Abstract
Mutations that cause reduced expression of the full-length Survival Motor Neurons (SMN) protein are a major cause of spinal muscular atrophy (SMA), a disease characterized by degeneration of the α-motor neurons in the anterior horn of the spinal cord. The severity of SMA may be influenced by the actions of modifier genes. One potential modifier gene is represented by ZPR1, which is down-regulated in patients with SMA and encodes a zinc finger protein that interacts with complexes formed by SMN. To test the functional significance of ZPR1 gene down-regulation, we examined a mouse model with targeted ablation of the Zpr1 gene. We report that ZPR1-deficient mice exhibit axonal pathology and neurodegeneration. These data identify ZPR1 deficiency as a contributing factor in neurodegenerative disorders.
Keywords: spinal muscular atrophy, SMN, axonopathy, Wallerian degeneration
Spinal muscular atrophy (SMA) is the leading cause of infant death in the U.S. that results from an inherited genetic defect and is characterized by the degeneration of α-motor neurons in the anterior horn of the spinal cord (1). SMA is caused by mutation of the Survival Motor Neurons (SMN) 1 gene that results in low level expression of the full-length SMN protein (2, 3). This genetic locus includes two copies of the SMN gene, SMN1 (telomeric) and SMN2 (centromeric) located in an inverted repeat on chromosome 5q13 (2). In 5q-linked SMA patients, the SMN1 gene is deleted or mutated, and the SMN2 gene expresses transcripts that undergo alternative splicing due to a translationally silent nucleotide difference (C → T, codon 280) in exon 7 (4). Alternative splicing of transcripts from the SMN2 gene causes skipping of exon 7 and predominant expression of a truncated SMNΔexon7 protein (4) that does not interact with many of the components of the SMN complex, including ZPR1 (5, 6). This loss of expression of full-length SMN protein is a major cause of SMA.
Although it is established that the severity of SMA negatively correlates with the amount of full-length SMN protein (3), the severity of SMA may also be influenced by the actions of modifier genes (7–9). Thus, the 5q13 locus also includes the Neuronal Apoptosis Inhibitory Protein gene (NAIP) and p44 (a gene that encodes a subunit of the TFIIH transcription factor), and homozygous deletion of these genes has been observed in 55% and 73% of patients with severe SMA type I, respectively (2, 10, 11). The ZPR1 gene also represents a potential modifier of SMA because ZPR1 is expressed at low levels in patients with severe SMA (9) and it is known that reduced ZPR1 expression causes defects in the subcellular localization of SMN complexes (5, 12).
The purpose of this study was to test whether reduced expression of ZPR1 contributes to neurodegeneration. Our approach was to examine mice with targeted ablation of the Zpr1 gene. We report that ZPR1-deficient mice exhibit neurodegeneration.
Results and Discussion
To test the hypothesis that the reduced expression of ZPR1 observed in SMA patients (9) may contribute to disease severity, we examined the effect of targeted ablation of the Zpr1 gene in mice. Homozygous Zpr1−/− mice exhibited an early embryonic lethal phenotype (12). In contrast, heterozygous Zpr1+/− mice were viable. However, these mice exhibited occasional seizures, fatigue, and an abnormal gait with increased paw abduction compared with wild-type mice. The Zpr1+/− mice showed longer strides and an irregular step pattern (Fig. 1A). The overlap between forepaw and hindpaw was reduced, and the angle between the left and right paws was increased. Similar observations were obtained in studies of both male and female mice.
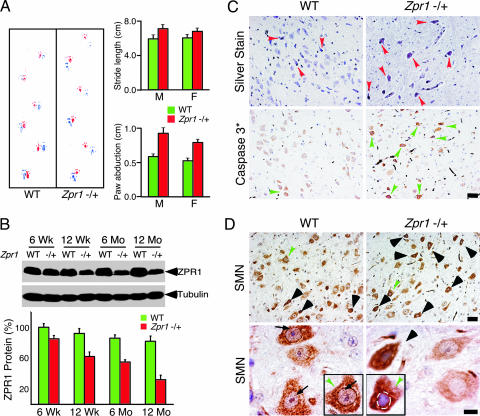
Fig. 1.
