Abstract
The concentration of GABA increases rapidly in wounded plant tissues, but the implication of this GABA pulse for plant–bacteria interactions is not known. Here we reveal that GABA stimulated the inactivation of the N-(3-oxooctanoyl)homoserine lactone (OC8-HSL) quorum-sensing signal (or “quormone”) by the Agrobacterium lactonase AttM. GABA induced the expression of the attKLM operon, which was correlated to a decrease in OC8-HSL concentration in Agrobacterium tumefaciens cultures. The Agrobacterium GABA transporter Bra was required for this GABA-signaling pathway. Furthermore, transgenic tobacco plants with elevated GABA levels were less sensitive to A. tumefaciens C58 infection than were wild-type plants. These findings indicate that plant GABA may modulate quorum sensing in A. tumefaciens, thereby affecting its virulence on plants. Whereas GABA is an essential cell-to-cell signal in eukaryotes, here we provide evidence of GABA acting as a signal between eukaryotes and pathogenic bacteria. The GABA signal represents a potential target for the development of a strategy to control the virulence of bacterial pathogens.
Keywords: phytopathology, plant signal, lactonase, quorum quenching
GABA is a nonprotein amino acid that is present in a large range of organisms including bacteria, yeasts, plants, and animals (1). In animals, GABA acts as a cell-to-cell signal during embryonic and adult neurogenesis and is an essential neurotransmitter in mature neurons. Alteration of its synthesis or degradation can cause severe clinical disorders (1, 2). In plants, disruption of enzymes for GABA degradation results in abnormal development (3). Moreover, the GABA gradient in female tissues is implicated in the growth and guidance of the pollen tube (4). In plants and bacteria, GABA synthesis and degradation are also associated with biotic and abiotic stresses, including acid conditions (5) and mechanical damage or stimulation (6). Typically, the synthesis of GABA occurs rapidly in wounded plant tissues because of stimulation of glutamate decarboxylase (GAD) activity by H+ or the Ca2+/calmodulin complex (6, 7). Although the enzymatic and regulatory steps for GABA synthesis are well known, the implication of GABA accumulation in the plant response to wounding or to bacterial/fungal infection at wounding sites remains unclear.
Plant wounding is required for development of tumors in tissues infected by Agrobacterium tumefaciens. Molecules such as acetosyringone that are associated with wounding in plants can activate the transfer of the T-DNA from the tumor-inducing (Ti) plasmid of A. tumefaciens to plant cells (8). The transformed plant tissues produce some opines that positively control the synthesis of the quorum-sensing (QS) signal N-(3-oxooctanoyl)homoserine lactone (OC8-HSL) (9). In A. tumefaciens, the OC8-HSL signal is implicated in the control of the conjugation of the Ti plasmid (10, 11), the amplification of the Ti plasmid copy number (12, 13), and the severity of tumoral symptoms (12). In addition to the sophisticated control of OC8-HSL synthesis by plant opines, A. tumefaciens C58 harbors two lactonases, AttM (14, 15) and AiiB (16), which may open the γ-butyrolactone (GBL) ring of the OC8-HSL QS signal (or “quormone”). The lactonase AttM, as well as AttK and AttL, is encoded by the attKLM operon, which is controlled at the transcriptional level by the repressor AttJ (14). The attKLM operon of A. tumefaciens C58 is involved in an assimilative pathway for GBL, γ-hydroxybutyrate (GHB), and succinate semialdehyde (SSA). In the presence of these compounds, the expression of the attKLM promoter is activated, and A. tumefaciens C58 does not accumulate OC8-HSL (15). Recently, proteome analysis revealed that this operon is also induced after exposure to tomato root segments (17), suggesting that the expression of the lactonase AttM may be controlled by as yet uncharacterized plant signal(s).
Here we show that GABA, which is structurally similar to the known attKLM-inducers GHB and SSA, activates the expression of the lactonase AttM, which in turn inactivates the QS signal. We propose GABA as a plant signal in the A. tumefaciens–plant interaction.
Results
GABA Stimulated the Expression of the attKLM Operon.
