPY−) (Fig. 6, which is published as supporting information on the PNAS web site). WT Runx3 was significantly more active than Runx3VWRPY− in reducing CD4 silencer-derived luc activity (Fig. 6), indicating that in the context of the CD4 silencer, Runx3-mediated transcriptional repression largely depends on its ability to tether Gro/TLE.
Besides silencing of CD4, Runx3 also regulates other CD8-T cell-specific genes (39, 40). Hence, in the absence of Runx3 function, proliferative capacity of peripheral Runx3−/− CD8+ T cells was impaired, resulting in a profound reduction in the proportion of splenic CD8+ T cells (Fig. 3A; refs. 11 and 12). In contrast, no significant difference in proportion of CD8+ T cells (Fig. 3A) or in cell proliferation ability (Fig. 3D) was noted between Runx3VWRPY−/− and WT. These data show that in developing CD8+ T cells, silencing of CD4 and cell proliferative capacity are two independent Runx3-mediated processes. When Runx3 ability to recruit Gro/TLE is lost, repression of CD4 is impaired, but cell proliferation remains intact, underscoring the fact that Runx3 can function as a positive regulator in one context and as a negative regulator in another.
Runx3 in Conjunction with Gro/TLE Down-Regulates αE/CD103 in Peripheral CD8+ T Cells.
Grueter et al. (27) have recently shown that Runx3 positively regulates the expression of the αE/CD103 integrin during T cell development and that knockdown of Runx3 markedly reduced the frequency of αE/CD103-expressingCD8+ T cells. αE/CD103 mediates the interaction with E-cadherin on epithelial cells and is normally expressed on 80–90% of mature CD8+ thymocytes and on a much lower proportion (40–50%) of CD8+ splenocytes (27, 41).
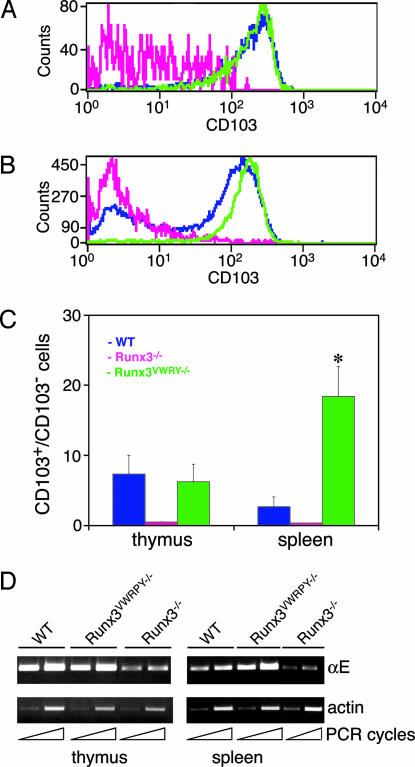
We assessed the ability of Runx3VWRPY− to positively regulate expression of αE/CD103. More than 90% of SP CD8+ thymocytes in WT and Runx3VWRPY−/− mice expressed αE/CD103 compared with <10% in Runx3−/− mice (Fig. 4A). This finding is not only consistent with data obtained by Grueter et al. (27), but it also implies that similarly to WT, Runx3VWRPY− can positively regulate the expression of αE/CD103. Compared with thymocytes, there was a lower proportion of CD8+CD103+ splenocytes in WT mice and even lower proportion in Runx3−/− mice (Fig. 4B), in full agreement with the published data (27). Strikingly, however, in Runx3VWRPY−/− mice, the proportion of CD8+CD103+ splenocytes was not reduced compared with thymocytes, and >90% of Runx3VWRPY−/− splenocytes expressed CD103 (Fig. 4B). In fact, the ratio of CD103+/CD103− splenocytes in Runx3VWRPY−/− mice was ≈9-fold higher compared with WT, whereas in thymocytes, the ratio was similar (Fig. 4C). Of note, the fact that Runx3VWRPY−/− splenocytes express higher-than-WT levels of αE/CD103 implies that the normally reduced expression of αE/CD103 in splenic CD8+ T cells (27, 41) is not due to a pause in transcriptional activation but rather to a down-regulation of αE/CD103 expression. These data show that in peripheral CD8+ T cells, Runx3, in conjunction with Gro/TLE, down-regulates αE/CD103 expression. This conclusion is further supported by the marked decline of αE/CD103 expression that was observed in proliferating cultured splenocytes from WT but not from Runx3VWRPY−/− mice (Fig. 7, which is published as supporting information on the PNAS web site). Thus, during development of CD8-lineage T cells, Runx3 functions both as a positive and negative regulator of the same gene and must recruit Gro/TLE to accomplish the transition.