ZPR1 deficiency causes motor defects and apoptotic degeneration of facial motor neurons. (A) Abnormal gait of Zpr1+/− mice. Footprint patterns of 12-month-old wild-type (WT) and Zpr1+/− littermates (red, forepaws; blue, hindpaws) were examined. Quantitative analysis of stride length and paw abduction (26) for males (M) and females (F) was performed three times for each mouse (six mice per group). The results are presented as the mean ± SD. (B) Reduced Zpr1 gene dosage causes decreased expression of ZPR1 protein in the brain. Quantitation of immunoblots of ZPR1 expression in the brain was performed by using metamorph software (mean ± SD; four mice per group). (C) Low levels of ZPR1 cause apoptotic degeneration of facial motor neurons. Histochemical detection of facial motor neurons in sagittal sections of the brainstem from 12-month-old WT and Zpr1+/− mice. Brainstem section were examined by using silver stain to detect degenerating facial motor neurons (arrowheads) and counterstained with hematoxylin (Upper). Immunohistochemical detection of apoptosis used a monoclonal antibody to cleaved caspase 3 (arrowheads) in brainstem sections of 12-month-old mice (Lower). (Scale bar is 50 μm.) (D) ZPR1 deficiency causes degeneration of facial motor neurons. Immunohistochemical detection of SMN in sagittal sections of the brainstem from WT and Zpr1+/− mice. Degenerating neurons were found to be chromolytic (arrowheads) due to increased cytoplasmic accumulation of SMN. Neurons (green arrowheads) are also shown at higher magnification (Lower Inset). Arrows show the presence of prominent punctate intranuclear staining of SMN in WT neurons. (Scale bars, 50 μm in Upper and 10 μm in Lower.)
Reduction of gene dosage by mutation of one allele of Zpr1 caused decreased expression of ZPR1 protein during mouse development (Fig. 1B). Quantitation of ZPR1 protein in total brain extracts of Zpr1+/+ and Zpr1+/− mice showed a gradual loss of ZPR1 from 15 ± 3.1% to 50 ± 5.6% (mean ± SD; n = 4) with increasing age of mice from 6 weeks to 12 months (Fig. 1B). Major morphological differences between the brains of 12-month-old wild-type and Zpr1+/− mice were not detected by histological analysis. However, analysis of facial motor neurons in the brainstem by histochemical staining (silver) and immunohistochemical staining (cleaved caspase 3) indicated degeneration of facial motor neurons in Zpr1+/− mice (Fig. 1C). Degenerating facial motor neurons exhibited loss of punctate localization of SMN within the nucleus in Zpr1+/− mice (Fig. 1D). This observation is consistent with the finding that low levels of ZPR1 expression causes mislocalization of SMN (5, 12). These data suggest that reduced Zpr1 gene dosage in mice causes progressive ZPR1 deficiency and motor defects.
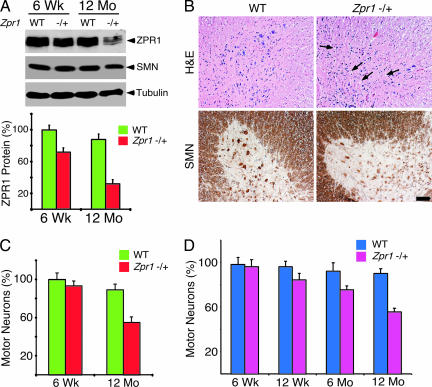
To determine whether ZPR1 deficiency causes loss of spinal cord motor neurons, we examined the expression of ZPR1 protein and the number of motor neurons in the spinal cord of wild-type and Zpr1+/− mice. Reduced ZPR1 protein expression was observed in the spinal cords of 6-week-old Zpr1+/− mice (23 ± 3.6%) and 12-month-old Zpr1+/− mice (55 ± 4.2%) compared with wild-type mice (mean ± SD; n = 4; Fig. 2A). The number of motor neurons detected in sections of the lumbar region of the spinal cord of 6-week-old wild-type and Zpr1+/− mice was similar (Fig. 2C). In contrast, the spinal cords of 12-month-old Zpr1+/− mice indicated reduced cell density, smaller size, and marked loss (43 ± 3.8%; n = 6) of anterior horn motor neurons compared with wild-type mice (Fig. 2 B and C). These data suggest that Zpr1+/− mice exhibit a progressive loss of motor neurons. To test this hypothesis, we examined serial sections of the thoracic region of spinal cords of wild-type and Zpr1+/− mice at different ages (Fig. 1D). Progressive loss of spinal cord motor neurons was observed with aging of Zpr1+/− mice starting from 2.0 ± 1.0% (6 weeks), 10 ± 2.2% (12 weeks), 18 ± 2.6% (6 months), to 38 ± 3.5% (12 months) (mean ± SD; n = 6 mice per group). This loss of motor neurons correlates with the age-dependent reduction in ZPR1 protein expression observed in Zpr1+/− mice (Figs. 1B and 2 A and D). The decreased number of spinal cord motor neurons in Zpr1+/− mice is similar to that observed in Smn+/− mice that exhibit a phenotype that resembles human SMA type III (13).