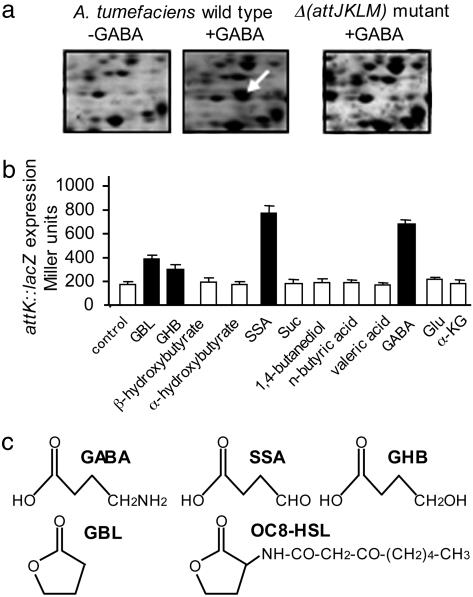
Proteome analysis was performed to investigate whether the plant pathogen A. tumefaciens responds to GABA, which is synthesized from glutamate (Glu) in wounded plant tissues. The addition of GABA to A. tumefaciens C58 cultures resulted in the accumulation of seven protein spots (undetected without GABA), five of which were identified by mass spectrometry and comparison with the reference map of the A. tumefaciens C58 proteome (18). The identified GABA-induced proteins were PnpA (a putative polyribonucleotide nucleotidyltransferase), FusA (a putative translational elongation factor), CyaA (a putative adenylate cyclase), and AttK and AttL, which are two proteins encoded by the same operon, attKLM. We verified that AttK and AttL did not accumulate in a previously constructed Δ(attJKLM) mutant (15), even in the presence of GABA (Fig. 1a illustrates the AttK protein spot). A transcriptional fusion, attK::lacZ (15), was used to demonstrate the transcriptional induction of the attKLM operon by GABA (Fig. 1b). GABA stimulated attK::lacZ expression at a level similar to that reached in the presence of SSA and at a higher level than those observed upon addition of GHB and GBL.
Fig. 1.
Expression of the A. tumefaciens operon attKLM in the presence of GABA. (a) The images show part of the two-dimensional gels around the expected position of AttK (indicated by an arrow), which was detected only in A. tumefaciens wild type in the presence of GABA (2 mM). (b) β-Galactosidase activity (in Miller units) was measured in cultures of A. tumefaciens C58 (attK::lacZ) 2 h after addition of the compounds tested (1 mM) Suc, succinate; α-KG, α-ketoglutarate. Values are given as means ± SD; black bars indicate means that are significantly different from that obtained with the addition of water as a blank control (Student’s t test with P < 0.05). (c) Structures of GABA, SSA, GHB, GBL, and OC8-HSL.
The A. tumefaciens bra Locus Was Required for attLKM Expression in the Presence of GABA.
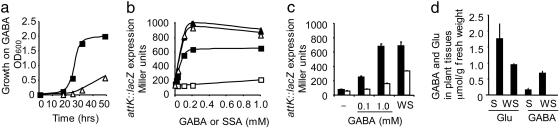
We investigated whether the GABA-signaling pathway in A. tumefaciens requires an active sensor/transport system for GABA. In the plant symbiotic bacterium Rhizobium leguminosarum, the braDEFG genes encode an active transport system for GABA (19). Based on sequence similarity to these genes, we identified the orthologous genes atu2427 (braD), atu2426 (braE), atu2425 (braF), and atu2424 (braG) on the circular chromosome of A. tumefaciens C58 (20). A braE mutant was constructed in A. tumefaciens C58 by a marker-exchange procedure. Growth of the A. tumefaciens braE mutant in a medium containing GABA as the sole nitrogen source was reduced (Fig. 2a). Moreover, the expression of the attK::lacZ fusion in the presence of GABA (Fig. 2b) or wounded stem tissues from tomato (Fig. 2c) was lower in the braE mutant than in the A. tumefaciens C58 wild type. We confirmed that wounding rapidly affected the concentrations of two amino acids, Glu and GABA, in the tomato stem (Fig. 2d). These changes are coherent with their respective substrate and product roles in GAD activity (6). However, the expression of attK::lacZ in the A. tumefaciens braE mutant, as compared with that in the wild-type strain, was higher in the presence of wounded plant tissues than in the presence of 1 mM GABA (Fig. 2c). A part of this Bra-independent induction of attKLM could be explained by an alternative GABA transporter in A. tumefaciens, which would specifically operate in wounded plant tissues. One cannot exclude the presence of a plant metabolite such as SSA, which would not require the transporter Bra to stimulate the expression of attKLM (Fig. 2b).
Fig. 2.
