Abstract
Primate tripartite motif 5α (TRIM5α) proteins mediate innate intracellular resistance to retroviruses. In humans, TRIM5 is located in a paralogous cluster that includes TRIM6, TRIM34, and TRIM22. Although TRIM6 and TRIM34 orthologs are found in other mammals, TRIM5 has to date been identified only in primates. Cow cells exhibit early blocks to infection by several retroviruses. We identify a cytoplasmic TRIM protein encoded by LOC505265 that is responsible for the restriction of infection by several lentiviruses and N-tropic murine leukemia virus in cow cells. Susceptibility of N-tropic murine leukemia virus to 505265-mediated restriction is determined primarily by residue 110 of the viral capsid protein. Phylogenetically, cow LOC505265 segregates with the TRIM5/TRIM6/TRIM34 group, but is not an ortholog of known TRIM genes. The B30.2/SPRY domain of 505265 exhibits long variable regions, a characteristic of the proteins encoded by this paralogous group, and shows evidence of positive selection. Apparently, cows have independently evolved a retroviral restriction factor from the same TRIM family that spawned TRIM5 in primates. Particular features of this subset of cytoplasmic TRIM proteins may be conducive to the convergent evolution of virus-restricting factors.
Keywords: capsid, convergent evolution, restriction factor, retrovirus, ungulates
Retroviruses initiate infection by fusion of the viral and target cell membranes and entry of the viral capsid into the cytosol (1, 2). Uncoating of the capsid into smaller subunits occurs before or concomitantly with reverse transcription. At this early phase of infection, certain retroviruses encounter blocks in the cells of particular mammalian species. These blocks are mediated by dominant host factors that can be competed by virus-like particles containing capsids related to that of the restricted virus (3–14). The study of recombinant retroviruses indicates that the viral capsid protein is the major determinant of susceptibility to these early restrictions (3–14). For example, the identity of residue 110 on the N-tropic murine leukemia virus (N-MLV) capsid protein strongly influences the ability of this virus to negotiate the early events of infection in human cells (7, 9, 15). HIV-1 and simian immunodeficiency virus of macaques (SIVmac) encounter postentry blocks in the cells of Old World monkeys and New World monkeys, respectively (16–18). A genetic screen identified a tripartite motif (TRIM) protein, TRIM5α, as the major factor restricting HIV-1 during the early phase of infection in Old World monkey cells (19). Differences among TRIM5α orthologs in primate species account for the patterns of restriction observed for particular retroviruses (15, 20–25).
TRIM proteins contain RING, B-box, and coiled coil domains and constitute a large family of proteins with poorly understood functions (26, 27). Many cytoplasmic TRIM proteins, like TRIM5α, contain a C-terminal B30.2(SPRY) domain that is thought to mediate binding to specific ligands (26, 27). Indeed, studies of recombinants of TRIM5α from different primate species indicate that sequences in the B30.2 domain dictate the potency and specificity of the restriction of particular retroviruses (24, 28–37). Comparison of TRIM5 sequences among primate species indicates that the TRIM5α B30.2 domain, but not the RING and B-box 2 domains, has been subjected to strong positive selection during primate evolution (30, 34). The source of such selection may have been ancient retroviral epidemics, which studies of endogenous retroviral sequences indicate have plagued mammals repeatedly over millions of years of evolution (38–42).
Genetic lability characterizes the subset of TRIM genes related to TRIM5. In humans, TRIM5 is located in a paralogous cluster at 11p15.4 that includes TRIM6, TRIM34, and TRIM22 (30). Unlike most cytoplasmic TRIM proteins, the proteins encoded by these genes exhibit longer B30.2 domain variable regions than those of the putative ancestral TRIM protein (30). Presumably, these expansions were driven by requirements for binding particular ligands. Equally striking is the labile nature of TRIM5-related genes in the genomes of different mammalian species. Rodents and dogs have no TRIM5 ortholog, yet both groups retain TRIM6 and TRIM34 orthologs (30). Thus, TRIM5 appears to have arisen relatively recently in mammalian evolution (perhaps only in primates), probably by duplication of an ancestor of TRIM6 or TRIM34.
Given the long history of exposure of many vertebrate species to retroviruses (38–42) and the potential benefit of expressing virus-restricting elements, the evolution of proteins with TRIM5-like functions in nonprimate species might be expected. Although TRIM6 and TRIM34 genes are found in the genomes of a number of mammalian species, neither has been shown to encode a protein with antiretroviral activity (ref. 30 and X.L., unpublished work). Although rodents lack TRIM5, their genomes contain TRIM6/TRIM34 paralogs not found in the human genome (e.g., TRIM12, TRIM30, 9230105E10Rik in the mouse). However, there is currently no evidence that the proteins encoded by these genes restrict retrovirus infection (refs. 26 and 30 and B.S., unpublished observations).
Surveys of the susceptibility of cells from different mammalian species to retroviral vectors have suggested that early blocks to N-MLV or feline immunodeficiency virus (FIV) might exist in nonprimate lineages (9, 25). The sporadic distribution pattern of restriction on the phylogenetic tree of vertebrate species hints that the ability to encode retroviral restriction factors may have been convergently acquired multiple times during mammalian evolution (9). Here, we investigate retroviral restriction in cow cells, which have been reported to be poor hosts for N-MLV and FIV vectors (9, 25). We identify a bovine TRIM protein that exhibits antiretroviral activity and investigate its relatedness to the proteins encoded by the TRIM5/TRIM6/TRIM22/TRIM34 cluster.
