Abstract
The primate TRIM5α proteins have recently been defined as cellular restriction factors, preventing primate infection by retroviruses from different species. For instance, rhesus TRIM5α (rhTRIM5α) restricts infection by HIV-1. Virtually all TRIM5α proteins block the early replication of retroviruses by preventing the accumulation of reverse transcription products, but the underlying mechanism remains unclear. In this article, we find that disrupting proteasome function alters rhTRIM5α localization and allows the normal generation of HIV-1 late reverse transcription products, even though HIV-1 infection and the generation of nuclear 1-LTR and 2-LTR viral cDNA forms remain impaired. This finding suggests rhTRIM5α restricts HIV infection in two distinct phases: (i) altering the normal passage of the reverse-transcribing viral genome to the nucleus and (ii) targeting the reverse transcription complex to be disrupted by the proteasome. Because proteasome inhibitor blocks the second phase, accumulation of a nonfunctional viral DNA genome can be readily observed. Defining each phase may reveal HIV-1 targets for future antiviral therapy in which dual blockade may be equally as effective as naturally occurring rhTRIM5α protein in preventing HIV-1 infection in vivo.
Keywords: HIV, innate immunity
The species-specific tropism of many retroviruses is determined by cellular restriction factors present in different organisms. An early characterized restriction factor is the murine Fv1 gene, which prevents infection by particular strains of murine leukemia virus (MLV) (reviewed in ref. 1). Fv1 restriction involves the recognition of a capsid determinant in MLV (2, 3). Furthermore, MLV strains restricted by Fv1 can generate reverse transcription (RT) products but do not successfully integrate into the host-cell chromosome (4). This restriction to infection is also saturable because high levels of affected virus can overcome restriction (5, 6).
Similar restrictions to HIV infection in non-human primates (7–12) and non-human retroviruses in human cells (13–15) have also been characterized. The cellular TRIM5α protein was recently identified as underlying HIV-1 restriction in Old World monkey cells (16). Furthermore, a TRIM5-cyclophilin A fusion protein in owl monkey cells (TRIM-Cyp) also prevents infection by HIV-1 (17). Hence, the TRIM5α proteins constitute a major class of restriction factors regulating cellular infection by retroviruses from different species (reviewed in refs. 1 and 18).
TRIM5α belongs to a large family of proteins containing a tripartite motif (TRIM) comprising a RING domain, one or two B-box domains, and a coiled coil region (19). TRIM family members form high-order molecular-weight structures via their coiled coil regions, generating intracellular structures or bodies (19). The RING domain also has E3 ubiquitin ligase activity in some TRIM members like human TRIM5δ (20), fostering speculation that TRIM5α restriction may also involve proteasome activity (16). Many TRIM proteins also contain a carboxy-terminal motif specific for the TRIM family member. For instance, TRIM5α contains a characteristic SPRY domain at its carboxy terminus critical for determining the species-specific restriction of HIV-1 by Old World monkeys (21–23).
HIV-1 restriction by Old World monkey TRIM5α is similar to MLV restriction by Fv1 in that restriction can be saturated by excess virus (8, 10, 16), is governed by the retroviral capsid (8, 9, 13, 24, 25), and acts early during viral replication in target cells. However, TRIM5α and Fv1 have different outcomes when restricting early viral replication, with TRIM5α preventing RT products from accumulating (8, 26–28), whereas Fv1 acts later, preventing integrated provirus formation (4). In all instances, the precise mechanism allowing these cellular factors to obstruct early retroviral replication remains unclear. Therefore, to characterize the mechanism used by rhesus monkey TRIM5α (rhTRIM5α) to restrict early HIV-1 replication in target cells, we analyzed the effect of proteasome inhibitors on the cell biology of TRIM5α and its ability to restrict various steps early in HIV-1 infection. We found that proteasome inhibitors altered rhTRIM5α subcellular localization and prevented the rhTRIM5α block to HIV-1 RT, allowing HIV-1 late RT products to now accumulate even though viral infection and nuclear 1-LTR and 2-LTR cDNA forms remained impaired. This finding suggests that rhTRIM5α restriction involves two phases in which rhTRIM5α first interacts with the viral capsid and obscures its normal trafficking (analogous to Fv1), followed by proteasomal degradation of the viral core that blocks accumulation of RT products.
Results
MG132 Alters rhTRIM5α Intracellular Localization.