Fig. 4.
Runx3VWRPY− activates expression of αE/CD103 in CD8+ thymocytes but fails to down-regulate αE/CD103 expression in peripheral CD8+ T cells. (A) αE/CD103 expression in thymocytes from 2-month-old WT (blue), Runx3VWRPY−/− (green), and Runx3−/− (pink) mice. Cells were stained with anti-CD8 and anti-CD103 mAb and analyzed by FACS. CD103 expression is shown in SP CD8 thymocytes. (B) αE/CD103 expression in CD8+ splenocytes from 2-month-old WT, Runx3VWRPY−/−, and Runx3−/− mice, color-coded and analyzed as in A. (C) The average ratio and SE of CD103+/CD103− in CD8+ thymocytes (n > 3) and splenocytes (n > 5) of WT (blue), Runx3VWRPY−/− (green), and Runx3−/− (pink) mice. The average proportion of CD103+ in CD8+ splenocytes of Runx3VWRPY−/− mice was significantly (∗) higher than in WT mice (P = 0.003). (D) Runx3 regulates αE/CD103 expression through transcriptional regulation of the αE gene. RT-PCR (Upper) representative results of two and three independent experiments by using independently derived cDNAs of thymus and spleen RNA, respectively, are shown. In thymocytes, the level of αE transcript was similar in Runx3VWRPY−/− and WT, whereas in splenocytes, it was higher in Runx3VWRPY−/− compared with WT.
We next asked whether Runx3 directly regulates αE/CD103 transcription. αE/CD103 is a heterodimer composed of the αE and β7 chains. Because the β7 chain is highly expressed in T cells, the limiting factor in αE/CD103 expression is the transcription of the αE gene (42). We used RT-PCR to monitor αE transcription in thymocytes and splenocytes of WT, Runx3VWRPY−/−, and Runx3−/− mice (Fig. 4D). The loss of Runx3 did not completely abolish αE transcription in CD8+ thymocytes or splenocytes (Fig. 4D), as also evidenced by the low, yet detectable, surface expression of αE/CD103 in Runx3−/− thymocytes and splenocytes (Fig. 4 B and C). However, in comparison with WT splenocytes, the αE transcription level in Runx3VWRPY−/− splenocytes was significantly higher, whereas in thymocytes, the level was similar (Fig. 4D). These αE transcription levels correlated well with αE/CD103 surface expression (Fig. 4 B and C). These results indicate that in CD8-lineage T cells, Runx3 controls αE/CD103 expression through transcriptional regulation of the heterodimeric subunit αE. When Runx3 function is lost, αE/CD103 expression in CD8+ thymocytes and splenocytes diminished. However, when only the ability to recruit Gro/TLE is lost, αE/CD103 expression in thymocytes remains normal, but down-regulation in peripheral CD8+ T cells is impaired, which attests to the crucial role of Runx3-Gro/TLE in mediating the switch from transcriptional activation to repression.