Requirement of GABA transporter Bra for the attKLM expression in A. tumefaciens. (a) Growth of the A. tumefaciens wild type (squares) and the mutant braE (triangles) in the presence of GABA as the sole nitrogen source. (b and c) The expression of the attK::lacZ fusion (β-galactosidase activity) in cultures of A. tumefaciens wild type (filled symbols in b and filled bars in c) and braE mutant (open symbols in b or open bars in c) was measured (in b) 2 h after the addition of SSA (triangles) or GABA (squares) and (in c) 20 h after the addition of GABA or wounded 6-week-old tomato stems (WS) (0.25 g of fresh weight per milliliter). (d) GABA and Glu concentrations in unwounded (S) and wounded (WS) 6-week-old tomato stems; values are given as means ± SD.
A. tumefaciens Did Not Accumulate OC8-HSL in the Presence of GABA.
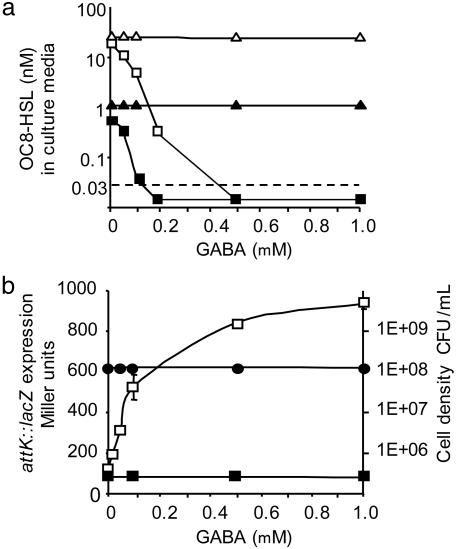
During the infection process, A. tumefaciens C58 synthesizes the QS signal OC8-HSL at two rates, depending on the absence or presence of agrocinopines that are produced by the A. tumefaciens C58-induced tumors. Agrocinopines stimulate the expression of the transcriptional regulator traR, which in turn activates the transcription of the traI gene encoding OC8-HSL synthase (9). Therefore, the agrocinopine-dependent synthesis of OC8-HSL may be mimicked in A. tumefaciens cultures that constitutively express the traR gene. In the absence of GABA, OC8-HSL reached a concentration of 0.5 nM in cultures of A. tumefaciens C58, as compared with 20 nM in those of A. tumefaciens C58 that overexpress traR. In the presence of 0.5 and 1.0 mM GABA, the OC8-HSL quormone was not detected in the cultures of either A. tumefaciens C58 or A. tumefaciens C58 overexpressing traR (Fig. 3a). We verified that, under the same experimental conditions, cell density remained unaffected by the addition of GABA and that the decrease in OC8-HSL concentration correlated with an increase in attK::lacZ expression (Fig. 3b). Addition of GABA, however, had no effect on the concentration of OC8-HSL in cultures of the Δ(attJKLM) mutant (Fig. 3a), which was previously shown to be unable to inactivate OC8-HSL even in the presence of the attK-inducers GBL, GHB, and SSA (15). These results demonstrate that, despite the positive control that may be exerted by traR on the synthesis of the QS signal, the induction of the attKLM operon by 0.5–1.0 mM GABA correlates to a dramatic decrease in the level of OC8-HSL. Wounded plant tissues stimulated attKLM expression at a level similar to that observed in the presence of 1.0 mM GABA (Fig. 2c); thus, the obliteration of the OC8-HSL signal is likely to occur in planta.
Fig. 3.
Effect of GABA on the concentration of OC8-HSL in A. tumefaciens cultures at the end of the exponential growth phase. (a) The OC8-HSL concentration was measured in cultures of A. tumefaciens C58 wild type (squares) and its Δ(attJKLM) derivative (triangles) overexpressing (open symbols) or not overexpressing (filled symbols) the traR gene. The dashed line indicates the detection limit of OC8-HSL in this experiment. (b) Under the same conditions described in a, the expression of the attK::lacZ (open squares) and attJ::lacZ (filed squares) fusions and the cell density [in colony-forming units (CFU) per milliliter; filled circles] of the A. tumefaciens C58 cultures were measured. Values are given as means ± SD.
Virulence of A. tumefaciens Was Attenuated in Plants Overexpressing GAD.