Results
Susceptibility of a Bovine Cell Line to Retroviral Infections.
Previous studies have found that certain cells of bovine origin do not support infection by some retroviruses (7, 9, 18, 25, 43, 44). To examine the ability of several different gammaretroviruses and lentiviruses to negotiate the early phase of infection in bovine cells, the infectivity of 10 different vesicular stomatitis virus G-pseudotyped retrovirus vectors expressing GFP was evaluated in Madin-Darby bovine kidney (MDBK) cells. First, each single-round vector was titrated on canine Cf2Th cells, which have been shown to be susceptible to infection by many retroviral vectors (18). Doses of each virus that allowed efficient infection of Cf2Th cells were then incubated with MDBK cells, and the percentage of GFP-positive cells was measured (Table 1). In general, infection of MDBK cells was less efficient than that of the Cf2Th cells. B-tropic MLV (B-MLV) efficiently infected MDBK cells, whereas infection by N-MLV was very inefficient. Infection by the BNBB-MLV chimera, which is identical to B-MLV except that capsid residue 110 is changed from glutamic acid to arginine, was very inefficient in MDBK cells, similar to that of N-MLV. By contrast, the NBNN-MLV chimera, which is identical to N-MLV except that capsid residue 110 is glutamic acid, infected MDBK cells, although not as efficiently as B-MLV. Thus, the same changes in residue 110 of the capsid protein that have been shown to alter MLV susceptibility to Fv1- and human TRIM5α-imposed restrictions (7, 9, 15, 47) also influence the efficiency of MLV infection of MDBK cells.
Table 1.
Susceptibility of canine and bovine cells to infection by different retroviral vectors
| Virus vector | % GFP-positive cells* |
MDBK/Cf2Th ratio, % | |
|---|---|---|---|
| Cf2Th (canine) | MDBK (bovine) | ||
| Gammaretrovirus | |||
| N-MLV | 90 ± 9 | 0.7 ± 0.2 | 0.8 |
| B-MLV | 84 ± 15 | 43 ± 10 | 51.1 |
| BNBB-MLV | 74 ± 11 | 0.8 ± 0.5 | 1.1 |
| NBNN-MLV | 93 ± 4 | 11 ± 2 | 11.8 |
| Lentivirus | |||
| HIV-1 | 99 ± 1 | 0.9 ± 1 | 0.9 |
| SIVmac | 95 ± 6 | 1.4 ± 1 | 1.5 |
| SIVagm† | 65 ± 8 | 1.3 ± 1 | 2.0 |
| FIV | 82 ± 1 | 1.5 ± 1 | 1.8 |
| EIAV | 99 ± 1 | 15 ± 1 | 15.2 |
| BIV | 99 ± 1 | 70 ± 5 | 71.0 |
*Approximately 2 × 104 cells were incubated with the indicated GFP-expressing retroviral vectors. After 48 h, cells were analyzed for GFP expression by fluorescence-activated cell sorting.
†Cytotoxicity of the SIVagm-GFP vector preparation limited the amount of virus that could be used.
Of the lentivirus vectors, only those derived from two ungulate lentiviruses, bovine immunodeficiency virus (BIV) and equine infectious anemia virus (EIAV), infected MDBK cells (Table 1). Infection by the EIAV vector was less efficient than that of the BIV vector. None of the primate lentivirus vectors [HIV-1, SIVmac, SIV derived from African green monkeys (SIVagm)] infected MDBK cells efficiently; likewise, infection of MDBK cells by FIV was inefficient. Thus, although all of the retroviral vectors tested infected Cf2Th canine cells efficiently, particular vectors (N-MLV, BNBB-MLV, HIV-1, SIVmac, SIVagm, and FIV) did not efficiently infect MDBK cow cells.
Identification of Candidate Bovine TRIM Genes.