Discussion
Through comparisons of biogenesis and function of TrkC neurons, LC/DC and CD8+ T lymphocytes in Runx3VWRPY−/−, Runx3−/−, and WT mice, we obtained unique insights into the bifunctional nature of Runx3 and into the mechanism by which it represses target genes. Whereas Runx3 could function as either transcriptional activator or transcriptional repressor (14, 28), the phenotypic consequences of these opposing regulatory modes and how the transition from activation to repression occurs in the in vivo context of the animal have not been addressed. We found that in developing TrkC neurons, the ability of Runx3 to mediate transcriptional repression, in conjunction with Gro/TLE, was not essential for the development of functional TrkC neurons. In contrast, in the LC/DC compartment, where Runx3 functions as a component of TGF-β signaling cascade, recruitment of Gro/TLE is required for proper maturation of DC but not for normal development of skin LC.
Analysis of Runx3VWRPY− function in the CD8 T cell-lineage provided evidence that Runx3 regulates transcription of αE/CD103 in opposing regulatory modes and that repression of both αE/CD103 and CD4 requires the binding of the corepressor, Gro/TLE. Given that CD4 transcription in CD8+ T cells is epigenetically repressed (11, 28, 30), our data indicate that Runx3 involvement in epigenetic silencing of CD4 requires recruitment of Gro/TLE. When Runx3 is lost or is unable to tether Gro/TLE, CD4 is derepressed. But why is derepression of CD4 significantly higher in Runx3VWRPY−/− splenic T cells as compared with Runx3−/− cells? A possible explanation could be that while in Runx3−/− cells, the loss of Runx3 is partially compensated by Runx1 activity (11, 30), in Runx3VWRPY−/− cells, such compensation is less efficient. We hypothesize that this phenomenon occurs because the mutant protein, which is unable to elicit repression but can still bind to the CD4 silencer’s RUNX sites, outcompetes Runx1. As a result, Runx1 compensation diminishes, leading to the observed higher derepression of CD4 in Runx3VWRPY−/− T cells compared with Runx3−/− cells. This hypothesis is supported by findings that in a compound mutant mouse Runx3−/−/Runx1−/+, i.e., null for Runx3 and hemizygous for Runx1, CD4 expression in CD8+ T cells is completely derepressed and all peripheral CD8+ T cells also expressed CD4 (12).
How tethering of Gro/TLE to target promoters/silencers evokes transcriptional repression is not yet fully understood. However, as Gro/TLE interacts with histones and histone deacetylases (18, 43), it is likely that recruitment of Gro/TLE by the DNA-bound Runx3 modulates local chromatin structure at the CD4 locus, resulting in a repressed transcriptional state (44). Our data provide evidence for a Runx3-Gro/TLE-mediated epigenetic silencing and pertain to the mechanism of Runx3-mediated repression and silencing of other genes (14). Interestingly, repression of both αE/CD103 and CD4 occurs in mature CD8+ T cells and may reflect a cell stage-specific availability of components, such as chromatin modifications enzymes (18) required for Runx3-Gro/TLE-mediated repression.
Previous studies have shown that the chromatin remodeling complexes BRG1-associated factor (BAF) are involved in transcriptional repression of CD4 during thymopoiesis (45, 46). The available information on molecular events that lead to silencing of CD4 during CD8-lineage differentiation was recently integrated into a hypothetical model (30). It predicts that CD4 repression in DP thymocytes is initiated by a step, which precedes the engagement of BAF and involves binding of Runx3 to the CD4 silencer and recruitment of another “as-yet-unknown” component, which facilitates histone deacetylation. Our data not only support this model but are also consistent with the possibility that Gro/TLE constitutes the missing link.
Materials and Methods
Generation of Runx3VWRPY−/− Mice.
