Abstract
The quaking viable mouse mutation (qkv) is a deletion including the 5′ regulatory region of the quaking gene (Qki), which causes body tremor and severe dysmyelination in mouse. The function of the human quaking gene, called quaking homolog KH domain RNA-binding (mouse) (QKI), is not well known. We have previously shown that QKI is a new candidate gene for schizophrenia. Here we show that human QKI mRNA levels can account for a high proportion (47%) of normal interindividual mRNA expression variation (and covariation) of six oligodendrocyte-related genes (PLP1, MAG, MBP, TF, SOX10, and CDKN1B) in 55 human brain autopsy samples from individuals without psychiatric diagnoses. In addition, the tightly coexpressed myelin-related genes (PLP1, MAG, and TF) have decreased mRNA levels in 55 schizophrenic patients, as compared with 55 control individuals, and most of this difference (68–96%) can be explained by variation in the relative mRNA levels of QKI-7kb, the same QKI splice variant previously shown to be down-regulated in patients with schizophrenia. Taken together, our results suggest that QKI levels may regulate oligodendrocyte differentiation and maturation in human brain, in a similar way as in mouse. Moreover, we hypothesize that previously observed decreased activity of myelin-related genes in schizophrenia might be caused by disturbed QKI splicing.
Keywords: myelin, quaking, splice variant
The quaking gene (Qki) is a member of the signal transduction and activation of RNA (STAR) protein family, also called GSG (GRP33, Sam68, and GLD-1) or SGQ (Sam68, GLD-1, and Qk1), following the name of the initial protein members (1–3). The gene is highly conserved over different species and is important for normal development in both vertebrates and invertebrates (4, 5). The function of the Qki gene has been well studied in mouse by using an autosomal recessive mutant (qkv), characterized by body tremor and severe dysmyelination of the CNS (6). The mutation consists of a 1-Mb deletion including the 5′ regulatory region of Qki (1) that causes oligodendrocyte dysfunction and reduced expression of myelin components in CNS (7). The Qki gene contains an RNA-binding domain (KH domain) that binds directly to cellular RNA (8). Several different sequence motifs have been identified as targets for the Qki protein (9–12). One of these Qki response elements (QREs) consists of a bipartite consensus sequence that has been shown to bind the Qki protein in vivo in at least 23 different mouse mRNA species (9). This mouse protein directly regulates the expression of at least one myelin-specific gene, e.g., myelin basic protein (Mbp) (1), and controls the alternative splicing of the myelin-associated glycoprotein (Mag) (12). In addition, Qki protein regulates the cell cycle inhibitor cyclin-dependent kinase inhibitor 1B (Cdkn1b) (10), which is involved in the terminal differentiation of oligodendrocytes (13). Cdkn1b also activates the mouse Mbp promoter through interaction with the transcription factor sex-determining region Y-box10 (Sox10) (14, 15).
The human homolog of the mouse Qki gene is called the quaking homolog KH domain RNA-binding (mouse) (QKI). The genetic structure of the human QKI gene is well defined, and four different mRNA splice variants, QKI-5kb, QKI-6kb, QKI-7kb, and QKI-7kb-B, are observed. The splice variants are produced by differential splicing of the exon 6, 7a-c, 8, and different parts of the untranslated region of the gene (16).