Fig. 2.
Progressive loss of spinal cord motor neurons in Zpr1+/− mice. (A) Reduced Zpr1 gene dosage causes decreased expression of ZPR1 protein in the spinal cord. The expression of ZPR1 in the spinal cords of 6-week-old and 12-month-old WT and Zpr1+/− mice was examined by immunoblot analysis and quantitated by using metamorph software (mean ± SD; four mice per group). (B) Staining with hematoxylin and eosin (Upper) and an antibody to SMN (Lower) of the anterior horns of the lumbar region of spinal cords from 12-month-old WT and Zpr1+/− mice (scale bar, 100 μm). Void spaces are indicated (arrows). (C) ZPR1 deficiency causes loss of spinal cord motor neurons. Quantitation of the number of spinal motor neurons in serial sections of spinal cords from 6-week-old and 12-month-old WT and Zpr1+/− littermates was performed. Motor neurons were counted in every fifth section of the lumbar (L1–L5) region of the spinal cords (mean ± SD; six mice per group). The loss of motor neurons was calculated by using number of motor neurons in 6-week-old mice (100%) as reference point. (D) Progressive loss of spinal cord motor neurons in Zpr1+/− mice. Quantitation of motor neuron numbers in serial sections of the spinal cords from WT and Zpr1+/− littermates. Motor neurons were counted in every fifth section of the thoracic (T9–T12) region of spinal cords (mean ± SD; six mice per group). The loss of motor neurons was calculated by using number of motor neurons in 6-week-old mice as the reference point (100%).
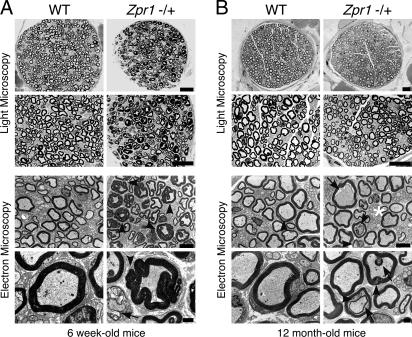
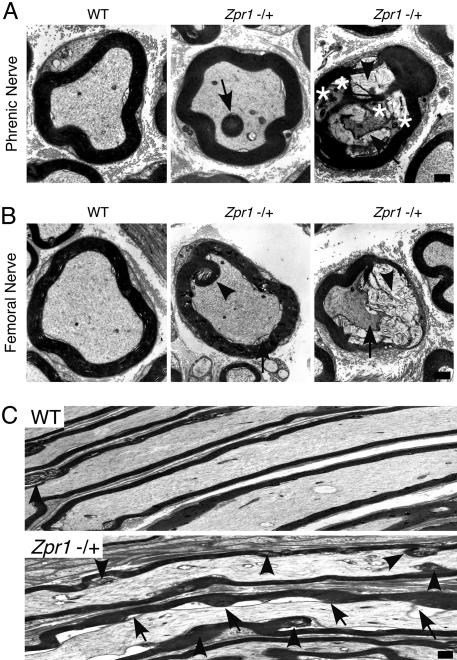
Loss of spinal cord motor neurons can be associated with defects in the peripheral nervous system (PNS) of patients with motor neuron diseases, including SMA. Therefore, we examined the effect of ZPR1 deficiency on the PNS by examining the phrenic and femoral nerves. Phrenic nerves from 6-week-old Zpr1+/+ and Zpr1+/− mice did not show any striking differences when examined by light microscopy, but analysis by electron microscopy indicated some hypomylenation and myelin degeneration in Zpr1+/− mice (see Fig. 6, which is published as supporting information on the PNAS web site). In marked contrast, large differences between the phrenic nerves of 12-month-old wild-type and Zpr1+/− mice were detected. Many axons with myelin foci (tomaculi) were present in Zpr1+/− mice compared to wild-type mice (Fig. 6B Upper) and increased myelin and axon degeneration was observed in the 12-month-old Zpr1+/− mice (Fig. 6B Lower). Axonopathy was also detected in the femoral nerve of Zpr1+/− mice, but the effect of ZPR1 deficiency was different in the femoral and phrenic nerves of young animals. In 6-week-old Zpr1+/− mice, the phrenic nerve was hypomyelinated (Fig. 6A), but the femoral nerve exhibited hypermyelination and acute myelin degeneration (Fig. 3A). At 12 months of age, the femoral nerves of Zpr1+/− mice showed a reduction in the number of myelinated axons (18 ± 2%) compared to wild-type mice (mean ± SD; n = 6; Fig. 3B). Together, these data demonstrate that reduced expression of ZPR1 causes progressive PNS axonopathy.