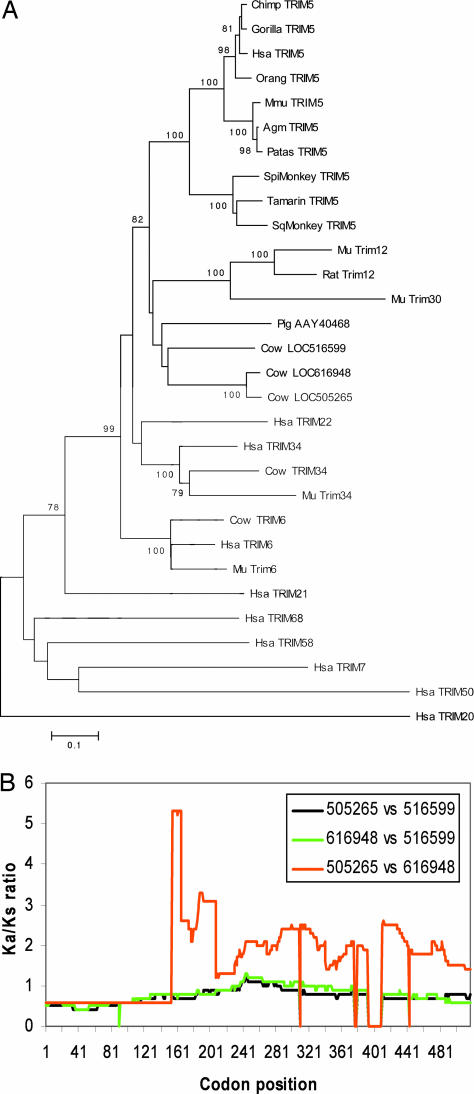
To identify TRIM5-like proteins that might contribute to the poor infectivity of certain retroviruses in cow cells, we queried the emerging bovine genome database for sequences related to TRIM5. Five cow genes of interest (LOC514492, LOC539820, LOC516599, LOC616948, and LOC505265) were identified. The predicted amino acid sequences of the encoded proteins were aligned with those of several TRIM5/6/12/22/34 family members, and the alignment was used to build phylogenetic trees. Four different methods supported the same basic tree (Fig. 1A) and placed all five cow sequences within the TRIM5/6/12/22/34 group. LOC514492 and LOC539820 are the bovine orthologs of TRIM6 and TRIM34, respectively; as expected, the predicted protein products of these genes clustered with the respective TRIM6 and TRIM34 proteins from other mammalian species. Moreover, these cow proteins exhibit B30.2 variable region lengths typical of TRIM6 and TRIM34 (Table 2) (30). The proteins encoded by the other three loci (LOC516599, LOC616948, and LOC505265) clustered significantly with each other, but are not orthologs of any known TRIM protein. A related protein, AAY40468, was identified in pigs (Fig. 1A). These predicted proteins exhibit expanded v2 and v3 variable regions in the B30.2 domain, compared with those of the putative ancestral TRIM protein; by contrast, their B30.2 v1 region length is that expected for the TRIM ancestor (Table 2 and Fig. 5, which is published as supporting information on the PNAS web site). This pattern of B30.2 variable region length is also seen for TRIM6, TRIM22, and TRIM12 (30). The B30.2 v2 region of the 505265 protein contains 43 residues; only rat TRIM12 has such an expanded v2 region. Imperfect tandem repeats can be detected in the LOC505265 sequence encoding this v2 region, reminiscent of those that are associated with the long v1 region of African green monkey TRIM5α and the very long v3 region of spider monkey TRIM5α (30, 34). Thus, the cow LOC516599, LOC616948, and LOC505265 genes encode TRIM proteins that are distinct members of the TRIM5/6/12/22/34 subfamily of TRIM proteins.
Fig. 1.
Identification of candidate bovine restrictions factors. (A) The predicted amino acid sequences of the proteins encoded by bovine LOC516599, LOC616948, LOC505265, LOC514492 (cow TRIM6) and LOC539820 (cow TRIM34) were aligned with those of other TRIM proteins by using clustal x (50). The alignment was used to build trees in mega3.1 by using neighbor joining, maximum parsimony, the Unweighted Pair Group Method with Authentic Mean (upgma), and maximum evolution methods with 1,000 bootstrap iterations. Values of 100 represent 99–100% concordance in the bootstrap analysis. The scale bar represents evolutionary distance in substitutions/amino acid residue. Hsa, Homo sapiens; Mmu, Macaca mulatta; Spi, spider monkey; Sq, squirrel monkey; Mu, mouse. (B) The plot shows the Ka/Ks ratios at various codon positions for pairwise comparisons of LOC516599, LOC616948, and LOC505265. The alignment is shown in Fig. 5. The Ka/Ks ratio across gaps in the alignment was arbitrarily set to 0. The Ka/Ks ratios, calculated as described (45), were estimated as rolling averages for a window of 200 codons.
Table 2.
Lengths of the B30.2 domain variable regions in TRIM proteins
| TRIM protein | B30.2 variable region length, aa | |||
|---|---|---|---|---|
| v1 | v2 | v3 | v4 | |
| 505265 (cow) | 20 | 43 | 28 | 5 |
| 616948 (cow) | 20 | 29 | 29 | 5 |
| 516599 (cow) | 20 | 26 | 29 | 5 |
| AAY40468 (pig) | 20 | 20 | 29 | 5 |
| TRIM6 (human, mouse) | 19 | 23 | 25 | 5 |
| 514492/TRIM6 (cow) | 18 | 23 | 25 | 5 |
| TRIM12 (rat) | 19 | 43 | 30 | 5 |
| TRIM12 (mouse) | 19 | 30 | 30 | 5 |
| TRIM22 (human) | 19 | 29 | 29 | 5 |
| TRIM34 (human, mouse, rat, dog) | 13 | 23–25 | 29 | 5 |
| 539820/TRIM34 (cow) | 13 | 25 | 31 | 5 |
| TRIM5 (Old World primates) | 26–46 | 17 | 32 | 5 |
| TRIM5 (New World primates) | 17 | 15–17 | 41–96 | 5 |
| TRIM20 (human, chimpanzee) | 20 | 12 | 23 | 10 |
| TRIM50 (human, mouse, rat, pig) | 20 | 12 | 23 | 9 |
| Consensus | 19–21 | 11–13 | 20–23 | 4–10 |
The boundaries of the TRIM B30.2 domain variable regions are defined as in Song et al. (30). The variable region lengths in amino acid residues are reported, along with those expected of the putative ancestral TRIM protein (Consensus). Variable region lengths greater than those of the consensus are in bold.