A fragment of Runx3, spanning almost the entire intron 5 and exon 6 (Fig. 1), was cloned from a 129/Sv mouse genomic library (Stratagene). To generate the mutant VWRPY− allele, the nucleotides encoding the C-terminal end VWRPY, which serves as a TLE-binding site, were modified to encode a stop codon (UAG) followed by codons for the amino acids arginine (R) and proline (P) (Fig. 5). A loxP-flanked neo cassette was inserted into the unique AccI site in intron 5 (Fig. 1). Correctly targeted R1 ES clones were identified (Fig. 1 and Fig. 5), taking advantage of the newly created NotI site. Several chimeric males were generated from targeted ES cells and crossed to ICR mice to establish germ-line transmission. Homozygotes then were crossed onto the transgenic Cre-deleter mouse strain PGK-Cre (32) to generate Runx3VWRPY−/− mice lacking the neo cassette. Cre-mediated excision of the neo was confirmed by PCR. Mice were bred and maintained in a pathogen-free facility. Mouse experiments were approved by the Institutional Animal Care and Use Committee of The Weizmann Institute.
RT-PCR Analysis.
RNA was isolated from thymocytes and splenocytes of WT, Runx3VWRPY−/−, and Runx3−/− mice, and PCR products were derived by using the following sets of primers: For Runx3, exon 2 (5′GGCAAGATGGGCGAGAACAG) and exon 6 (5′CGTAGGGAAGGAGCGGTCAA), detected both P1 and P2 promoter-derived transcripts (47), yielding a 673-bp fragment. For αE, 5′CCAGAAGGCCAAAAATTTCA (sense) and 5′TCAGCAGCGACTCCTTTTCCGCTT (antisense) yielded a 197-bp fragment. mRNA levels within samples were assessed by PCR analysis of actin cDNA.
Immunohistochemistry and Histology.
Paraffin sections from embryos at embryonic day 14.5 and 17.5 for immunostaining with anti-Runx3 and anti-PV, respectively, were processed as described in ref. 9. Primary antibodies were rabbit anti-Runx3 (1:1,000) and anti-PV (1:1,500) (Swant, Bellinzona, Switzerland). Biotinylated secondary antibodies and the ABC complex from Vectastain kit (Vector Laboratories) were used for detection.
Flow Cytometry.
Preparation and analysis of LC, bone marrow-derived DC and T lymphocytes was carried out as described in refs. 7 and 12. Single-cell suspensions were prepared in FACS buffer (7, 12), incubated with antibodies, and analyzed by using a FACSCalibur (Becton Dickinson) and cellquest software (Becton Dickinson). mAbs included CD4-biotinylated, CD8α-Percp, CD8β-PE/Percp, TCRβ-FITC, HSA-PE, CD11c-APC, CD11b-PE, IA/IE (MHC II)-PE, CD3-bioitinylated, CD103-biotinylated, and streptavidin-APC (Pharmingen). Differences between average values of WT and mutant mice (either Runx3VWRPY−/− or Runx3−/−) were evaluated by using Student’s t test.
Cell Proliferation Assays.
Splenic CD8+ cells were isolated by using a magnetic cell sorting separation system (Miltenyi Biotec, Auburn, CA), labeled by incubation with 5 μM 5,6-carboxyfluorscein diacetate, succinimidyl ester (Molecular Probes) and further processed as detailed in ref. 12. Cells were stimulated with anti-CD3 mAb [(2 μg/ml; Pharmingen) plus IL-2 (20u/ml; PeproTech, Rocky Hill, NJ] and at 0, 48, and 72 h of incubation time, cells were stained with anti-CD8β and anti-CD4 mAb and analyzed by FACS.
Cell Transfection and Reporter Gene Assays.
Reporter gene assays were conducted with HEK-293 and COS-7 cells by using vectors described in Fig. 6. Cells were transfected by using lipofectamin (Invitrogen) (COS-7) and CaPO4 (HEK-293) and luciferase measured by the Luciferase Assay System (Promega) according to the manufacturer’s instructions. Difference between average values was evaluated statistically by using a paired Student’s t test.
Supplementary Material
Acknowledgments
We thank Judith Chermesh and Rafi Saka for help in animal husbandry; Dorit Nathan and Tamara Berkuzki for technical assistance; and Drs. Ori Brenner, Joseph Lotem, and Ze’ev Paroush for helpful discussions and insightful comments. The work was supported by grants from the Commission of the European Union, Philip Morris USA, Inc., Philip Morris International, and Israel Science Foundation.