Fig. 3.
Defects in the development of the peripheral nervous system caused by ZPR1 deficiency. (A) Semithin and ultrathin sections of femoral nerves of 6-week-old WT and Zpr1+/− littermates were examined by light microscopy (Upper; scale bar, 20 μm) and transmission electron microscopy (Lower; scale bar, 5 μm in Upper and 1 μm in Lower). Arrowheads indicate hypermyelination and myelin folding (tomaculi). The myelin defects indicate Wallerian degeneration (arrow). (B) Semithin and ultrathin sections of femoral nerves of 12-month-old WT and Zpr1+/− littermates were examined by light microscopy (Upper; scale bar, 20 μm) and transmission electron microscopy (Lower; scale bar, 5 μm in Upper and 1 μm in Lower). Arrows indicate axon retraction and degeneration. Arrowheads indicate myelin degeneration.
Comparison of the phrenic and femoral nerves of wild-type and Zpr1+/− mice demonstrated that ZPR1 deficiency caused axonal and myelin defects (Wallerian degeneration) that are hallmarks of neurodegenerative disorders, including SMA (Fig. 4). Dark electron dense bodies in the axoplasm (Fig. 4A Right), axon retraction (Fig. 4C), hypomyelination (Fig. 3B), and acute myelin degeneration (Fig. 4 A Right and B Right) are pathological features observed in peripheral nerves of SMA patients (14–16). Wallerian degeneration and myelin defects can be caused by disruption of axon/glial signaling mediated by microvilli (17, 18). Indeed, ZPR1 deficiency does cause loss of microvilli (12) and may therefore contribute to defects in axon–Schwann cell signaling that can cause axonal pathology (19). Interestingly, some of the myelin defects observed in ZPR1-deficient mice have also been associated with other neurodegenerative disorders. For example, the hypermyelination (tomaculi) in the femoral nerve of 6-week-old mice (Fig. 3A) resembles the hypermyelination caused by axon shrinkage found in patients with Hereditary Neuropathy with Liability to Pressure Palsies (20) and adrenomyeloneuropathy (21). Altered myelination (Figs. 3 and 4) is also observed in patients with myelin disorders, including Charcot–Marie–Tooth (CMT) disease, also known as Hereditary Motor Sensory Neuropathy (22). However, major pathological features of CMT, caused by mutation of PMP22 gene (22, 23), including formation of onion bulbs by Schwann cells (22), were not observed in Zpr1+/− mice.
Fig. 4.
Axonal pathology in Zpr1+/− mice. (A) Transverse ultrathin sections of the phrenic nerve of 12 month-old WT and Zpr1+/− littermates were examined by transmission electron microscopy. Myelin foci are indicated (arrow). The presence of electron dense bodies (asterisks) in degenerating axons (arrow) and myelin degeneration (arrowhead) is indicated. (Scale bar, 1 μm.) (B) Transverse ultrathin sections of the femoral nerve of 12 month-old WT and Zpr1+/− littermates were examined by transmission electron microscopy. Green arrowheads indicate myelin foci and myelin degeneration (Center). Myelin degeneration (arrowhead) and axonal defects (arrow) indicate Wallerian degeneration (Right). (Scale bar, 1 μm.) (C) Longitudinal ultrathin sections of the femoral nerve of 12-month-old WT and Zpr1+/− littermates were examined by transmission electron microscopy. Myelin degeneration was observed at different locations in axons (arrowhead). Axon retraction (shrinking) and myelin degeneration (arrows) was caused by ZPR1 deficiency. (Scale bar, 1 μm.)