Analysis of Nonsynonymous/Synonymous (Ka/Ks) Variation in Cow TRIM Genes.
Analysis of Ka/Ks variations (Ka/Ks ratios) can provide insight into selection for or against a change in the coding capacity of a gene (45). The Ka/Ks ratios were calculated for pairwise comparisons of cDNAs corresponding to cow LOC516599, LOC616948, and LOC505265, excluding indels from the analysis (Fig. 1B). For the sequences encoding the RING and B-box 2 domains, the Ka/Ks ratios were <1, indicating selection against amino acid changes. For the sequences encoding the C-terminal portion of the cow TRIM proteins (the coiled coil and B30.2/SPRY domains), the Ka/Ks ratio increased. Particularly high Ka/Ks ratios in these sequences were observed when LOC616948 and LOC505265 were compared, suggesting that strong positive selection has been operative since the divergence of these two genes.
Necessity of Cow TRIM Expression for N-MLV Restriction in MDBK Cells.
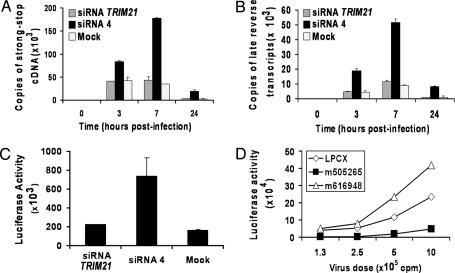
Studies of RNA extracted from MDBK cells and searches of cow EST databases indicated that expression of LOC516599 was very low compared with that of LOC616948 or LOC505265 (data not shown). Thus, we focused on the 616948 and 505265 proteins as candidate restriction factors in MDBK cells. MDBK cells transfected with short interfering RNA (siRNA) targeting these genes were tested for susceptibility to the retroviral vectors described above. Because of the high degree of sequence identity between LOC616948 and LOC505265, it is not possible to design siRNAs to down-regulate these genes individually (data not shown). Therefore, siRNAs directed against both LOC616948 and LOC505265 were designed and tested. Transfection of two of these siRNAs, siRNA 3 and siRNA 4, but not a control siRNA directed against cow TRIM21, resulted in a marked increase in the efficiency of N-MLV infection of MDBK cells (Fig. 2 and data not shown). Transfection of siRNA 4, but not the control TRIM21 siRNA, also resulted in an increase in BNBB-MLV and FIV infection of MDBK cells. (Fig. 2 and Fig. 6, which is published as supporting information on the PNAS web site). Transfection of siRNA 4 had less of an effect on the infectivity of NBNN-MLV, and had no significant effect on the susceptibility of MDBK cells to B-MLV, EIAV, or BIV vectors. These results suggest that the proteins encoded by LOC616948 and LOC505265, either alone or together, contribute to restriction of N-MLV and FIV infection in MDBK cells.
Fig. 2.
Implication of bovine 505265 in retroviral restriction in MDBK cells. (A–D) MDBK cells were mock-transfected or transfected with 100 pmol of siRNA directed against TRIM21 or siRNA 4. The cells were then incubated with increasing amounts of the indicated GFP-expressing viruses. Forty-eight hours later, GFP-positive cells were counted by fluorescence-activated cell sorting. The results shown are typical of those obtained in three independent experiments. (E) MDBK cells were transduced with the empty LPCX vector or LPCX vectors expressing cow LOC505265 or LOC616948 cDNAs with mutations that render them resistant to siRNA 4. Cells were then transfected with siRNA 4 and incubated with increasing amounts of the GFP-expressing N-MLV vector. GFP-positive cells were counted by fluorescence-activated cell sorting. The experiment was performed twice with similar results. (F) MDBK cells transduced with the empty LPCX vector or LPCX vectors expressing the mutant cDNAs (m505265 or m616948) described in E were lysed. Cell lysates were Western-blotted with an antibody directed against the hemagglutinin epitope tag at the C terminus of the protein.
To test whether N-MLV restriction could be rescued in the siRNA 4-transfected MDBK cells, we established MDBK cells stably expressing mutated LOC616948 or LOC505265 cDNAs. These cDNAs were modified with identical silent nucleotide changes that are predicted to render them resistant to siRNA 4. The encoded TRIM proteins contain a C-terminal epitope tag from influenza hemagglutinin. In MDBK cells stably expressing the mutated LOC505265 cDNA, transfection of siRNA 4 did not relieve the restriction to N-MLV infection (Fig. 2E). By contrast, stable expression of the mutated LOC616948 cDNA did not attenuate the ability of siRNA 4 to increase the susceptibility of MDBK cells to N-MLV infection. Differences in the steady-state levels of expression of the protein products of the mutant LOC505265 and LOC616948 cDNAs did not explain the observed differences in the rescue of restriction (Fig. 2F). These results indicate that LOC505265 encodes a protein that restricts N-MLV infection in MDBK cells.