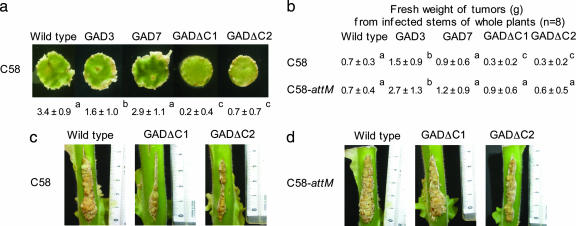
In A. tumefaciens, the OC8-HSL signal modulates the severity of the tumoral symptoms (12). It is tempting, therefore, to speculate that GABA-rich plants may affect the emergence of tumors because of the GABA-induced expression of the lactonase AttM. Virulence assays were performed on four homozygous transgenic tobacco lines that had an enhanced capacity to accumulate GABA (21). The lines GAD3 and GAD7 express a full-length tobacco GAD enzyme, whereas the lines GADΔC1 and GADΔC2 express a truncated enzyme, GADΔC, that lacks the autoinhibitory Ca2+/calmodulin binding domain (21). In the GADΔC lines, the synthesis of GABA by the truncated GAD is insensitive to the Ca2+/calmodulin complex. A virulence assay, performed on plant leaf discs by using A. tumefaciens C58, revealed that disease symptoms were less severe in GADΔC lines than in GAD lines and wild-type tobacco (Fig. 4a). Another virulence assay, performed on the stem of whole plants, confirmed that tumor weight (reflecting the severity of symptoms along a given 4-cm incision) was generally less in GADΔC plants than in GAD and wild-type plants (Fig. 4 b and c), suggesting that the Ca2+/calmodulin-independent synthesis of GABA in plant tissues affects the emergence of A. tumefaciens tumors. By contrast, the A. tumefaciens attM isogenic mutant was virulent in all mutant lines, as well as in the wild type (Fig. 4 b and d). The lactonase AttM, therefore, is strictly required for the attenuation of A. tumefaciens C58 virulence in GABA-accumulating plants.
Fig. 4.
Virulence assays on a wild-type tobacco plant and its transgenic derivatives GAD3, GAD7, GADΔC1, and GADΔC2. (a) Virulence assay on disks of tobacco leaves (n = 16) inoculated with A. tumefaciens C58. The scale of symptoms (0- 4) is described in Methods. (b) Virulence assays on stem of whole plants inoculated with A. tumefaciens C58 wild type or its attM mutant. The fresh weight (in grams) of tumors from infected stems of whole plants (n = 8) is given. (a and b) Values are given as means ± SD. Values on the same horizontal line that do not possess the same letter in superscript (a, b, or c) are statistically different (Student’s t test with P < 0.05). (c and d) Photographs illustrating the virulence assays described in b with wild-type, GADΔC1, and GADΔC2 tobacco plants infected with A. tumefaciens C58 (c) or attM mutant (d).
Expression of attKLM Was Not Regulated by Acetosyringone.
During the infection process, A. tumefaciens responds to other molecules from wounded plants, including phenolic compounds related to acetosyringone (8). These plant molecules induce the expression of the virulence (vir) genes of the Ti plasmid, thereby enabling transfer of a part of this plasmid (T-DNA) to a host plant. We investigated the possibility of cross-talk between the vir pathway of signal transduction and the GABA pathway. The addition of acetosyringone did not induce the expression of the attK::lacZ fusion and had no effect on the expression level of attK::lacZ in the presence of GABA (data not shown). Furthermore, no Vir proteins were identified among the seven GABA-induced proteins resulting from the proteomic analysis described above. Finally, in the presence of GABA, attK::lacZ expression reached a similar level in A. tumefaciens C58 and in an A. tumefaciens C58 derivative (C58.C1) that lacks the Ti plasmid (data not shown), confirming that the GABA-signaling pathway and vir-signaling pathway are independent. Remarkably, the genes regulated by these two plant inducible pathways are encoded by two independent replicons: the Ti plasmid for the vir genes and the At plasmid for the GABA-induced attKLM operon.