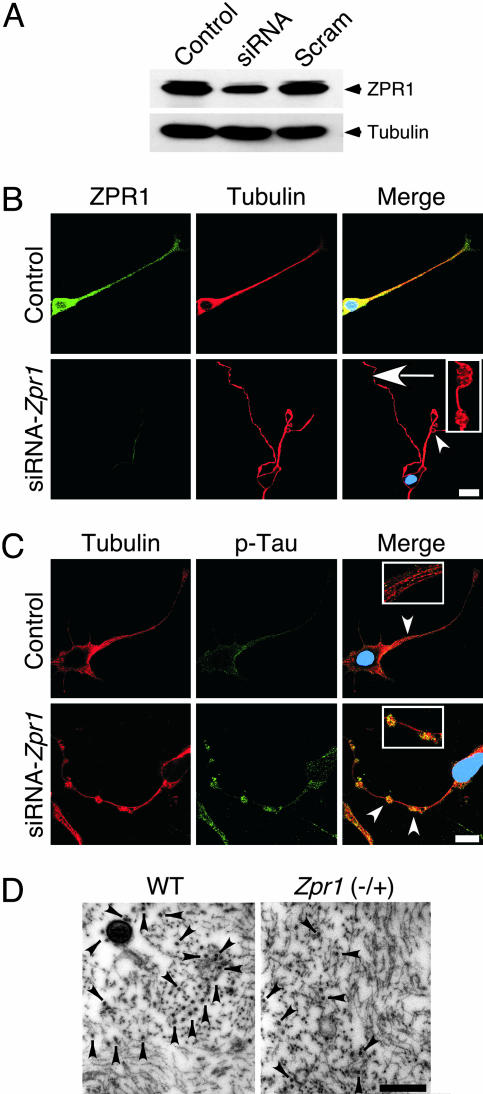
To gain insight into the effect of ZPR1 deficiency on axon degeneration, we performed experiments with an RNAi-based approach using a cultured cell line that can be induced to differentiate into motor neuron-like cells. Zpr1 gene suppression caused axon retraction and microtubule disruption in cultured cells (Fig. 5A). This observation indicates that a myelin defect is not required for the axonal degeneration that is caused by ZPR1 deficiency. However, the finding that ZPR1 deficiency causes microtubule disruption may be mechanistically relevant to the process of axonal degeneration because it is established that the integrity of the microtubule cytoskeleton is essential for maintaining axon stability (24). Regulatory phosphorylation of Tau proteins is thought to play a critical role in the stabilization of microtubules (24, 25) and accumulated hyperphosphorylated (Ser-262) Tau was found at sites of microtubule disruption caused by ZPR1 deficiency (Fig. 5B). Consistent with the hypothesis that microtubule disorganization may result from ZPR1 deficiency, femoral nerve axons from 12-month-old Zpr1+/− mice showed a reduced density of microtubules compared to wild-type mice (Fig. 5C). Quantitation of microtubules showed a loss of 30 ± 2% (mean ± SD; n = 10) in 12-month-old ZPR1-deficient mice compared with wild-type mice. Thus, the reduced number of microtubules caused by ZPR1 deficiency may be responsible for axon retraction and neurodegeneration.
Fig. 5.