Blockade of HIV-1 Infection in MDBK Cells Before Reverse Transcription.
We wanted to examine whether the poor infectivity of HIV-1 vectors in MDBK cells resulted from a block before reverse transcription and whether the 505265/616948 proteins were involved. MDBK cells were mock-transfected or transfected with siRNA 4 or TRIM21 siRNA. A real-time PCR assay was used to monitor viral cDNA synthesis at various times after incubating these MDBK cells with the vesicular stomatitis virus G-pseudotyped HIV-1-Luc vector. Two sets of PCR primers and Taqman probes were used. One set measures minus-strand strong-stop DNA, the initial DNA product of reverse transcription. The second set detects later DNA products that rely on the second template switch of reverse transcription. Synthesis of strong-stop DNA was inefficient in the mock- or TRIM21 siRNA-transfected MDBK cells at all time points examined. By contrast, significant increases in the amount of strong-stop DNA were observed in cells transfected with siRNA 4 (Fig. 3A). Similar results were obtained when late reverse transcripts were studied (Fig. 3B). The luciferase activities in these cells, which reflect the efficiency of HIV-1-Luc infection, paralleled the increases in viral cDNA synthesis (Fig. 3C). The block to HIV-1-Luc infection in MDBK cells transfected with siRNA 4 was rescued by expression of an siRNA 4-resistant LOC505265 cDNA, but not by an LOC616948 cDNA (Fig. 3D). The protein products of both of these mutated cDNAs were expressed efficiently in the MDBK cells (see Fig. 2F). We conclude that at least part of the block to HIV-1 infection in MDBK cells is mediated by the 505265 protein and involves a decrease in the efficiency of early events before or concurrent with the initiation of reverse transcription.
Fig. 3.
The block to HIV-1 infection in MDBK cells occurs before reverse transcription. (A and B) MDBK cells were mock-transfected or transfected with siRNA 4 or siRNA directed against TRIM21. Cells were then incubated with vesicular stomatitis virus G-pseudotyped, DNase-treated HIV-1-Luc virus. Extrachromosomal DNA was extracted at the indicated times and used to detect early, strong-stop (A) and late (B) reverse transcripts by quantitative PCR. Incubation of cells with control HIV-1-Luc viruses lacking envelope glycoproteins resulted in very low signals; these background signals were subtracted from those obtained with entry-competent HIV-1-Luc viruses. Error bars indicate the variation observed between two parallel infections assayed in duplicate. (C) A portion of the MDBK cells transfected as described above was incubated with 2 × 106 reverse transcriptase-cpm of HIV-1-Luc viruses. Forty-eight hours later, luciferase activity was measured in the cell extracts. Error bars indicate the variation in luciferase activity between duplicate infections. Similar results were obtained in two independent experiments. (D) MDBK cells were transduced with the empty LPCX vector or LPCX vectors expressing LOC505265 or LOC616948 cDNAs with mutations that render them resistant to siRNA 4. The level of expression of the 505265 and 616948 proteins in these cells is shown in Fig. 2F. Cells were transfected with siRNA 4 and incubated with increasing amounts of vesicular stomatitis virus G-pseudotyped, HIV-1-Luc virus. Forty-eight hours later, luciferase was measured in the cell extracts. Similar results were obtained in two independent experiments.
Effects of 505265 Expression on Retroviral Infection.
To examine directly the effects of the cow 505265 protein on the efficiency of retroviral infection in permissive cells, Cf2Th canine thymocytes stably expressing this protein were established by transduction with LPCX vectors containing the LOC505265 cDNA. Control cells were transduced with the empty LPCX vector. Staining with an antibody directed against the C-terminal hemagglutinin epitope tag revealed that the 505265 protein was located in the cytoplasm and excluded from the nuclei of these cells (Fig. 7, which is published as supporting information on the PNAS web site). The 505265-expressing Cf2Th cells were incubated with different amounts of the retroviral vectors described above and the efficiency of infection was monitored by GFP fluorescence. Expression of 505265 significantly inhibited infection by N-MLV, BNBB-MLV, FIV, HIV-1, and SIVmac, compared with the infection of cells transduced with the empty LPCX vector (Fig. 4 and Fig. 8, which is published as supporting information on the PNAS web site). By contrast, infections by B-MLV, NBNN-MLV, BIV, and SIVagm were not significantly affected by 505265 expression. 505265 partially inhibited EIAV infection. We conclude that the expression of cow 505265 is sufficient to account for most of the specific restrictions to retroviral infection observed in MDBK cells.
Fig. 4.
Specific inhibition of retroviral infection by cow 505265. Cf2Th cells transduced with the empty LPCX vector or an LPCX vector expressing cow 505265 protein were incubated with various amounts of the indicated viruses expressing GFP. GFP-positive cells were counted by fluorescence-activated cell sorting. The data shown are representative of those obtained in three independent experiments.