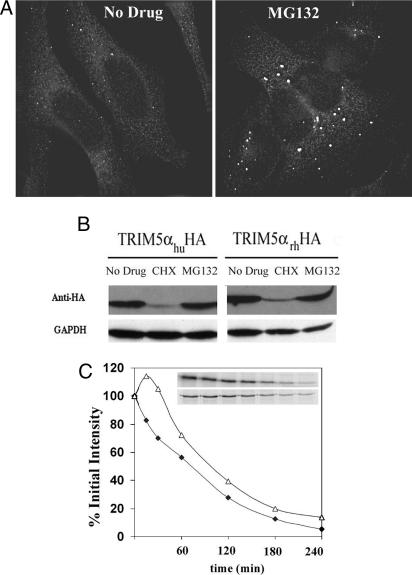
To gain insights into rhTRIM5α function and localization, we treated HeLa cells stably expressing GFP-rhTRIM5α with drugs affecting protein turnover and proteasome function. Inhibiting protein synthesis with cyclohexamide decreased GFP-rhTRIM5α expression (data not shown). Additionally, disrupting proteasome function with MG132 caused the GFP-rhTRIM5α cytoplasmic bodies to increase in size but decrease in number (data not shown). To verify that these changes were not due to the GFP fusion, the localization of epitope-tagged rhTRIM5α was tested. MG132 treatment of HeLa cells stably expressing hemagglutinin (HA)-tagged rhTRIM5α also caused cytoplasmic body size to increase relative to untreated cells (Fig. 1A). Western blot analysis of cells expressing HA-tagged rhTRIM5α or human TRIM5α (huTRIM5α) and treated with cyclohexamide revealed a large decrease in both TRIM5α protein levels (Fig. 1B), suggesting that these TRIM5α proteins turnover rapidly in the cell. In contrast, MG132 treatment had little effect on rhTRIM5α or huTRIM5α levels (Fig. 1B), indicating that the different localization of rhTRIM5α in MG132-treated cells (Fig. 1A) was not due to a large increase in rhTRIM5α expression. To gain further insight into the rate of rhTRIM5α turnover in cells, rhTRIM5α decline was analyzed by pulse–chase analysis (Fig. 1C). From this analysis, the half-life of rhTRIM5α was near 75 min. However, when MG132 was included in a parallel experiment to disrupt proteasome function, the half-life of rhTRIM5α remained similar (near 100 min; Fig. 1C). A small increase in rhTRIM5α expression was observed early after proteasome disruption, suggesting that a subset of rhTRIM5α was being degraded by the proteasome.
Fig. 1.
MG132 alters rhTRIM5α cytoplasmic localization. (A) rhTRIM5α.HA forms cytoplasmic bodies in stable HeLa cells (Left) that accumulate into larger bodies after 5 h of MG132 treatment (Right). (B) Western blot analysis reveals huTRIM5α.HA (Left) or rhTRIM5α.HA (Right) expression remains similar in HeLa cells after 6 h of MG132 treatment, despite declining with the translation inhibitor cyclohexamide (CHX). (C) Similar rhTRIM5α.HA stability without (Upper Inset; ♦) or with (Lower Inset; ▵) MG132 by pulse–chase analysis. Immunoprecipitated rhTRIM5α.HA bands are shown over time, with their percent intensity relative to the 0-min chase graphed.
MG132 Prevents Exogenous rhTRIM5α Restriction of HIV-1 Late RT Products but Not 2-LTR Circle Formation or Infection.
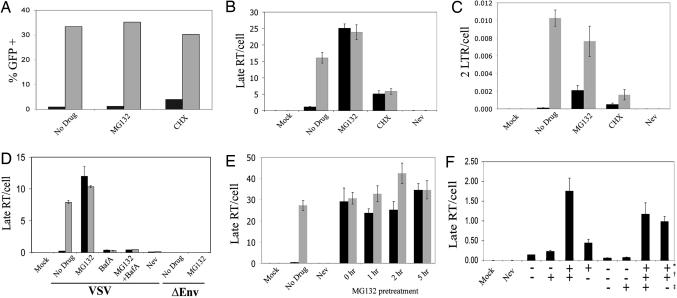
Next, we determined whether cyclohexamide or MG132 treatment influenced rhTRIM5α restriction of HIV infection using GFP reporter HIV-1. HeLa cells stably expressing rhTRIM5α or normal HeLa control cells were pretreated with either drug and infected in parallel. After 14 h, the virus and drugs were removed, and GFP expression was determined 48 h later. Stable rhTRIM5α expression in the HeLa cells restricted HIV-1 infection 50-fold (Fig. 2A). MG132 treatment caused a small equivalent increase in HIV-1 infection in both the rhTRIM5α expressing and control cells, as reported in refs. 29–31. In contrast, cyclohexamide treatment provided greater relief of rhTRIM5α restriction, increasing infection of the rhTRIM5α stable cells 6-fold compared with only a 1.3-fold increase in the control HeLa cells (Fig. 2A), probably reflecting the short half-life of rhTRIM5α observed in Fig. 1.
Fig. 2.
MG132 relieves exogenous rhTRIM5α restriction of HIV-1 late RT products but not 2-LTR circles. (A–C) Untransfected (gray bars) or rhTRIM5α.HA stable HeLa (black bars) were pretreated 5 h with MG132, cyclohexamide (CHX), or nevirapine (Nev) and infected 14 h with VSV-g-pseudotyped R7-GFPHIV-1 plus drug. Cells were analyzed for infection at 48 h (A; percentage of cells expressing GFP shown) or viral late RT products (B), or 2-LTR circles (C) per cell at 14 h by real-time PCR. Error bars reflect the SD of triplicate values in a representative experiment. (D) A similar experiment except cells were pretreated 2 h with drugs, including BafA, before infection with either VSV-g-pseudotyped (VSV) or envelope minus (ΔEnv) R7-GFPHIV-1 plus drug. (E) A similar experiment except the MG132 pretreatment time was varied before infection with VSV-g-pseudotyped R7-GFPHIV-1. (F) Washing MG132 in or out at 4 or 22 h into infection reveals rhTRIM5α restricts late RT product formation early but not late in infection. MG132 added: ∗, pretreatment + 4 h infection; †, 4–22 h postinfection; ‡, 22–44 h postinfection.