Discussion
Here we demonstrate that A. tumefaciens C58 has evolved a complex control of QS in response to plant signals. Although opines have previously been described as signals that stimulate OC8-HSL synthesis (9), here we reveal GABA as a signal for inducing AttM-mediated inactivation of this quormone. The Agrobacterium braDEFG locus, which encodes a putative ATP-binding cassette (ABC) transporter, was required for this GABA-signaling pathway. A model summarizing the regulation of the operon attKLM by GABA (Fig. 5) as well as a paradigm for signal exchange between a compatible plant host and A. tumefaciens (Fig. 6a) are proposed. Plant wounding is required to achieve complete expression of the vir functions that control the transfer of T-DNA from the Ti plasmid to plant cells, as well as tumor formation (8). At the same time, in wounded tissues GABA is accumulated to a high level, thereby preventing the accumulation of OC8-HSL via the GABA-induced lactonase-silencing pathway described here. Thereafter, in plant tumors some opines stimulate the synthesis of the OC8-HSL signal to a level that is sufficient to activate QS signal-regulated functions of A. tumefaciens (10–13). Here we show that this QS signaling may be disturbed in planta by the increased synthesis of GABA in genetically modified plants (Fig. 6b) in which GABA synthesis is independent of the Ca2+ calmodulin complex.
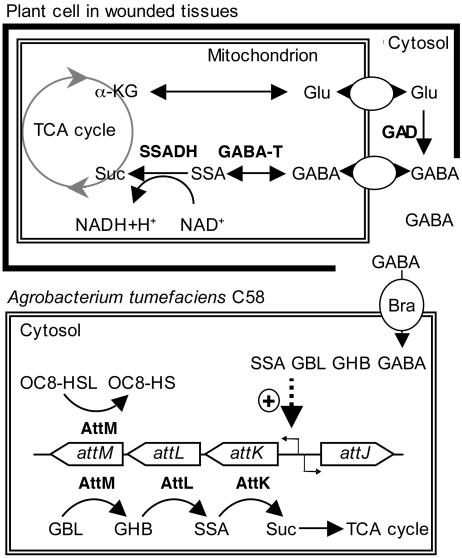
Fig. 5.
Scheme of attKLM regulation in the presence of wounded plant tissues. (Upper) The stress-induced synthesis and degradation of GABA in plants (6, 7). (Lower) Summary of the knowledge on the catabolic and QS signal-silencing functions of attKLM operon (14, 15), as well as its induction in the presence of SSA, GHB, and GBL (15) and GABA. α-KG, α-ketoglutarate; TCA cycle, tricarboxylic acid cycle; SSADH, SSA dehydrogenase; GABA-T, GABA transaminase; Suc, succinate; OC8-HS, N-(3-oxooctanoyl)homoserine.
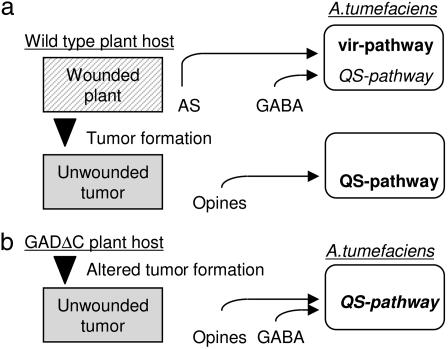
Fig. 6.
Model illustrating control of vir and QS pathways by plant signals. In the wild-type (a) and GADΔC transgenic plant lines (b), pathways activated by acetosyringone (AS) and opines are in bold face and that repressed by GABA is in italics; the QS pathway, which is simultaneously activated and repressed in the particular case of the GADΔC transgenic plant lines, is in bold italics.
Three nonexclusive hypotheses could explain the advantages to A. tumefaciens of such GABA-induced control of the N-acylhomoserine lactone (acyl-HSL) level. First, the silencing of QS signal-regulated functions, such as conjugation of the Ti plasmid and/or still undiscovered functions, would improve the availability of bacterial metabolites, including the Ti plasmid, to facilitate the transfer of T-DNA to the plant. Second, pure acyl-HSLs activate some plant defense responses (22), which may affect plant infectivity and transformation by A. tumefaciens (23). During the critical phase of T-DNA transfer to the plant at wounded sites, it is possible that A. tumefaciens cells obliterate acyl-HSL molecules and prevent the stimulation of some plant defense mechanisms. Third, because the GABA-induced lactonase AttM can cleave a range of acyl-HSLs (15, 16), this acyl-HSL-degrading activity may inactivate QS signals produced by other bacteria, thereby reducing competition for plant colonization at the early infection stage. The GABA-induced degradation of OC8-HSL would, however, constitute part of the plant defense mechanisms against bacterial infection. Such a role has already been assigned to GABA during infestation or feeding by invertebrate pests, whose neuromuscular junctions are very sensitive to GABA (21, 24). The transgenic GAD and GADΔC tobacco lines used in this work were previously described as more resistant than wild-type tobacco to infestation by the northern root-knot nematode (21).