Growing evidence from histological, genetic, and gene expression studies suggests that myelin and oligodendrocyte dysfunction contributes to the development of schizophrenia [see e.g., Stewart and Davis (17) for a recent review]. For example, several genes expressed in oligodendrocytes are reported as down-regulated in schizophrenic patients (18–20). Among these are genes that are expressed only in mature myelin-producing oligodendrocytes, such as MAG, proteolipid protein 1 (PLP1), and MBP, but also genes involved in the differentiation and maturation of oligodendrocytes, like SOX10 and transferrin (TF) (17). We have recently presented evidence for association between a locus including the human QKI gene and schizophrenia in two unrelated sample sets. In addition, we showed that all four QKI splicing variants are expressed in frontal cortex of human brain, and that the relative mRNA expression levels of the QKI splice-variant QKI-7kb is down-regulated in schizophrenic patients (21). Because Qki regulates differentiation and maturation of oligodendrocytes in mouse, we hypothesize that the gene has a similar function in humans. Thus, we predict that expression patterns of QKI will coincide with the expression pattern of genes involved in oligodendrocyte differentiation and myelination. We use 55 frontal cortex autopsy samples from normal human brains to test whether the mRNA expression level of the QKI gene (or the relative amount of specific splice variants) can predict mRNA expression (and coexpression) of the six oligodendrocyte-related (OR) genes CDKN1B, SOX10, MBP, MAG, PLP1, and TF. In addition, we searched in silico for potential QKI-binding motifs within these six genes. Finally, we compare brain mRNA levels between 55 schizophrenic and 55 control individuals and examine whether disturbed gene expression in the patients and differences between patient subgroups could be explained by the variation in QKI mRNA expression levels that we have described (21).
Results
Correlation Between mRNA Expression in OR Genes Suggests QKI as a Common Regulator.
The expression levels of six OR genes, MAG, MBP, PLP1, TF, SOX10, and CDKN1B, in the 55 brain autopsies investigated were all positively correlated (r > 0.5, Table 1), and MAG, PLP1, and TF were particularly strongly correlated (r > 0.9, Table 1). Principal component analysis indicated that 78% of the total expression variation of the six genes could be explained by an underlying principal component [principal component 1 (PC1)]. All genes loaded similarly on the PC1, with loadings ranging from 0.37 (CDKN1B and MBP) to 0.44 (MAG). Thus, PC1 appears to reflect normal interindividual variation in the proportion of immature and mature oligodendrocytes in brain autopsy samples. Higher-order principal components were disregarded, because they had eigen values <1 (<0.52) and explained little additional variation (<9%).
Table 1.
Correlation (r2) between mRNA levels of OR genes in brain autopsy samples from individuals not affected by schizophrenia (controls)
| Gene name | MAG | PLP1 | TF | MBP | SOX10 | CDKN1B |
|---|---|---|---|---|---|---|
| MAG | — | 0.90 | 0.95 | 0.67 | 0.79 | 0.71 |
| PLP1 | — | — | 0.92 | 0.73 | 0.78 | 0.74 |
| TF | — | — | — | 0.63 | 0.67 | 0.68 |
| MBP | — | — | — | — | 0.73 | 0.51 |
| SOX10 | — | — | — | — | — | 0.67 |
| CDKN1B | — | — | — | — | — | — |
The total expression level of QKI could explain 19–55% of the expression variation of individual OR genes and 47% of their coordinated variation, as assessed by PC1 (Table 2). In addition to the total QKI mRNA level, the relative mRNA amount of specific splice variants also appeared to modify expression of OR genes in human brain autopsies (Table 2). In particular, the relative mRNA level of splice variant QKI-7kb could explain significant (P < 0.0001) additional expression variation in MAG, PLP1, TF, and PC1 (41%, 31%, 46%, and 25%, respectively). The mRNA expression of SOX10 and CDKN1B was affected by QKI-7kb, but the variation was marginal (6%, P = 0.005; and 8%, P = 0.008 for the two genes, respectively). The balance between the Qki splice variants has previously been suggested as an important factor for the control of myelin-specific genes in mouse (11). In this study, we noted that the importance of QKI-7kb for the expression of the two MAG splice variants differed: the relative amount of QKI-7kb explained 53% (P < 0.0001) of the variation of S-MAG, but only 22% (P < 0.0001) of L-MAG. On the other hand, the total QKI expression level affected the two MAG splice variants equally (Table 2).
Table 2.