Abbreviations
- DC
dendritic cells
- DRG
dorsal root ganglia
- Gro
Groucho
- LC
Langerhans cells
- SP
single-protein
- TLE
transducin-like Enhancer-of-split.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.van Wijnen A. J., Stein G. S., Gergen J. P., Groner Y., Hiebert S. W., Ito Y., Liu P., Neil J. C., Ohki M., Speck N. Oncogene. 2004;23:4209–4210. doi: 10.1038/sj.onc.1207758. [DOI] [PubMed] [Google Scholar]
- 2.Levanon D., Groner Y. Oncogene. 2004;23:4211–4219. doi: 10.1038/sj.onc.1207670. [DOI] [PubMed] [Google Scholar]
- 3.Cameron E. R., Neil J. C. Oncogene. 2004;23:4308–4314. doi: 10.1038/sj.onc.1207130. [DOI] [PubMed] [Google Scholar]
- 4.de Bruijn M. F., Speck N. A. Oncogene. 2004;23:4238–4248. doi: 10.1038/sj.onc.1207763. [DOI] [PubMed] [Google Scholar]
- 5.Alarcon-Riquelme M. E. Arthritis Res. Ther. 2004;6:169–173. doi: 10.1186/ar1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brenner O., Levanon D., Negreanu V., Golubkov O., Fainaru O., Woolf E., Groner Y. Proc. Natl. Acad. Sci. USA. 2004;101:16016–16021. doi: 10.1073/pnas.0407180101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fainaru O., Woolf E., Lotem J., Yarmus M., Brenner O., Goldenberg D., Negreanu V., Bernstein Y., Levanon D., Jung S., Groner Y. EMBO J. 2004;23:969–979. doi: 10.1038/sj.emboj.7600085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inoue K., Ozaki S., Shiga T., Ito K., Masuda T., Okado N., Iseda T., Kawaguchi S., Ogawa M., Bae S. C., et al. Nat. Neurosci. 2002;5:946–954. doi: 10.1038/nn925. [DOI] [PubMed] [Google Scholar]
- 9.Levanon D., Bettoun D., Harris-Cerruti C., Woolf E., Negreanu V., Eilam R., Bernstein Y., Goldenberg D., Xiao C., Fliegauf M., et al. EMBO J. 2002;21:3454–3463. doi: 10.1093/emboj/cdf370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Q. L., Ito K., Sakakura C., Fukamachi H., Inoue K., Chi X. Z., Lee K. Y., Nomura S., Lee C. W., Han S. B., et al. Cell. 2002;109:113–124. doi: 10.1016/s0092-8674(02)00690-6. [DOI] [PubMed] [Google Scholar]
- 11.Taniuchi I., Osato M., Egawa T., Sunshine M. J., Bae S. C., Komori T., Ito Y., Littman D. R. Cell. 2002;111:621–633. doi: 10.1016/s0092-8674(02)01111-x. [DOI] [PubMed] [Google Scholar]
- 12.Woolf E., Xiao C., Fainaru O., Lotem J., Rosen D., Negreanu V., Bernstein Y., Goldenberg D., Brenner O., Berke G., et al. Proc. Natl. Acad. Sci. USA. 2003;100:7731–7736. doi: 10.1073/pnas.1232420100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fainaru O., Shseyov D., Hantisteanu S., Groner Y. Proc. Natl. Acad. Sci. USA. 2005;102:10598–10603. doi: 10.1073/pnas.0504787102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Durst K. L., Hiebert S. W. Oncogene. 2004;23:4220–4224. doi: 10.1038/sj.onc.1207122. [DOI] [PubMed] [Google Scholar]
- 15.Stifani S., Blaumueller C. M., Redhead N. J., Hill R. E., Artavanis-Tsakonas S. Nat. Genet. 1992;2:119–127. doi: 10.1038/ng1092-119. [DOI] [PubMed] [Google Scholar]