Overall, the present work argues for a function of the ubiquitous, nonprotein amino acid GABA, which has previously been described as an essential signal for neurogenesis in animals (1, 2) and for pollen tube guidance in higher plants (4). The evidence that GABA acts as a signal between eukaryotes and bacteria offers an opportunity to develop strategies for defense against bacterial pathogens. Finally, because GABA synthesis and degradation are closely associated with the plant mitochondrion, it would be fascinating were GABA to play a role in communication between the ancient α-proteobacterium mitochondrion and the free-living α-proteobacterium A. tumefaciens.
Methods
Bacterial Strains, Plasmids, and Culture Conditions.
All of the A. tumefaciens strains used in this work were A. tumefaciens C58 derivatives. C58.C1 lacks the Ti plasmid (25). The attM::acc1 (GmR) and braE::aphA (KmR) mutants were constructed according to the same marker-exchange procedure described for mutant Δ(attJKLM)::aphA (15). In these mutants, the GmR and KmR cassettes (26) were inserted at the unique sites NarI and EcoRV of the attM and braE genes, respectively. A. tumefaciens NT1(pZLR4) was used as a biosensor for OC8-HSL (27). The plasmid p6000 (28) was used to drive constitutive expression of traR in A. tumefaciens C58. All plasmids, including those harboring the attK::lacZ and attJ::lacZ fusions (15), were introduced into A. tumefaciens strains by electroporation. A. tumefaciens C58 and its derivatives were cultivated at 30°C in Agrobacterium broth (AB) minimal medium (29) in the presence of mannitol and ammonium, except where alternative carbon and nitrogen sources are indicated. When appropriate, antibiotics were used at the following concentrations: ampicillin, 50 mg/liter; carbenicillin, 100 mg/liter; gentamycin, 20 mg/liter; kanamycin, 50 mg/liter; and tetracycline, 5 mg/liter.
Protein Analysis and β-Galactosidase Activity and OC8-HSL Measurements.
Two-dimensional gel electrophoresis and protein identification were performed according to established protocols (17), with cells grown for 24 h in the presence of glucose as the sole carbon source. Briefly, soluble proteins were separated in the first dimension on an 18-cm immobilized pH gradient (IPG) strip with a pH 4–7 gradient (Amersham Pharmacia Biosciences) for a total of 50 kV·h. For the second dimension, SDS/PAGE was performed on 12.5% total monomer (12.5% T), 2.6% cross-linker (2.6% C) gels, using the PROTEAN Plus Dodeca cell (Bio-Rad). Proteins of interest were excised from Coomassie blue-stained gels, digested with trypsin, and identified by peptide mass fingerprinting by using the Voyager-DE STR MALDI-TOF mass spectrometer (Applied Biosystems) and ms-fit software (http://prospector.ucsf.edu/ucsfhtml4.0/msfit.htm).
All measurements of transcriptional fusions were performed in the presence of mannitol and ammonium as carbon and nitrogen sources. The cells expressing the transcriptional fusion attK::lacZ or attJ::lacZ were cultivated in Agrobacterium broth (AB) mannitol medium for 24 h. These cultures were then supplemented with the compounds to be tested. After 2 or 24 h at 25°C, the expression of the fusion attK::lacZ was measured. The methods for quantification of β-galactosidase activity and the concentration of OC8-HSL were previously described (15, 27); these measurements were performed on three independent cultures.
Plant Material and Virulence Assays.
Leaf disks (1 cm in diameter) from 8-week-old Nicotiana tabacum L. cv. ‘Delgold’ wild-type plants and transgenic derivatives (21) were used in virulence assays as previously described (30). Tumors emerged around the leaf disk, and the level of virulence of A. tumefaciens was estimated according to the following symptom scale: 0, no tumors; 1, fewer than five tumors; 2, more than five tumors, but none contiguous; 3, contiguous tumors; 4, contiguous and large tumors, which modify the shape of the disk. Virulence assays were also conducted on stems of the 12-week-old tobacco plants, which were incised with a scalpel (incision of 4 cm in length) and inoculated with A. tumefaciens (108 colony-forming units per incision); the weights of tumors were measured 5 weeks after bacterial inoculation, as previously described (31). Unwounded and wounded stems of 6-week-old Solanum lycopersicum L. cv. ‘Monique’ were immediately frozen in liquid nitrogen and extracted in aqueous methanol as previously described (32); the composition of individual amino acids (including GABA) was determined by ion-exchange chromatography using the AminoTac JLC-500/V amino acid analyzer (33). Wounded tissues were generated by cutting 0.1-cm sections from tomato stems and then stored for 10 min at 25°C before amino acids analysis and assay of attK::lacZ induction.