Loss of microtubules and axon degeneration caused by ZPR1 deficiency. (A) Differentiated NSC-34 cells were transfected with mock (Control), control RNA duplex Scramble II (Scram), and Zpr1 siRNA (siRNA). Levels of expression of ZPR1 and tubulin proteins (72 h after transfection) were examined by immunoblot analysis using antibodies to ZPR1 and tubulin. (B) Zpr1 gene suppression causes axonal defects in differentiated NSC-34 cells that resemble motor neurons. Cells transfected with scrambled siRNA (Control) and ZPR1 specific siRNA (siRNA-Zpr1) were cultured for 72 h and stained with antibodies to Tubulin (red) and ZPR1 (green). Arrowhead indicates axon retraction after Zpr1 gene suppression. Arrow indicates axonal swelling and microtubule disruption (see Inset). (Scale bar, 20 μm.) (C) ZPR1 deficiency causes microtubule disruption in differentiated NSC-34 cells. Cells were stained with antibodies to Tubulin (red) and phospho(Ser-262)-Tau (green). Arrowheads indicate normal microtubules in control cells (see Upper Inset) and accumulated phospho-Tau at the sites of microtuble disruption (see Lower Inset). (Scale bar, 20 μm.) (D) ZPR1 deficiency causes loss of microtubules in axons of peripheral nerves. Transverse ultrathin sections of the femoral nerve of 12-month-old WT and Zpr1+/− littermates were examined by transmission electron microscopy. Neurofilaments and microtubules were detected in the axoplasm. Arrowheads indicate microtubules. (Scale bar, 1 μm.)
The results of this study demonstrate that ZPR1 deficiency causes motor neuron degeneration in mice. Because the expression of ZPR1 is suppressed in humans with severe SMA (9), our data indicate that decreased ZPR1 expression may contribute to SMA pathogenesis. One outstanding question relates to the mechanism of suppression of ZPR1 gene expression in humans with SMA. Sequence analysis of genomic DNA has not identified mutations in the ZPR1 gene that correlate with SMA. Nevertheless, ZPR1 expression is decreased in patients with severe SMA (9). These data suggest that epigenetic changes in the ZPR1 gene or an uncharacterized mutation in a regulatory region of the ZPR1 gene may contribute to the changes in ZPR1 gene expression that are associated with severe SMA (9). The identification of ZPR1 as a modifier gene that may contribute to SMA pathogenesis provides a foundation for the design of novel therapies for the treatment of severe SMA.
Methods
Mice.
The murine Zpr1 gene was disrupted by replacing exon 1 with a NeoR cassette using homologous recombination (12). The mice were genotyped by PCR using tail DNA (12). The Zpr1+/− mice were backcrossed to the C57BL/6J strain for eight generations and were housed in a facility accredited by the American Association for Laboratory Animal Care, and the animal studies were approved by the Institutional Animal Care and Use Committee of the University of Massachusetts Medical School.
Footprint Analysis.
Gait was examined by applying nontoxic paint to the hind (blue) and fore (red) feet of the mice. The animals were placed on a clean sheet of white paper. Footprint patterns were analyzed for two parameters: stride length and paw abduction. Mean values were measured from three different tests per mouse, and six mice were examined in each group (26).
Spinal Cord Morphology.
The tissue was fixed in 4% paraformaldehyde 24 h before processing and embedding in paraffin wax (27). Sections were cut at 7 μm and stained with hematoxylin and eosin. Motor neurons were counted in every fifth section of the lumbar (L1–L5) and thoracic (T9–T12) regions of the spinal cord (13, 27). Neurodegeneration was examined using a silver stain kit (Sigma). Immunohistochemical staining was performed by using monoclonal antibodies to SMN (Transduction Laboratories, Lexington, KY) and cleaved caspase 3 (Cell Signaling Technology, Beverly, MA). Immune complexes were detected by using a biotinylated secondary antibody, streptavidin-conjugated horseradish peroxidase (Biogenex Laboratories, San Ramon, CA), and the substrate 3,3′-diaminobenzene (Vector Laboratories) followed by brief counterstaining with hematoxylin.
Nerve Morphology.
The tissue was washed with 0.5 M Na cacodylate-HCl buffer (pH 7.0) and fixed with 5 ml of 1.25% glutaraldehyde (30 min at 30°C) and overnight (4°C) with 5 ml of 2.5% glutaraldehyde in cacodylate buffer. The tissue was postfixed (1 h) in 1% osmium tetraoxide (wt/vol) in 0.1 M phosphate buffer (pH 7.2). The fixed nerves were embedded in epoxy resin (12). Semithin sections for light microscopy and ultrathin sections for transmission microscopy were cut on a Reichart–Jung ultramicrotome using a diamond knife. The semithin sections were stained with toluidine blue. The ultrathin sections were mounted on copper support grids in serial order, contrasted with lead citrate and uranyl acetate, and examined on a Philips CM 10 transmission electron microscope at 80 kV accelerating voltage.
Mammalian Cell Culture and siRNA Studies.