- 16.Paroush Z., Finley R. L., Jr, Kidd T., Wainwright S. M., Ingham P. W., Brent R., Ish-Horowicz D. Cell. 1994;79:805–815. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- 17.Gasperowicz M., Otto F. J. Cell. Biochem. 2005;95:670–687. doi: 10.1002/jcb.20476. [DOI] [PubMed] [Google Scholar]
- 18.Chen G., Courey A. J. Gene. 2000;249:1–16. doi: 10.1016/s0378-1119(00)00161-x. [DOI] [PubMed] [Google Scholar]
- 19.Levanon D., Goldstein R. E., Bernstein Y., Tang H., Goldenberg D., Stifani S., Paroush Z., Groner Y. Proc. Natl. Acad. Sci. USA. 1998;95:11590–11595. doi: 10.1073/pnas.95.20.11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imai Y., Kurokawa M., Tanaka K., Friedman A. D., Ogawa S., Mitani K., Yazaki Y., Hirai H. Biochem. Biophys. Res. Commun. 1998;252:582–589. doi: 10.1006/bbrc.1998.9705. [DOI] [PubMed] [Google Scholar]
- 21.Ehlers M., Laule-Kilian K., Petter M., Aldrian C. J., Grueter B., Wurch A., Yoshida N., Watanabe T., Satake M., Steimle V. J. Immunol. 2003;171:3594–3604. doi: 10.4049/jimmunol.171.7.3594. [DOI] [PubMed] [Google Scholar]
- 22.Kawazu M., Asai T., Ichikawa M., Yamamoto G., Saito T., Goyama S., Mitani K., Miyazono K., Chiba S., Ogawa S., et al. J. Immunol. 2005;174:3526–3533. doi: 10.4049/jimmunol.174.6.3526. [DOI] [PubMed] [Google Scholar]
- 23.Nishimura M., Fukushima-Nakase Y., Fujita Y., Nakao M., Toda S., Kitamura N., Abe T., Okuda T. Blood. 2004;103:562–570. doi: 10.1182/blood-2003-06-2109. [DOI] [PubMed] [Google Scholar]
- 24.Telfer J. C., Hedblom E. E., Anderson M. K., Laurent M. N., Rothenberg E. V. J. Immunol. 2004;172:4359–4370. doi: 10.4049/jimmunol.172.7.4359. [DOI] [PubMed] [Google Scholar]
- 25.Banchereau J., Steinman R. M. Nature. 1998;392:245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 26.Steinman R. M., Hawiger D., Liu K., Bonifaz L., Bonnyay D., Mahnke K., Iyoda T., Ravetch J., Dhodapkar M., Inaba K., et al. Ann. N.Y. Acad. Sci. 2003;987:15–25. doi: 10.1111/j.1749-6632.2003.tb06029.x. [DOI] [PubMed] [Google Scholar]
- 27.Grueter B., Petter M., Egawa T., Laule-Kilian K., Aldrian C. J., Wuerch A., Ludwig Y., Fukuyama H., Wardemann H., Waldschuetz R., et al. J. Immunol. 2005;175:1694–1705. doi: 10.4049/jimmunol.175.3.1694. [DOI] [PubMed] [Google Scholar]
- 28.Taniuchi I., Littman D. R. Oncogene. 2004;23:4341–4345. doi: 10.1038/sj.onc.1207671. [DOI] [PubMed] [Google Scholar]
- 29.Zou Y. R., Sunshine M. J., Taniuchi I., Hatam F., Killeen N., Littman D. R. Nat. Genet. 2001;29:332–336. doi: 10.1038/ng750. [DOI] [PubMed] [Google Scholar]
- 30.Bosselut R. Nat. Rev. Immunol. 2004;4:529–540. doi: 10.1038/nri1392. [DOI] [PubMed] [Google Scholar]
- 31.Levanon D., Brenner O., Negreanu V., Bettoun D., Woolf E., Eilam R., Lotem J., Gat U., Otto F., Speck N., et al. Mech. Dev. 2001;109:413–417. doi: 10.1016/s0925-4773(01)00537-8. [DOI] [PubMed] [Google Scholar]