Acknowledgments
We thank Y. Dessaux and C. Elmerich for critical reading of the manuscript, A. Raffoux for technical assistance in Agrobacterium genetics, O. Jambon for plant husbandry, Nathalie Mansion for photographs [Centre National de la Recherche Scientifique (CNRS), Gif-sur-Yvette, France], and S. Boutet for amino acid measurements (Institut National de la Recherche Agronomique, Versailles, France). This work was supported by grants from the CNRS (to D.F.), the United States–Israel Binational Agricultural Research and Development Fund (to R.R. and E.R.), the Natural Science and Engineering Research Council of Canada (to B.J.S.), and a Valachi Pikovsky fellowship (to R.R.).
Abbreviations
- GAD
glutamate decarboxylase
- Ti
tumor-inducing
- QS
quorum-sensing
- OC8-HSL
N-(3-oxooctanoyl)homoserine lactone
- GBL
γ-butyrolactone
- GHB
γ-hydroxybutyrate
- SSA
succinate semialdehyde
- acyl-HSL
N-acylhomoserine lactone.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Bouché N., Lacombe B., Fromm H. Trends Cell Biol. 2003;13:607–610. doi: 10.1016/j.tcb.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Kiegstein A. R. Nat. Neurosci. 2005;8:1132–1133. doi: 10.1038/nn0905-1132. [DOI] [PubMed] [Google Scholar]
- 3.Bouché N., Fait A., Bouchez D., Moller S. G., Fromm H. Proc. Natl. Acad. Sci. USA. 2003;100:6843–6848. doi: 10.1073/pnas.1037532100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palanivelu R., Brass L., Edlund A. F., Preuss D. Cell. 2003;114:47–59. doi: 10.1016/s0092-8674(03)00479-3. [DOI] [PubMed] [Google Scholar]
- 5.Metzner M., Germer J., Hengge R. Mol. Microbiol. 2004;51:799–811. doi: 10.1046/j.1365-2958.2003.03867.x. [DOI] [PubMed] [Google Scholar]
- 6.Shelp B. J., Bown A. W., McLean M. D. Trends Plant Sci. 1999;4:446–452. doi: 10.1016/s1360-1385(99)01486-7. [DOI] [PubMed] [Google Scholar]
- 7.Bouché N., Fromm H. Trends Plant Sci. 2004;9:110–115. doi: 10.1016/j.tplants.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 8.Stachel S. E., Messens E., van Montagu M., Zambryski P. Nature. 1985;318:624–629. [Google Scholar]
- 9.Piper K. R., Beck von Bodman S., Hwang I., Farrand S. K. Mol. Microbiol. 1999;32:1077–1089. doi: 10.1046/j.1365-2958.1999.01422.x. [DOI] [PubMed] [Google Scholar]
- 10.Zhang L., Murphy P. J., Kerr A., Tale M. E. Nature. 1993;362:446–448. doi: 10.1038/362446a0. [DOI] [PubMed] [Google Scholar]
- 11.Piper K. R., Beck von Bodman S., Farrand S. K. Nature. 1993;362:448–450. doi: 10.1038/362448a0. [DOI] [PubMed] [Google Scholar]
- 12.Pappas K. M., Winans S. C. Mol. Microbiol. 2003;48:1059–1073. doi: 10.1046/j.1365-2958.2003.03488.x. [DOI] [PubMed] [Google Scholar]
- 13.Li P. L., Farrand S. K. J. Bacteriol. 2000;182:179–188. doi: 10.1128/jb.182.1.179-188.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang H. B., Wang L. H., Zhang L. H. Proc. Natl. Acad. Sci. USA. 2002;99:4638–4643. doi: 10.1073/pnas.022056699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carlier A., Chevrot R., Dessaux Y., Faure D. Mol. Plant–Microbe Interact. 2004;17:951–957. doi: 10.1094/MPMI.2004.17.9.951. [DOI] [PubMed] [Google Scholar]