Discussion
TRIM proteins appeared with the metazoans and dramatically expanded in number during vertebrate evolution (27, 30). Approximately 70 TRIM genes have been identified in mammals (26), but the function of most of the encoded TRIM proteins is unknown. Some TRIM family members have been reported to exhibit antiviral activity, but to date only primate TRIM5α variants have demonstrated high potency and specificity for particular retroviruses (26). Here, we identify a second TRIM protein, cow 505265, that exhibits substantial ability to restrict infection by several lentiviruses and a gammaretrovirus. Like TRIM5α (15, 19), 505265 blocks infection before the initiation of reverse transcription. As changes in the viral capsid protein influence susceptibility to 505265-mediated restriction, 505265 likely targets the incoming capsid complex. The pattern of retroviruses blocked by 505265 parallels the susceptibility of MDBK cells to infection; one exception is SIVagm, which did not efficiently infect MDBK cells, yet was not blocked by 505265. Treatment of MDBK cells with siRNA 4 did not increase the efficiency of SIVagm infection, and expression of 616948 in Cf2Th cells did not block SIVagm infection (data not shown). Further studies will be required to dissect the blocks encountered in MDBK cells by SIVagm. Of note, 505265 did not inhibit infection by BIV, which naturally infects cows. The effects of 505265 on infection by EIAV, another ungulate lentivirus, were minimal. A similar situation applies to primate TRIM5α proteins, which at best partially restrict their cognate lentiviruses (26). Retroviruses have apparently evolved capsids that are only moderately restricted by the TRIM protein(s) expressed by the natural host.
The LOC516599, LOC616948, and LOC505265 genes are not orthologs of any known gene, but clearly belong to the TRIM5/6/12/22/30/34 group. These three TRIM genes are thus paralogs of cow TRIM6 and TRIM34 and probably arose as ungulates diverged from other mammals. In this respect, LOC516599, LOC616948, and LOC505265 resemble the TRIM12 and TRIM30 genes, which have been found only in rodent species, and TRIM5 and TRIM22, found only in primates. Apparently, a number of novel genes have arisen within the TRIM5/6/12/22/30/34 group in different mammalian species. The acquisition of specific antiretroviral activity by LOC505265 in cows and TRIM5 in primates represents an example of convergent evolution. It is of interest that this group of TRIM genes was used in at least two separate instances to create antiviral factors. Certain features of the TRIM proteins in this group, including intracellular association with particular host molecules, oligomerization state, or ligand-binding characteristics, may be conducive to the acquisition of antiviral activity.
Ancient retroviral epidemics probably imposed sporadic, but powerful, selective pressure on any TRIM proteins that could modulate the efficiency of infection. TRIM5 genes exhibit evidence of positive selection, particularly in the sequences encoding the C-terminal B30.2 domain (30, 34). Sequence divergence in the variable regions of the B30.2 domain, which is critical for capsid recognition (46), determines the potency and viral specificity of restriction by TRIM5α proteins from different primate species. Similarly, the B30.2 domains of other TRIM proteins with antiviral activity might be expected to exhibit hallmarks of selection imposed by the requirement to coevolve with rapidly changing viruses. The LOC516599, LOC616948, and LOC505265 genes are closely related to each other, supporting the idea that they arose by successive duplication events, first of a common ancestor and later of an LOC505265 and LOC616948 ancestor. The B30.2 variable regions of the encoded proteins are expanded, compared with those of the putative TRIM ancestor. Our analysis of Ka/Ks ratios suggests that, after the divergence of LOC505265 and LOC616948, positive selection has operated on the coiled coil and B30.2 domains of the encoded proteins, whereas purifying selection has preserved the RING and B-box 2 amino acid residues. This pattern is reminiscent of that seen for TRIM5 variants in primates (30, 34).
As seen for susceptibility to Fv1 and human TRIM5α (7, 9, 15, 47), the identity of the amino acid at residue 110 in the MLV capsid protein strongly influences the sensitivity of MLV variants to the cow 505265 protein. Residue 110 is located at the outer tip of the spokes that compose the hexameric unit of retroviral capsids (48). The spokes of adjacent hexamers flank a trimeric depression on the capsid surface that has been hypothesized to represent a potential binding site for the trimeric TRIM5α proteins (49). The charged side chain of residue 110 is well positioned to influence the interactions of restriction factors with this putative binding site. Future studies should test this hypothesis.
Materials and Methods
cDNA Cloning, Analysis, and Expression.
Generation of cDNA clones for cow LOC616948 and LOC505265, phylogenetic analysis, and expression of these cDNAs is described in Supporting Materials and Methods, which is published as supporting information on the PNAS web site. The sequences of LOC505265 and LOC616948 cDNAs from MDBK cells have been deposited in GenBank under accession nos. DQ381150 and DQ381151, respectively.
Quantitative Real-Time PCR.
Levels of strong-stop and late reverse transcription products were assessed as described (refs. 15 and 23 and Supporting Materials and Methods).
Single-Round Infections.
Retroviral vectors expressing GFP or luciferase were made and used as described (refs. 15 and 23 and Supporting Materials and Methods).
RNA Interference.
The sequences of the siRNAs and the transfection of these siRNAs into MDBK cells are described in detail in Supporting Materials and Methods.