A concern with these experiments is the possible deleterious effects of long-term drug treatment on normal cell function. Therefore, the experiment was repeated, except that DNA was harvested 14 h into infection and late viral RT products were analyzed by real-time PCR to gauge effects on early HIV-1 replication. As previously reported, late RT product levels were greatly inhibited in cells expressing rhTRIM5α (Fig. 2B). However, disrupting proteasome function with MG132 rescued late RT products to levels similar to control cells (Fig. 2B), even though HIV-1 infection remained restricted (Fig. 2A). Equivalent levels of late RT products were also observed in the rhTRIM5α-expressing and control cells after cyclohexamide treatment. However, these levels were 3-fold below the untreated control cells, indicating although cyclohexamide relieved the rhTRIM5α block to late RT products, this treatment caused general inhibition of early HIV-1 replication.
The effect of these drugs on the nuclear localization of the viral cDNA was measured by using viral 2-LTR circles as a surrogate for nuclear localization. Although disrupting proteasome function restored late RT product levels in the rhTRIM5α-stable HeLa cells to those measured in control HeLa cells lacking rhTRIM5α (Fig. 2B), the levels of 2-LTR circles in the rhTRIM5α expressing HeLa remained ≈4-fold below control levels, even though 2-LTR circles were partially rescued by MG132 treatment (Fig. 2C). Similarly, 2-LTR circle levels in the rhTRIM5α cells were not restored to the same level in unrestricted HeLa cells after cyclohexamide treatment, even though 2-LTR levels increased 5-fold relative to the untreated restricted cells (Fig. 2C). Notably, cyclohexamide treatment decreased 2-LTR circle output 6.4-fold in the unrestricted HeLa cells, suggesting that new protein synthesis is necessary for efficient nuclear import (Fig. 2C). Taken together, these results suggest that MG132 treatment does not relieve rhTRIM5α restriction but rather arrests the restriction at a point after late RT products but before complete nuclear entry, potentially revealing an intermediate step in the rhTRIM5α restriction mechanism of HIV-1. Interestingly, after proteasome disruption by MG132, the molecular consequences of restriction by rhTRIM5α become similar to that seen for murine Fv1 in which late RT products accumulate normally, but nuclear localization of the viral genome is impaired (13).
To confirm that the rescue of HIV-1 late RT products by MG132 in the rhTRIM5α-expressing cells was specific for rhTRIM5α restriction and not a general consequence of MG132 treatment, similar experiments were performed by using HIV-1 defective for entry (Fig. 2D). To prevent entry, cells were infected with either HIV-1 lacking viral envelope protein (ΔEnv; Fig. 2D) or VSV-g-pseudotyped HIV-1 in the presence of bafilomycin A1 (BafA), which blocks endosome acidification and thus VSV-g-mediated fusion. Both scenarios prevented our GFP reporter HIV-1 from infecting control HeLa cells as measured by flow cytometry for GFP, confirming their entry defect (data not shown). Importantly, adding MG132 did not rescue late RT product output in either cell type when viral entry was blocked by either method (Fig. 2D). Therefore, MG132 specifically rescues HIV-1 late RT products from rhTRIM5α restriction in the cytosol because the same phenomenon could not be recapitulated by MG132 even when HIV-1 was trapped in endosomes by BafA.
MG132 Acts Rapidly to Relieve Exogenous rhTRIM5α Restriction of HIV-1 Late RT Products.
To determine how quickly MG132 functioned to recover late RT products in the rhTRIM5α expressing cells, the MG132 pretreatment time before infection was varied (Fig. 2E). Adding MG132 at the same time as infection without any pretreatment still increased late RT products in rhTRIM5α expressing HeLa to a level similar to control HeLa cells (Fig. 2E; 0 h), indicating MG132 acts quickly on rhTRIM5α restriction to restore late RT products. This finding is consistent with MG132 relieving rhTRIM5α restriction by rapidly inhibiting proteasome-mediated degradation rather than influencing ubiquitin-mediated protein trafficking, which is disrupted through depleting free ubiquitin pools after hours of proteasome inhibition (32).
To explore the time frame that HIV-1 late RT products were sensitive to rhTRIM5α restriction, an MG132 washout experiment was performed (Fig. 2F). Here, cells were infected with virus for 4 h only. MG132 was included in some infections to prevent rhTRIM5α restriction of late RT products, and the drug was washed out 4 or 22 h into infection. The reverse experiment of infecting cells with no MG132 but then adding drug at 4 and 22 h was also performed. Washing out MG132 4 h into infection caused late RT products to decline in the rhTRIM5α cells, demonstrating that the ability of MG132 to rescue HIV late RT product production is reversible. However, washing out MG132 at 22 h did not decrease late RT product levels relative to cultures in which MG132 was maintained, indicating that the viral late RT products eventually reach a state in which it is no longer susceptible to rhTRIM5α restriction. Adding MG132 4 or 22 h into infection could not rescue late RT product production in rhTRIM5α cells, indicating that the point that MG132 relieved restriction of late RT products had passed in these cells and occurred early in replication.
Proteasome Inhibitors Prevent Endogenous rhTRIM5α Restriction of HIV-1 Late RT Products but Not 2-LTR Circles or Infection.