- 16.Carlier A., Uroz S., Smadja B., Fray R., Latour X., Dessaux Y., Faure D. Appl. Environ. Microbiol. 2003;69:4989–4993. doi: 10.1128/AEM.69.8.4989-4993.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosen R., Sacher A., Shechter N., Becher D., Buttner K., Biran D., Hecker M., Ron E. Z. Proteomics. 2004;4:1061–1073. doi: 10.1002/pmic.200300640. [DOI] [PubMed] [Google Scholar]
- 18.Rosen R., Matthysse A. G., Becher D., Biran D., Yura T., Hecker M., Ron E. Z. FEMS Microbiol. Ecol. 2003;44:355–360. doi: 10.1016/S0168-6496(03)00077-1. [DOI] [PubMed] [Google Scholar]
- 19.Hosie A. H., Allaway D., Galloway C. S., Dunsby H. A., Poole P. S. J. Bacteriol. 2002;184:4071–4080. doi: 10.1128/JB.184.15.4071-4080.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wood D. W., Setubal J. C., Kaul R., Monks D. E., Kitajima J. P., Okura V. K., Zhou Y., Chen L., Wood G. E., Almeida N. F., Jr, et al. Science. 2001;294:2317–2323. doi: 10.1126/science.1066804. [DOI] [PubMed] [Google Scholar]
- 21.McLean M. D., Yevtushenko D. P., Deschene A., Van Cauwenberghe O. R., Makhmoudova A., Potter J. W., Bown A. W., Shelp B. J. Mol. Breed. 2003;11:277–285. [Google Scholar]
- 22.Mathesius U., Mulders S., Gao M., Teplitski M., Caetano-Anolles G., Rolfe B. G., Bauer W. D. Proc. Natl. Acad. Sci. USA. 2003;100:1444–1449. doi: 10.1073/pnas.262672599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ditt R. F., Nester E., Comai L. FEMS Microbiol. Lett. 2005;247:207–213. doi: 10.1016/j.femsle.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 24.MacGregor K. E., Shelp B. J., Peiris S. E., Bown A. W. J. Chem. Ecol. 2003;29:2177–2182. doi: 10.1023/a:1025650914947. [DOI] [PubMed] [Google Scholar]
- 25.Vaudequin-Dransart V., Petit A., Chilton W. S., Dessaux Y. Mol. Plant–Microbe Interact. 1998;11:583–591. doi: 10.1094/mpmi-8-0311. [DOI] [PubMed] [Google Scholar]
- 26.Dennis J., Zylstra G. Appl. Environ. Microbiol. 1998;64:2710–2715. doi: 10.1128/aem.64.7.2710-2715.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cha C., Gao P., Chen Y. C., Shaw P. D., Farrand S. K. Mol. Plant–Microbe Interact. 1998;11:1119–1129. doi: 10.1094/MPMI.1998.11.11.1119. [DOI] [PubMed] [Google Scholar]
- 28.Maurhofer M., Reimann C., Schimidli-Sacherer P., Heeb S., Haas D., Défago G. Phytopathology. 1998;88:678–684. doi: 10.1094/PHYTO.1998.88.7.678. [DOI] [PubMed] [Google Scholar]
- 29.Chilton M. D., Currier T. C., Farrand S. K., Bendich A. J., Gordon M. P., Nester E. W. Proc. Natl. Acad. Sci. USA. 1974;71:3672–3676. doi: 10.1073/pnas.71.9.3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Banta L. M., Joerger R. D., Howitz V. R., Campbell A. M., Binns A. N. J. Bacteriol. 1994;176:3242–3249. doi: 10.1128/jb.176.11.3242-3249.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chesnokova O., Coutinho J. B., Khan I. H., Mikhail M. S., Kado C. I. Mol. Microbiol. 1997;23:579–590. doi: 10.1046/j.1365-2958.1997.d01-1875.x. [DOI] [PubMed] [Google Scholar]
- 32.Zhang G., Bown A. W. Phytochemistry. 1997;44:1007–1009. [Google Scholar]
- 33.Diaz C., Purdy S., Christ A., Morot-Gaudry J. F., Wingler A., Masclaux-Daubresse C. Plant Physiol. 2005;138:898–908. doi: 10.1104/pp.105.060764. [DOI] [PMC free article] [PubMed] [Google Scholar]