Variation of expression levels in CDKN1B, SOX10, MBP, PLP1, TF, MAG, and PC1 explained by total QKI mRNA levels and additional variation explained by the relative mRNA amount of each of four QKI splice variants
| Gene name |
QKI |
QKI-5kb |
QKI-6kb |
QKI-7kb |
QKI-7kbB |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Variation, % | P value | Variation, % | P value | Variation, % | P value | Variation, % | P value | Variation, % | P value | |
| CDKN1B | 41 | <0.0001 | 10 | 0.0025 | 1 | 0.2803 | 8 | 0.0076 | 10 | 0.0024 |
| SOX10 | 55 | 0.0002 | 0 | 0.8047 | 0 | 0.7782 | 6 | 0.0050 | 2 | 0.1243 |
| MBP | 52 | 0.0015 | 1 | 0.3164 | 0 | 0.8597 | 3 | 0.0494 | 1 | 0.3243 |
| TF | 19 | 0.0016 | 9 | 0.0129 | 7 | 0.0357 | 46 | <0.0001 | 15 | 0.0012 |
| PLP1 | 40 | 0.0012 | 8 | 0.0065 | 1 | 0.3546 | 31 | <0.0001 | 9 | 0.0036 |
| MAG | 27 | 0.1264 | 4 | 0.0869 | 4 | 0.0865 | 41 | <0.0001 | 10 | 0.0047 |
| S-MAG | 14 | 0.1277 | 4 | 0.1231 | 5 | 0.0953 | 53 | <0.0001 | 9 | 0.0149 |
| L-MAG | 13 | 0.1266 | 4 | 0.1149 | 3 | 0.1806 | 22 | 0.0001 | 3 | 0.1663 |
| PC1 | 47 | 0.0001 | 5 | 0.0192 | 2 | 0.1671 | 25 | <0.0001 | 9 | 0.0021 |
Human Myelin-Related Genes Contain a Potential QKI-Binding Site Located in the 3′ UTR of the Transcripts.
In mouse, one type of a QRE has been identified as a bipartite consensus sequence, consisting of a core element (NACUAAY) and a half site (YAAY) positioned within 20 bp up- or downstream of the core element (9). To identify potential QKI-binding target motifs of this type in the six investigated human genes, their sequences were retrieved from the public databases and scanned for a core QKI-binding element in combination with a half site, as described in Materials and Methods. We also aligned the homologous genes to investigate whether the sequence of putative and experimentally verified QREs was conserved between mice and humans.
We first identified putative QKI-binding elements in mRNA sequences of three of the six investigated genes, namely in MBP, PLP1, and SOX10 (Fig. 1). For these genes, the putative site was located in the 3′ UTR of the mRNA sequence. In MBP, the putative human-binding site resided in a portion of the mRNA that corresponded to an experimentally verified QRE in mice, with high affinity for QKI (QRE-2) (9), and we noted that the sequence of the core and the half site had been particularly well conserved between mouse and humans. In PLP1 and SOX10, the putative human QREs were not described previously in mouse. However, the sequence of these putative elements was also well conserved between the two species, suggesting that this portion of the 3′ UTR might be functional. Because other types of QRE elements have also been described (9–12), we therefore attempted to find other QREs for the remaining three genes (CDKN1B, MAG, and TF). An experimentally verified QRE that contains the bipartite consensus sequence has been identified in the coding sequence of Cdkn1b in mouse (10). In human CDKN1B, the sequence of the mouse QRE is fairly well conserved (Fig. 1). However, we noted three substitutions in the half site and two substitutions in the core site, making it dubious that this portion of the QRE is functionally important in humans. A quaking alternative splicing element (QASE) has previously been identified and experimentally verified downstream of the 5′ splice site of mouse MAG exon 12 (12). We found the QASE sequence to be highly conserved in the latest assemblies of the human and mouse genome sequences (22) (Fig. 1). In summary, we have detected potential QKI-binding sites in five of the six studied genes, indicating that these genes might be directly regulated by QKI.
Fig. 1.
Alignment of human and mouse sequences. The aligned reference sequence numbers or the positions in the human genome are indicated on the left, and the end position of each sequence is indicated on the right. Asterisks indicate sequence identity. Bold text indicates quaking-binding elements previously described in the mouse. Boxed regions highlight the core and half sites of the QRE consensus sequence.
Expression of Myelin-Specific Genes in Schizophrenia Patients.