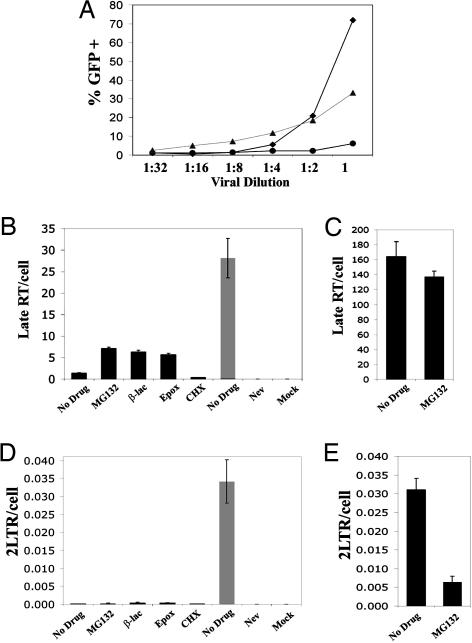
Next, we sought to determine whether inhibiting proteasome function could restore the production of HIV late RT products in rhesus monkey cells expressing endogenous levels of rhTRIM5α. For these studies, we used primary rhesus monkey lung fibroblasts (PRL) cells kindly provided by Joseph Sodroski (Harvard Medical School, Boston), who cloned the rhTRIM5α restriction factor from these cells (16). First, we determined the concentration of VSV-g-pseudotyped HIV that was sensitive to rhTRIM5α restriction in these cells by analyzing the infectivity of a dilution series of GFP reporter HIV-1. As seen in Fig. 3A, undiluted virus infected >70% of the cells, whereas a 1:4 dilution of this virus stock only infected 5% of the PRL cells, consistent with the previously described multiple hit model of infection that defines this saturable restriction factor when expressed in endogenous cells (7, 14). Furthermore, at a 1:4 dilution, viral infection in the PRL cells dropped below infection levels in the HeLa control cells lacking rhTRIM5α (Fig. 3A). We therefore examined infection of PRL cells under restricted (1:4 dilution; Fig. 3 B and D) and saturating (undiluted; Fig. 3 C and E) conditions. Three different types of proteasome inhibitors (MG132, β-lactone, and epoxomicin) were tested. We found that all three inhibitors increased late RT product levels under conditions in which the rhesus monkey PRL cells restricted HIV infection (Fig. 3B). In contrast, MG132 treatment of undiluted HIV-1, which saturates restriction in the PRL cells (Fig. 3A), slightly decreased the levels of late RT products (Fig. 3C). Therefore, proteasome inhibition only elevates late RT product production under circumstances when HIV-1 is restricted by rhTRIM5α. Furthermore, the proteasome inhibitors had little impact on the 2-LTR circle output in the PRLs infected with restricted amounts of HIV-1 (Fig. 3D). Therefore, the proteasome inhibitors influenced restriction of HIV-1 by endogenous and exogenous rhTRIM5α in the same trend.
Fig. 3.
Proteasome inhibitors relieve endogenous rhTRIM5α restriction of HIV-1 late RT products. (A) Percentage of PRL (♦), HeLa (▴), or rhTRIM5α.HA HeLa (•) cells infected by VSV-g-pseudotyped R7-GFPHIV-1 dilutions, measured using the GFP reporter. (B–E) PRL cells (black bars) were preincubated either with MG132, cyclohexamide (CHX), or nevirapine (Nev) for 2 h or β-lactone or epoxomicin for 6 h and infected 18 h with 1:4-diluted (B and D) or undiluted (C and E) R7-GFPHIV-1 plus drug. Quantitative real-time PCR analysis shows proteasome inhibitors prevent endogenous rhTRIM5α restriction of late RT products (B) but not 2-LTR circles (D). HeLa cells infected with 1:4-diluted virus demonstrate unrestricted infection (gray bars).
MG132 Permits Extensive RT of HIV-1 cDNA Despite Exogenous rhTRIM5α.
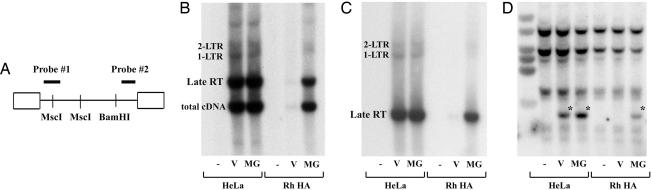
To gain further insights into the state of the reverse transcribed HIV-1 cDNA when proteasome function was disrupted in rhTRIM5α restricted cells, DNA present in the restricted cells was analyzed by Southern blot analysis 24 h into infection (Fig. 4). Two probes were used visualizing either the 5′ or 3′ end of the reverse transcribed viral cDNA (probe 1 and 2, respectively; Fig. 4A). HIV-1 late cDNA only accumulated to substantial levels in the rhTRIM5α cells after MG132 treatment, and detection by the 5′ and 3′ probes indicate this viral cDNA had nearly completed RT (Fig. 4 B and C). Furthermore, despite increases in this unintegrated viral late RT product with MG132, nuclear 1-LTR and 2-LTR circles remained low in rhTRIM5α cells compared with HeLa control cells. This finding indicates the viral cDNA intermediate detected in restricted cells after MG132 treatment has completely reverse transcribed but cannot efficiently gain access to the nucleus.
Fig. 4.
MG132 allows extensive RT of HIV-1 cDNA despite exogenous rhTRIM5α. (A) Location of DNA digest sites and probes for Southern blot analysis of HIV-1 5′ (probe 1) and 3′ (probe 2) cDNA ends. (B–D) Southern blot analysis of uninfected (−) or R7-GFPHIV-1 infected HeLa or rhTRIM5α.HA stable HeLa (Rh HA) cells without (V) or with MG132 (MG). The same blot probed for 5′ (B) or 3′ (C) HIV-1 cDNA or GAPDH for DNA loading (D) is shown. Asterisks mark residual probe 2. Although late RT products with complete ends escape rhTRIM5α restriction via MG132, 1-LTR and 2-LTR circles remain impaired.