Supplementary Material
Acknowledgments
We thank Ms. Yvette McLaughlin and Ms. Sheri Farnum for manuscript preparation. This work was supported by National Institutes of Health Grants AI63987 and AI52014, Center for AIDS Research Award AI60354, the International AIDS Vaccine Initiative, the Bristol-Myers Squibb Foundation, and the late William F. McCarty-Cooper. This research was supported in part by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research under Contract N01-CO-12400.
Abbreviations
- MLV
murine leukemia virus
- N-MLV
N-tropic MLV
- B-MLV
B-tropic MLV
- TRIM
tripartite motif
- SIVmac
simian immunodeficiency virus of macaques
- SIVagm
SIV derived from African green monkeys
- FIV
feline immunodeficiency virus
- BIV
bovine immunodeficiency virus
- EIAV
equine infectious anemia virus
- siRNA
short interfering RNA
- MDBK
Madin-Darby bovine kidney
- Ka/Ks
nonsynonymous/synonymous.
Footnotes
References
- 1.Arts E. J., Wainberg M. A. Adv. Virus Res. 1996;46:97–163. doi: 10.1016/s0065-3527(08)60071-8. [DOI] [PubMed] [Google Scholar]
- 2.Whitcomb J. M., Hughes S. H. Annu. Rev. Cell Biol. 1992;8:275–306. doi: 10.1146/annurev.cb.08.110192.001423. [DOI] [PubMed] [Google Scholar]
- 3.Cowan S., Hatziioannou T., Cunningham T., Muesing M. A., Gottlinger H. G., Bieniasz P. D. Proc. Natl. Acad. Sci. USA. 2002;99:11914–11919. doi: 10.1073/pnas.162299499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hatziioannou T., Cowan S., Von Schwedler U. K., Sundquist W. I., Bieniasz P. D. J. Virol. 2004;78:6005–6012. doi: 10.1128/JVI.78.11.6005-6012.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Owens C. M., Yang P. C., Gottlinger H., Sodroski J. J. Virol. 2003;77:726–731. doi: 10.1128/JVI.77.1.726-731.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Owens C. M., Song B., Perron M. J., Yang P. C., Stremlau M., Sodroski J. J. Virol. 2004;78:5423–5437. doi: 10.1128/JVI.78.10.5423-5437.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Towers G., Bock M., Martin S., Takeuchi Y., Stoye J. P., Danos O. Proc. Natl. Acad. Sci. USA. 2000;97:12295–12299. [Google Scholar]
- 8.Hatziioannou T., Cowan S., Goff S. P., Bieniasz P. D., Towers G. J. EMBO J. 2003;22:385–394. doi: 10.1093/emboj/cdg042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Besnier C., Ylinen L., Strange B., Lister A., Takeuchi Y., Goff S. P., Towers G. J. J. Virol. 2003;77:13403–13406. doi: 10.1128/JVI.77.24.13403-13406.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Munk C., Brandt S. M., Lucero G., Landau N. R. Proc. Natl. Acad. Sci. USA. 2002;99:13843–13848. doi: 10.1073/pnas.212400099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kootstra N. A., Munk C., Tonnu N., Landau N. R., Verma I. M. Proc. Natl. Acad. Sci. USA. 2003;100:1298–1303. doi: 10.1073/pnas.0337541100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Towers G., Collins M., Takeuchi Y. J. Virol. 2002;76:2548–2550. doi: 10.1128/jvi.76.5.2548-2550.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Towers G. J., Hatziioannou T., Cowan S., Goff S. P., Luban J., Bieniasz P. D. Nat. Med. 2003;9:1138–1143. doi: 10.1038/nm910. [DOI] [PubMed] [Google Scholar]
- 14.Besnier C., Takeuchi Y., Towers G. Proc. Natl. Acad. Sci. USA. 2002;99:11920–11925. doi: 10.1073/pnas.172384599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perron M. J., Stremlau M., Song B., Ulm W., Mulligan R. C., Sodroski J. Proc. Natl. Acad. Sci. USA. 2004;101:11827–11832. doi: 10.1073/pnas.0403364101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shibata R., Sakai H., Kawamura M., Tokunaga K., Adachi A. J. Gen. Virol. 1995;76:2723–2730. doi: 10.1099/0022-1317-76-11-2723. [DOI] [PubMed] [Google Scholar]
- 17.Himathongkham S., Luciw P. A. Virology. 1996;219:485–488. doi: 10.1006/viro.1996.0276. [DOI] [PubMed] [Google Scholar]
- 18.Hofmann W., Schubert D., LaBonte J., Munson L., Gibson S., Scammell J., Ferrigno P., Sodroski J. J. Virol. 1999;73:10020–10028. doi: 10.1128/jvi.73.12.10020-10028.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stremlau M., Owens C. M., Perron M. J., Kiessling M., Autissier P., Sodroski J. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 20.Hatziioannou T., Perez-Caballero D., Yang A., Cowan S., Bieniasz P. D. Proc. Natl. Acad. Sci. USA. 2004;101:10774–10779. doi: 10.1073/pnas.0402361101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keckesova Z., Ylinen L. M., Towers G. J. Proc. Natl. Acad. Sci. USA. 2004;101:10780–10785. doi: 10.1073/pnas.0402474101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yap M. W., Nisole S., Lynch C., Stoye J. P. Proc. Natl. Acad. Sci. USA. 2004;101:10786–10791. doi: 10.1073/pnas.0402876101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song B., Javanbakht H., Perron M., Park D. H., Stremlau M., Sodroski J. J. Virol. 