- 32.Lallemand Y., Luria V., Haffner-Krausz R., Lonai P. Transgenic Res. 1998;7:105–112. doi: 10.1023/a:1008868325009. [DOI] [PubMed] [Google Scholar]
- 33.Arber S., Ladle D. R., Lin J. H., Frank E., Jessell T. M. Cell. 2000;101:485–498. doi: 10.1016/s0092-8674(00)80859-4. [DOI] [PubMed] [Google Scholar]
- 34.Starr T. K., Jameson S. C., Hogquist K. A. Annu. Rev. Immunol. 2003;21:139–176. doi: 10.1146/annurev.immunol.21.120601.141107. [DOI] [PubMed] [Google Scholar]
- 35.Germain R. N. Nat. Rev. Immunol. 2002;2:309–322. doi: 10.1038/nri798. [DOI] [PubMed] [Google Scholar]
- 36.Leung R. K., Thomson K., Gallimore A., Jones E., Van den Broek M., Sierro S., Alsheikhly A. R., McMichael A., Rahemtulla A. Nat. Immunol. 2001;2:1167–1173. doi: 10.1038/ni733. [DOI] [PubMed] [Google Scholar]
- 37.Sawada S., Scarborough J. D., Killeen N., Littman D. R. Cell. 1994;77:917–929. doi: 10.1016/0092-8674(94)90140-6. [DOI] [PubMed] [Google Scholar]
- 38.Siu G., Wurster A. L., Duncan D. D., Soliman T. M., Hedrick S. M. EMBO J. 1994;13:3570–3579. doi: 10.1002/j.1460-2075.1994.tb06664.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kohu K., Sato T., Ohno S., Hayashi K., Uchino R., Abe N., Nakazato M., Yoshida N., Kikuchi T., Iwakura Y., et al. J. Immunol. 2005;174:2627–2636. doi: 10.4049/jimmunol.174.5.2627. [DOI] [PubMed] [Google Scholar]
- 40.Sato T., Ohno S., Hayashi T., Sato C., Kohu K., Satake M., Habu S. Immunity. 2005;22:317–328. doi: 10.1016/j.immuni.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 41.Kutlesa S., Wessels J. T., Speiser A., Steiert I., Muller C. A., Klein G. J. Cell Sci. 2002;115:4505–4515. doi: 10.1242/jcs.00142. [DOI] [PubMed] [Google Scholar]
- 42.Robinson P. W., Green S. J., Carter C., Coadwell J., Kilshaw P. J. Immunology. 2001;103:146–154. doi: 10.1046/j.1365-2567.2001.01232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palaparti A., Baratz A., Stifani S. J. Biol. Chem. 1997;272:26604–26610. doi: 10.1074/jbc.272.42.26604. [DOI] [PubMed] [Google Scholar]
- 44.Yang X. J., Seto E. Curr. Opin. Genet. Dev. 2003;13:143–153. doi: 10.1016/s0959-437x(03)00015-7. [DOI] [PubMed] [Google Scholar]
- 45.Chi T. H., Wan M., Zhao K., Taniuchi I., Chen L., Littman D. R., Crabtree G. R. Nature. 2002;418:195–199. doi: 10.1038/nature00876. [DOI] [PubMed] [Google Scholar]
- 46.Chi T. H., Wan M., Lee P. P., Akashi K., Metzger D., Chambon P., Wilson C. B., Crabtree G. R. Immunity. 2003;19:169–182. doi: 10.1016/s1074-7613(03)00199-7. [DOI] [PubMed] [Google Scholar]
- 47.Bangsow C., Rubins N., Glusman G., Bernstein Y., Negreanu V., Goldenberg D., Lotem J., Ben-Asher E., Lancet D., Levanon D., et al. Gene. 2001;279:221–232. doi: 10.1016/s0378-1119(01)00760-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.