Discussion
TRIM5α contains a RING domain, which is known to act as an E3 ubiquitin ligase and play a role in directing the ubiquitylation of specific proteins. Furthermore, a splice variant from the TRIM5 gene, TRIM5δ, can direct autoubiquitylation (20). The RING domain is required for efficient restriction by rhTRIM5α (33, 34), yet we show here, consistent with others, that disrupting proteasome function does not relieve rhTRIM5α-mediated restriction of HIV-1 infection (29). Instead, we found that disrupting proteasome function influences rhTRIM5α activity by preventing its ability to block the accumulation of HIV-1 late RT products. This change in rhTRIM5α action on HIV-1 by disrupting proteasome function has important ramifications, implying that rhTRIM5α restricts HIV-1 infection in more than one way and that ubiquitin/proteasome probably plays an important role in the cumulative action of rhTRIM5α to restrict HIV-1 infection.
Inhibiting proteasome function can affect cellular function in various ways, not only by blocking the degradation of polyubiquitylated proteins but also by disrupting monoubiquitination-mediated protein and vesicular trafficking by depleting the cytoplasmic-free pools of ubiquitin. An example in which inhibiting proteasome function perturbs protein trafficking is seen during release of retroviral Rous sarcoma virus (RSV) virions, where several hours exposure to proteasome inhibitors blocks virion release (32). However, we find here that the effect of proteasome inhibition to relieve rhTRIM5α restriction of HIV-1 late RT products was rapid (Fig. 2 E and F). This rapid effect suggests that the proteasome inhibitors are directly blocking proteasome-mediated degradation to allow HIV-1 late RT products to accumulate during rhTRIM5α restriction rather than effecting monoubiquitylation and protein trafficking. Furthermore, all three compounds (MG132, β-lactone, and epoxomicin) tested in this study, which inhibit proteasome function by different mechanisms (see ref. 35), had the same outcome on rhTRIM5α restriction of HIV-1, allowing late RT products to accumulate (Fig. 3B). This finding suggests that it is the direct inhibition of proteasome function by the proteasome drugs that allows HIV-1 late RT products to accumulate during rhTRIM5α restriction of HIV-1 infection.
Importantly, the proteasome inhibitors altered HIV-1 restriction by endogenously expressed rhTRIM5α in PRL cells and rhTRIM5α exogenously expressed in HeLa cells in a similar trend. Moreover, an increase in late RT products was not observed with MG132 treatment when restriction in PRL cells was saturated with a higher titer of virus. This finding indicates that proteasome disruption can only stimulate HIV-1 late RT product formation when the endogenous rhTRIM5α is decreasing HIV-1 infection in the PRL cells. Therefore, the smaller stimulatory effect of proteasome inhibitors on HIV-1 late RT products in the PRL cells is likely a consequence of the low endogenous expression of rhTRIM5α and the relatively smaller degree of restriction exhibited by PRL cells compared with HeLa cells expressing exogenous rhTRIM5α.
Although disrupting proteasome function altered rhTRIM5α localization, a major influence on rhTRIM5α protein levels was not detected here. Pulse–chase analysis revealed that the half-life of rhTRIM5α was ≈75 min (Fig. 1C), but only a minor increase in rhTRIM5α levels was observed after 6 h treatment with proteasome inhibitor by Western analysis (Fig. 1B). Furthermore, the rhTRIM5α half-life increased by <50% when proteasome function was blocked in pulse–chase studies (Fig. 1C). This finding indicates that proteasome function only plays a minimal role in regulating the constitutive levels of rhTRIM5α, consistent with a recent publication from Bieniasz and coworkers (29) in which disrupting ubiquitylation in a cell line with a temperature sensitive E1 ubiquitin ligase had little effect on expression levels of huTRIM5α or owl monkey TRIM-Cyp. Because MG132 induces rhTRIM5α to form larger cytoplasmic bodies, we note that these bodies might also preclude a subset of rhTRIM5α from being solubilized in our cell lysis buffer, thus preventing large increases in rhTRIM5α after MG132 treatment being observed in our analysis. However, this possibility seems unlikely given the short time course of the pulse–chase studies presented in Fig. 1C. Collectively, these data imply that the proteasome is not influencing rhTRIM5α turnover, but perhaps other factors, to influence HIV-1 late RT product accumulation.
That proteasome inhibition increased HIV-1 late RT product output but rhTRIM5α continued to restrict HIV-1 infection (Fig. 2) suggests rhTRIM5α-mediated loss of HIV-1 late RT products is not required for restriction and that another mechanism applies. This possibility is supported by a recent report from Towers and coworkers (12), who found that TRIM5α isolated from squirrel monkeys restricted infection by simian immunodeficiency virus (SIV) from rhesus macaques (SIVmac) without blocking the generation of late RT products (12). Therefore, perhaps the squirrel monkey TRIM5α analyzed by the Towers group has lost the ability to function as an E3 ubiquitin ligase and recruit proteasome activity, thereby allowing SIVmac late RT products to accumulate, analogous to the effect of disrupting proteasome function on rhTRIM5α restriction of HIV-1 reported here.