Differentiated NSC-34 cells with properties that resemble motor neurons (12, 28) were transfected by using Oligofectamine with 100 nM SMARTpool siRNA (Dharmacon) designed to silence the mouse Zpr1 gene or Scramble II (Control, 5′-GCGCGCTTTGTAGGATTCG-3′) (12). Cy3-labeled Luciferase GL2 duplex (Dharmacon) was used as a transfection control. Cells were harvested 72 h after transfection, and the level of ZPR1 expression was examined by immunoblot analysis. Coverslips were processed for immunofluorescence analysis at 72 h after transfection.
Immunofluorescence Analysis.
Cells cultured on glass coverslips were rinsed with PBS and fixed (−20°C) with methanol (5 min) and acetone (2 min) (12). Double labeling (ZPR1/Tubulin) was performed by sequential incubations (1 h) with anti-Tubulin (clone TUJ1, Covance), Texas red-conjugated anti-mouse IgG secondary antibody (Jackson ImmunoResearch) and FITC-conjugated anti-ZPR1 (clone LG1) (5) at 25°C. Double labeling (Tubulin/phospho-Tau) was performed by sequential incubations (1 h) with anti-Tubulin, Texas red-conjugated anti-mouse secondary antibody, rabbit anti-phospho(Ser-262)-Tau (Calbiochem), and FITC-conjugated anti-rabbit IgG secondary antibody at 25°C. The coverslips were mounted on slides by using Vectashield with DAPI (Vector Laboratories) and examined by immunofluorescence microscopy using a confocal microscope (Leica TCS SP2) equipped with 405-nm diode laser.
Supplementary Material
Acknowledgments
We thank Deepti Terela for technical assistance and Kathy Gemme for administrative assistance. This study was supported by grants from the Families of SMA (to L.G.), the Muscular Dystrophy Association (to L.G.), and the National Institute of Neurological Diseases and Stroke (to R.J.D.). R.J.D. and R.A.F. are investigators of the Howard Hughes Medical Institute.
Abbreviation
- SMA
spinal muscular atrophy.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Markowitz J. A., Tinkle M. B., Fischbeck K. H. J. Obstet. Gynecol. Neonatal Nurs. 2004;33:12–20. doi: 10.1177/0884217503261125. [DOI] [PubMed] [Google Scholar]
- 2.Lefebvre S., Burglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., et al. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 3.Lefebvre S., Burlet P., Liu Q., Bertrandy S., Clermont O., Munnich A., Dreyfuss G., Melki J. Nat. Genet. 1997;16:265–269. doi: 10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- 4.Lorson C. L., Hahnen E., Androphy E. J., Wirth B. Proc. Natl. Acad. Sci. USA. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gangwani L., Mikrut M., Theroux S., Sharma M., Davis R. J. Nat. Cell Biol. 2001;3:376–383. doi: 10.1038/35070059. [DOI] [PubMed] [Google Scholar]
- 6.Gubitz A. K., Feng W., Dreyfuss G. Exp. Cell. Res. 2004;296:51–56. doi: 10.1016/j.yexcr.2004.03.022. [DOI] [PubMed] [Google Scholar]
- 7.Zerres K., Wirth B., Rudnik-Schoneborn S. Neuromusc. Disord. 1997;7:202–207. doi: 10.1016/s0960-8966(97)00459-8. [DOI] [PubMed] [Google Scholar]
- 8.Rochette C. F., Surh L. C., Ray P. N., McAndrew P. E., Prior T. W., Burghes A. H., Vanasse M., Simard L. R. Neurogenetics. 1997;1:141–147. doi: 10.1007/s100480050021. [DOI] [PubMed] [Google Scholar]
- 9.Helmken C., Hofmann Y., Schoenen F., Oprea G., Raschke H., Rudnik-Schoneborn S., Zerres K., Wirth B. Hum. Genet. 2003;114:11–21. doi: 10.1007/s00439-003-1025-2. [DOI] [PubMed] [Google Scholar]