2005;79:3930–3937. doi: 10.1128/JVI.79.7.3930-3937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakayama E. E., Miyoshi H., Nagai Y., Shioda T. J. Virol. 2005;79:8870–8877. doi: 10.1128/JVI.79.14.8870-8877.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saenz D. T., Teo W., Olsen J. C., Poeschla E. M. J. Virol. 2005;79:15175–15188. doi: 10.1128/JVI.79.24.15175-15188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nisole S., Stoye J. P., Saib A. Nat. Rev. Microbiol. 2005;3:799–808. doi: 10.1038/nrmicro1248. [DOI] [PubMed] [Google Scholar]
- 27.Reymond A., Meroni G., Fantozzi A., Merla G., Cairo S., Luzi L., Riganelli D., Zanaria E., Messali S., Cainarca S., et al. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stremlau M., Perron M., Welikala S., Sodroski J. J. Virol. 2005;79:3139–3145. doi: 10.1128/JVI.79.5.3139-3145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yap M. W., Nisole S., Stoye J. P. Curr. Biol. 2005;15:73–78. doi: 10.1016/j.cub.2004.12.042. [DOI] [PubMed] [Google Scholar]
- 30.Song B., Gold B., O’hUigin C., Javanbakht H., Li X., Stremlau M., Winkler C., Dean M., Sodroski J. J. Virol. 2005;79:6111–6121. doi: 10.1128/JVI.79.10.6111-6121.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sayah D. M., Sokolskaja E., Berthoux L., Luban J. Nature. 2004;430:569–573. [Google Scholar]
- 32.Nisole S., Lynch C., Stoye J. P., Yap M. W. Proc. Natl. Acad. Sci. USA. 2004;101:13324–13328. doi: 10.1073/pnas.0404640101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sebastian S., Luban J. Retrovirology. 2005;2:40. doi: 10.1186/1742-4690-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sawyer S. L., Wu L. I., Emerman M., Malik H. S. Proc. Natl. Acad. Sci. USA. 2005;102:2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perez-Caballero D., Hatziioannou T., Yang A., Cowan S., Bieniasz P. D. J. Virol. 2005;79:8969–8978. doi: 10.1128/JVI.79.14.8969-8978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ylinen L. M., Keckesova Z., Wilson S. J., Ranasinghe S., Towers G. J. J. Virol. 2005;79:11580–11587. doi: 10.1128/JVI.79.18.11580-11587.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Javanbakht H., Diaz-Griffero F., Stremlau M., Si Z., Sodroski J. J. Biol. Chem. 2005;280:26933–26940. doi: 10.1074/jbc.M502145200. [DOI] [PubMed] [Google Scholar]
- 38.Boeke J. D., Stoye J. P. In: Retroviruses. Coffin J. M., Hughes S. H., Varmus H. E., editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 343–436. [PubMed] [Google Scholar]
- 39.Herniou E., Martin J., Miller K., Cook J., Wilkinson M., Tristem M. J. Virol. 1998;72:5955–5966. doi: 10.1128/jvi.72.7.5955-5966.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson W. E., Coffin J. M. Proc. Natl. Acad. Sci. USA. 1999;96:10254–10260. doi: 10.1073/pnas.96.18.10254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coffin J. M. In: Fundamental Virology. Fields B. N., Knipe D. M., editors. Philadelphia: Lippincott–Raven; 1996. pp. 763–844. [Google Scholar]
- 42.Tristem M. J. Virol. 2000;74:3715–3730. doi: 10.1128/jvi.74.8.3715-3730.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coskun A. K., Sutton R. E. J. Virol. 2005;79:4150–4158. doi: 10.1128/JVI.79.7.4150-4158.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ikeda Y., Collins M. K., Radcliffe P. A., Mitrophanous K. A., Takeuchi Y. Gene Ther. 2002;9:932–938. doi: 10.1038/sj.gt.3301708. [DOI] [PubMed] [Google Scholar]
- 45.Li W. H. J. Mol. Evol. 1993;36:96–99. doi: 10.1007/BF02407308. [DOI] [PubMed] [Google Scholar]
- 46.Stremlau M., Perron M., Lee M., Li Y., Song B., Javanbakht H., Diaz-Griffero F., Anderson D., Sundquist W. I., Sodroski J. Proc. Natl. Acad. Sci. USA. 2006;103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kozak C. A., Chakrabarti A. Virology. 1996;225:300–305. doi: 10.1006/viro.1996.0604. [DOI] [PubMed] [Google Scholar]
- 48.Mortuza G. B., Haire L. F., Stevens A., Smerdon S. J., Stoye J. P., Taylor I. A. Nature. 2004;431:481–485. doi: 10.1038/nature02915. [DOI] [PubMed] [Google Scholar]
- 49.Mische C. C., Javanbakht H., Song B., Diaz-Griffero F., Stremlau M., Strack B., Si Z., Sodroski J. J. Virol. 2005;79:14446–14450. doi: 10.1128/JVI.79.22.14446-14450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.