The ability of proteasome inhibitors to restore one aspect of the HIV-1 life cycle during rhTRIM5α restriction, where late RT products are generated, while HIV-1 infection is still blocked suggests that rhTRIM5α restricts HIV infection in two phases. In one phase, rhTRIM5α restriction appears to mimic the murine restriction factor, Fv1, allowing late RT products to accumulate but reducing their access to the nucleus (13), as measured by using 2-LTR circles as a marker for nuclear entry. Because TRIM5α and Fv1 recognize determinants in retroviral capsid for restriction (13), this may conceivably perturb the formation, composition, or trafficking of viral complexes housing the viral cDNA or mask their nuclear import motifs to obscure viral cDNA trafficking into the nucleus. In a second phase, rhTRIM5α either induces degradation or prevents late RT product formation via proteasome function. Perhaps, rhTRIM5α directs incoming early RT product complexes to be degraded by the proteasome, preventing late RT product formation. This theory is consistent with the MG132 washout experiment depicted in Fig. 2F in which rhTRIM5α could decrease late RT products when MG132 was washed out early in infection (4 h) but not late in infection (22 h).
Because the Fv1-like restriction phase is only seen if proteasome inhibitors block the late RT product degradation phase, we speculate that rhTRIM5α and Fv1 might initially interact with incoming viral cores with similar consequences, but only rhTRIM5α has the second ability to block late RT product formation via proteasome function. In this way, rhTRIM5α restricts retroviral infection as an evolutionarily improved version of Fv1. Ultimately, restricting HIV-1 infection in two phases would ensure that rhTRIM5α efficiently blocked viral infection. By preventing late RT product formation, infection is more efficiently blocked. Dissecting these phases may expand understanding of how cellular TRIM5α proteins block retroviruses in vivo and reveal dual targets for future antiviral therapies.
Materials and Methods
Cells and Pharmaceuticals.
293T and HeLa (American Type Culture Collection), HeLa cells stably expressing rhTRIM5α.HA or huTRIM5α.HA (16), and primary rhesus monkey lung fibroblasts [PRL; gifts from Dr. Joseph Sodroski] (36) were cultured at 37°C and 7% CO2 in DMEM (HyClone) supplemented with 10% FBS/100 units/ml penicillin/100 μg/ml streptomycin/292 μg/ml l-glutamine (Gibco). Ciprofloxacin (10 μg/ml; Cellgro; Mediatech, Washington, DC) was included in the medium plus 1 μg/ml puromycin (Sigma) for rhTRIM5α.HA or huTRIM5α.HA HeLa to maintain selection.
Microscopy.
rhTRIM5α stable cells were adhered to fibronectin-treated coverslips and cultured 5 h more with or without 1 μg/ml MG132. Cells were fixed with 3.7% formaldehyde (Polysciences) in 0.1 M Pipes, pH 6.8. Cells were stained with anti-HA monoclonal antibody (Sigma) and FITC-conjugated donkey anti-mouse antibody (Jackson ImmunoResearch). Images were collected and deconvolved with a Deltavision microscope and software (Applied Precision, Issquah, WA).
Protein Analysis.
For Western blot analysis, cells treated with or without MG132 or cyclohexamide for 6 h were lysed with laemmli buffer and proteins were resolved by SDS/PAGE, transferred to a nitrocellulose membrane (Whatman) and probed with anti-HA monoclonal antibody (Sigma) or anti-GAPDH monoclonal antibody (Chemicon International) plus anti-mouse peroxidase-conjugated goat antibody (Pierce). For pulse–chase studies, rhTRIM5α.HA stable cells were cultured overnight in complete medium, starved for 1 h at 37°C in methionine/cysteine-free DMEM containing 5% dialyzed FBS and labeled by adding 100 μCi/ml [35S]methionine/cysteine and incubating 1 h at 37°C. Cells were washed and chased for various times at 37°C in complete medium with or without MG132. After chase, cells were lysed in cold IVKA lysis buffer [50 mM Hepes, pH 7.4/150 mM NaCl/10% glycerol/1% Triton X-100/1 mM EGTA/1.5 mM MgCl2/1 mM Na-orthovanadate/1 mM phenylmethylsulfonyl fluoride/protease inhibitor mixture (Roche Biochemicals)]. Clarified cell lysates were incubated with anti-HA antibody and then immunoprecipitated with protein A Sepharose (Sigma). Subsequent immunoprecipitation of GAPDH from the same lysates allowed for normalization. Immunoprecipitated samples were resolved by SDS/PAGE, transferred to nitrocellulose, detected by autoradiography, and quantified on a PhosphoImager (Molecular Dynamics).
Infections.
To produce virus, a 10-cm plate of 293T cells was transfected with 12 μg of R7ΔEnvGFP and 8 μg of VSV-g expression plasmid by using polyethylenimine (molecular weight, 25,000; Polysciences). The VSV-g plasmid was omitted to make the R7ΔEnvGFP virus lacking envelope protein. Virus was harvested as described in ref. 37. To assess virus infectivity, equivalent numbers of cells in a 96- or 48-well plate were pretreated 5 h with drug where appropriate before adding virus plus drug if appropriate. Infections were typically performed for 14 h after which virus and drug were removed, normal medium added, and GFP expression determined 48–72 h after infection by using a FACSCalibur flow cytometer (Becton Dickinson).
Quantitative Real-Time PCR.