- 10.Roy N., Mahadevan M. S., McLean M., Shutler G., Yaraghi Z., Farahani R., Baird S., Besner-Johnston A., Lefebvre C., Kang X., et al. Cell. 1995;80:167–178. doi: 10.1016/0092-8674(95)90461-1. [DOI] [PubMed] [Google Scholar]
- 11.Burglen L., Seroz T., Miniou P., Lefebvre S., Burlet P., Munnich A., Pequignot E. V., Egly J. M., Melki J. Am. J. Hum. Genet. 1997;60:72–79. [PMC free article] [PubMed] [Google Scholar]
- 12.Gangwani L., Flavell R. A., Davis R. J. Mol. Cell. Biol. 2005;25:2744–2756. doi: 10.1128/MCB.25.7.2744-2756.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jablonka S., Schrank B., Kralewski M., Rossoll W., Sendtner M. Hum. Mol. Genet. 2000;9:341–346. doi: 10.1093/hmg/9.3.341. [DOI] [PubMed] [Google Scholar]
- 14.Rudnik-Schoneborn S., Goebel H. H., Schlote W., Molaian S., Omran H., Ketelsen U., Korinthenberg R., Wenzel D., Lauffer H., Kreiss-Nachtsheim M., et al. Neurology. 2003;60:983–987. doi: 10.1212/01.wnl.0000052788.39340.45. [DOI] [PubMed] [Google Scholar]
- 15.Hausmanowa-Petrusewicz I., Vrbova G. NeuroReport. 2005;16:657–661. doi: 10.1097/00001756-200505120-00001. [DOI] [PubMed] [Google Scholar]
- 16.Crawford T. O., Pardo C. A. Neurobiol. Dis. 1996;3:97–110. doi: 10.1006/nbdi.1996.0010. [DOI] [PubMed] [Google Scholar]
- 17.Michailov G. V., Sereda M. W., Brinkmann B. G., Fischer T. M., Haug B., Birchmeier C., Role L., Lai C., Schwab M. H., Nave K. A. Science. 2004;304:700–703. doi: 10.1126/science.1095862. [DOI] [PubMed] [Google Scholar]
- 18.Guertin A. D., Zhang D. P., Mak K. S., Alberta J. A., Kim H. A. J. Neurosci. 2005;25:3478–3487. doi: 10.1523/JNEUROSCI.3766-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bjartmar C., Yin X., Trapp B. D. J. Neurocytol. 1999;28:383–395. doi: 10.1023/a:1007010205037. [DOI] [PubMed] [Google Scholar]
- 20.Chance P. F. Ann. N.Y. Acad. Sci. 1999;883:14–21. [PubMed] [Google Scholar]
- 21.Pujol A., Hindelang C., Callizot N., Bartsch U., Schachner M., Mandel J. L. Hum. Mol. Genet. 2002;11:499–505. doi: 10.1093/hmg/11.5.499. [DOI] [PubMed] [Google Scholar]
- 22.Sander S., Nicholson G. A., Ouvrier R. A., McLeod J. G., Pollard J. D. Muscle Nerve. 1998;21:217–225. doi: 10.1002/(sici)1097-4598(199802)21:2<217::aid-mus9>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 23.Adlkofer K., Martini R., Aguzzi A., Zielasek J., Toyka K. V., Suter U. Nat. Genet. 1995;11:274–280. doi: 10.1038/ng1195-274. [DOI] [PubMed] [Google Scholar]
- 24.Ahmad F. J., Hughey J., Wittmann T., Hyman A., Greaser M., Baas P. W. Nat. Cell Biol. 2000;2:276–280. doi: 10.1038/35010544. [DOI] [PubMed] [Google Scholar]
- 25.Lee V. M., Goedert M., Trojanowski J. Q. Annu. Rev. Neurosci. 2001;24:1121–1159. doi: 10.1146/annurev.neuro.24.1.1121. [DOI] [PubMed] [Google Scholar]
- 26.Carter R. J., Lione L. A., Humby T., Mangiarini L., Mahal A., Bates G. P., Dunnett S. B., Morton A. J. J. Neurosci. 1999;19:3248–3257. doi: 10.1523/JNEUROSCI.19-08-03248.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masu Y., Wolf E., Holtmann B., Sendtner M., Brem G., Thoenen H. Nature. 1993;365:27–32. doi: 10.1038/365027a0. [DOI] [PubMed] [Google Scholar]
- 28.Cashman N. R., Durham H. D., Blusztajn J. K., Oda K., Tabira T., Shaw I. T., Dahrouge S., Antel J. P. Dev. Dyn. 1992;194:209–221. doi: 10.1002/aja.1001940306. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.