For real-time PCR studies, cells were seeded in 12-well plates at either 2 × 105 or 0.6 × 105 cells per well for the rhTRIM5α.HA HeLa (Fig. 2) or PRL (Fig. 3) experiments, respectively. The next day, monolayers were pretreated before infection with drug in the following amounts where appropriate: 20 μg/ml cyclohexamide, 10 μM nevirapine, 1 μg/ml MG132, 25 μM β-lactone, 3 μM epoxomicin, or 50 nM BafA. VSV-g-pseudotyped HIV-1 or envelope minus R7ΔEnvGFP HIV-1 was pretreated with 20 units/ml DNaseI (Roche) in 10 mM MgCl2 for 60 min at room temperature before addition to cell monolayers with drug if appropriate. After 2 h at 37°C, infections were typically supplemented with an equal volume of medium containing appropriate drug and returned to 37°C until harvest 12 h later. Cells were collected, treated with RNase A, and genomic DNA extracted as per manufacturer instructions (Qiagen DNeasy tissue kit; Valencia, CA). After digestion with 1 unit/μl DpnI (New England Biolabs) for 4 h at 37°C, 50 ng of DNA was analyzed for cellular β-actin or viral late RT product or 2-LTR circles by real-time PCR by using published primers (37, 38), iQ SYBR Green Supermix, and the iCycler iQ Real-Time PCR detection system (Bio-Rad). β-Actin in PRL cells was measured by using the following primers: forward, 5′-TCACCCACACTGTGCCCATCTATGA-3′; and reverse, 5′-CAGCGGAACCGCTCGTTGCCAATGG-3′. Dilutions of genomic DNA (10-fold), proviral plasmid, or plasmid containing cloned 2-LTR junction DNA, in 30 ng/μl tRNA, were used to generate standard curves for quantifying β-actin, viral late RT products, or 2-LTR circles, respectively.
Southern Blot Analysis.
HeLa or rhTRIM5α.HA stable HeLa cells were pretreated 2 h with 1 μg/ml MG132 where appropriate and infected at 24 h with DNaseI-treated, VSV-g-pseudotyped R7ΔEnvGFP HIV-1 plus drug where necessary. Genomic DNA was extracted from infected cells as above, and 10 μg of DNA was digested with MscI, BamHI, and DpnI overnight. After electrophoresis on 0.7% agarose and transfer, nylon membranes were probed with either 32P-labeled DNA or RNA probes. HIV-1 DNA probes were amplified and labeled with 32P by PCR using the R7ΔEnvGFP plasmid template and the following primers: probe 1, forward 5′-AGAAGAAATGATGACAGCATG-3′ and reverse 5′-TGCCAGTTCTAGCTCTG-3′ (39); probe 2, forward 5′-CCGGAATTCGTGAGCAAGGGCGAGGAGCTGTTC-3′ and reverse 5′-CGGGGTACCCTTGTACAGCTCGTCCAT-3′. The GAPDH 32P-labeled RNA probe was synthesized with a MAXIscript kit (Ambion, Austin, TX) using the pTRI-GAPDH-human template (Ambion). Probed blots were visualized by autoradiography.
Acknowledgments
We thank Dr. Joseph Sodroski (Harvard Medical School, Boston) for providing TRIM5α stable HeLa and PRL cells. This work was supported by National Institutes of Health Grant R01 AI47770 (to T.J.H.). T.J.H. is an Elizabeth Glaser Scientist.
Abbreviations
- MLV
murine leukemia virus
- rhTRIM5α
rhesus monkey TRIM5α
- huTRIM5α
human TRIM5α
- RT
reverse transcription
- ΔEnv
deletion of HIV-1 envelope protein
- BafA
bafilomycin A1
- PRL
primary rhesus monkey lung fibroblasts
- HA
hemagglutinin.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Bieniasz P. D. Nat. Immunol. 2004;5:1109–1115. doi: 10.1038/ni1125. [DOI] [PubMed] [Google Scholar]
- 2.Kozak C. A., Chakraborti A. Virology. 1996;225:300–305. doi: 10.1006/viro.1996.0604. [DOI] [PubMed] [Google Scholar]
- 3.DesGroseillers L., Villemur R., Jolicoeur P. J. Virol. 1983;47:24–32. doi: 10.1128/jvi.47.1.24-32.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pryciak P. M., Varmus H. E. J. Virol. 1992;66:5959–5966. doi: 10.1128/jvi.66.10.5959-5966.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pincus T., Hartley J. W., Rowe W. P. Virology. 1975;65:333–342. doi: 10.1016/0042-6822(75)90039-2. [DOI] [PubMed] [Google Scholar]
- 6.Duran-Troise G., Bassin R. H., Rein A., Gerwin B. I. Cell. 1977;10:479–488. doi: 10.1016/0092-8674(77)90035-6. [DOI] [PubMed] [Google Scholar]
- 7.Besnier C., Takeuchi Y., Towers G. Proc. Natl. Acad. Sci. USA. 2002;99:11920–11925. doi: 10.1073/pnas.172384599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cowan S., Hatziioannou T., Cunningham T., Muesing M. A., Gottlinger H. G., Bieniasz P. D. Proc. Natl. Acad. Sci. USA. 2002;99:11914–11919. doi: 10.1073/pnas.162299499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hatziioannou T., Perez-Caballero D., Yang A., Cowan S., Bieniasz P. D. Proc. Natl. Acad. Sci. USA. 2004;101:10774–10779. doi: 10.1073/pnas.0402361101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hatziioannou T., Cowan S., Goff S. P., Bieniasz P. D., Towers G. J. EMBO J. 2003;22:385–394. doi: 10.1093/emboj/cdg042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keckesova Z., Ylinen L. M., Towers G. J. Proc. Natl. Acad. Sci. USA. 2004;101:10780–10785. doi: 10.1073/pnas.0402474101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ylinen L. M., Keckesova Z., Wilson S. J., Ranasinghe S., Towers G. J. J. Virol. 2005;79:11580–11587. doi: 10.1128/JVI.79.18.11580-11587.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Towers G., Bock M., Martin S., Takeuchi Y., Stoye J. P., Danos O. Proc. Natl. Acad. Sci. USA. 2000;97:12295–12299. doi: 10.1073/pnas.200286297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Towers G., Collins M., Takeuchi Y. J. Virol. 2002;76:2548–2550. doi: 10.1128/jvi.76.5.2548-2550.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Besnier C., Ylinen L., Strange B., Lister A., Takeuchi Y., Goff S. P., Towers G. J. J. Virol. 2003;77:13403–13406. doi: 10.1128/JVI.77.24.13403-13406.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stremlau M., Owens C. M., Perron M. J., Kiessling M., Autissier P., Sodroski J. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 17.Sayah D. M., Sokolskaja E., Berthoux L., Luban J. Nature. 2004;430:569–573. doi: 10.1038/nature02777. [DOI] [PubMed] [Google Scholar]
- 18.Nisole S., Stoye J. P., Saib A. Nat. Rev. Microbiol. 2005;3:799–808. doi: 10.1038/nrmicro1248. [DOI] [PubMed] [Google Scholar]
- 19.Reymond A., Meroni G., Fantozzi A., Merla G., Cairo S., Luzi L., Riganelli D., Zanaria E., Messali S., Cainarca S., et al. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu L., Yang L., Moitra P. K., Hashimoto K., Rallabhandi P., Kaul S., Meroni G., Jensen J. P., Weissman A. M., D’Arpa P. Exp. Cell Res. 2003;288:84–93. doi: 10.1016/s0014-4827(03)00187-3. [DOI] [PubMed] [Google Scholar]
- 21.Sawyer S. L., Wu L. I., Emerman M., Malik H. S. Proc. Natl. Acad. Sci. USA. 2005;102:2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stremlau M., Perron M., Welikala S., Sodroski J. J. Virol. 2005;79:3139–3145. doi: 10.1128/JVI.79.5.3139-3145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yap M. W., Nisole S., Stoye J. P. Curr. Biol. 2005;15:73–78. doi: 10.1016/j.cub.2004.12.042. [DOI] [PubMed] [Google Scholar]
- 24.Owens C. M., Song B., Perron M. J., Yang P. C., Stremlau M., Sodroski J. J. Virol. 2004;78:5423–5437. doi: 10.1128/JVI.78.10.5423-5437.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Owens C. M., Yang P. C., Gottlinger H., Sodroski J. J. Virol. 2003;77:726–731. doi: 10.1128/JVI.77.1.726-731.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shibata R., Sakai H., Kawamura M., Tokunaga K., Adachi A. J. Gen. Virol. 1995;76:2723–2730. doi: 10.1099/0022-1317-76-11-2723. [DOI] [PubMed] [Google Scholar]
- 27.Munk C., Brandt S. M., Lucero G., Landau N. R. Proc. Natl. Acad. Sci. USA. 2002;99:13843–13848. doi: 10.1073/pnas.212400099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Himathongkham S., Luciw P. A. Virology. 1996;219:485–488. doi: 10.1006/viro.1996.0276. [DOI] [PubMed] [Google Scholar]
- 29.Perez-Caballero D., Hatziioannou T., Zhang F., Cowan S., Bieniasz P. D. J. Virol. 2005;79:15567–15572. doi: 10.1128/JVI.79.24.15567-15572.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwartz O., Marechal V., Friguet B., Arenzana-Seisdedos F., Heard J. M. J. Virol. 1998;72:3845–3850. doi: 10.1128/jvi.72.5.3845-3850.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Butler S. L., Johnson E. P., Bushman F. D. J. Virol. 2002;76:3739–3747. doi: 10.1128/JVI.76.8.3739-3747.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patnaik A., Chau V., Wills J. W. Proc. Natl. Acad. Sci. USA. 2000;97:13069–13074. doi: 10.1073/pnas.97.24.13069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Javanbakht H., Diaz-Griffero F., Stremlau M., Si Z., Sodroski J. J. Biol. Chem. 2005;280:26933–26940. doi: 10.1074/jbc.M502145200. [DOI] [PubMed] [Google Scholar]
- 34.Perez-Caballero D., Hatziioannou T., Yang A., Cowan S., Bieniasz P. D. J. Virol. 2005;79:8969–8978. doi: 10.1128/JVI.79.14.8969-8978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee D. H., Goldberg A. L. Trends Cell Biol. 1998;8:397–403. doi: 10.1016/s0962-8924(98)01346-4. [DOI] [PubMed] [Google Scholar]
- 36.Hofmann W., Schubert D., LaBonte J., Munson L., Gibson S., Scammell J., Ferrigno P., Sodroski J. J. Virol. 1999;73:10020–10028. doi: 10.1128/jvi.73.12.10020-10028.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Campbell E. M., Nunez R., Hope T. J. J. Virol. 2004;78:5745–5755. doi: 10.1128/JVI.78.11.5745-5755.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Butler S. L., Hansen M. S., Bushman F. D. Nat. Med. 2001;7:631–634. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- 39.Zennou V., Petit C., Guetard D., Nerhbass U., Montagnier L., Charneau P. Cell. 2000;101:173–185. doi: 10.1016/S0092-8674(00)80828-4. [DOI] [PubMed] [Google Scholar]