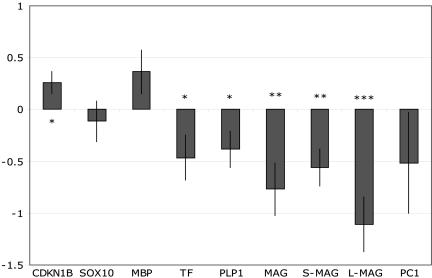
In schizophrenic patients, we observed a 1.7-fold down-regulation of total MAG (P = 0.003) as well as a down-regulation of both MAG splice variants (1.5-fold, P = 0.003; and 2.2-fold, P < 0.001 for S- and L-MAG, respectively). PLP1 and TF were also 1.3- and 1.4-fold down-regulated in the patients (P values were 0.031 and 0.036, respectively) (Fig. 2). In addition, we observed a 1.2-fold up-regulation of CDKN1B (P = 0.016). The difference in mRNA levels of S-MAG between patients and controls was less pronounced than the difference in L-MAG, and consequently the ratio of the two splice variants was significantly altered in schizophrenic patients (P < 0.001; data not shown). We have previously reported a reduction in relative expression of splice-variant QKI-7kb in schizophrenic patients compared with controls (21). To evaluate the possibility that this abnormal QKI expression may modify myelination in schizophrenia, we studied the covariation of expression levels of OR genes with splice-variant QKI-7kb. We found that the expression difference in MAG, PLP1, and TF between schizophrenic cases and controls (Fig. 2) was greatly reduced when the covariation of these three genes with the variation of QKI-7kb was removed (Table 3), demonstrating that the disturbed expression levels of these three genes in schizophrenia is strongly associated with a disturbed balance in the mRNA levels of QKI splice-variant 7kb.
Fig. 2.
Disease effects on mRNA expression of OR genes and PC1. Messenger RNA expression levels relative to the expression of reference genes (ACTB, GAPD) for schizophrenic patients are shown. The zero line indicates the average expression level for controls. Asterisk indicates significant deviation in mRNA levels between the patients and controls (P < 0.05, P < 0.01, and P < 0.001 for one, two, and three asterisks, respectively). Mean and standard errors are given.
Table 3.
Variation in expression levels of OR genes between schizophrenic patients and controls and between patient subgroups, when QKI expression levels are, or are not, accounted for
| Gene name | Analysis without QKI |
Analysis accounting for QKI levels |
||||
|---|---|---|---|---|---|---|
| Schizo. vs. Ctrl. (df = 1) |
Neuroleptic treatment (df = 2) |
Schizo. vs. Ctrl. |
Neuroleptic treatment |
|||
| SS | SS | SS−ΔQKI7kb | SSoverlap, % | SS−QKI | SSoverlap, % | |
| CDKN1B | 1.84* | 1.39 | 1.58 | 14 | 0.02 | 98 |
| SOX10 | 0.35 | 8.39* | 0.15 | 56 | 0.01 | 100 |
| MBP | 3.59 | 4.14 | 3.34 | 7 | 1.03 | 75 |
| TF | 5.81* | 18.12*** | 0.25 | 96 | 5.19 | 71 |
| PLP1 | 3.97* | 10.11** | 1.01 | 75 | 1.84 | 82 |
| MAG | 15.84** | 7.52 | 5.02 | 68 | 2.35 | 69 |
| S-MAG | 8.47** | 7.52** | 1.91 | 77 | 2.58 | 66 |
| L-MAG | 33.06*** | 7.03 | 19.23** | 42 | 3.97 | 44 |
| PC1 | 7.32 | 79.76** | 1.55 | 79 | 8.03 | 90 |
The variation in target gene expression explained by the disease status or the patient subgroup is listed for models excluding (left) or including (right) total QKI expression [schizophrenic (Schizo.) vs. control (Ctrl.)] or relative QKI-7kb (subgroup) levels. The reduction (or overlap) in variation is expressed as a percentage of the variation explained by models excluding QKI expression. Variation is estimated by sums of squares (SS). Asterisk (∗) indicates significant deviation in mRNA levels between patients and controls and between the three groups of patients (P < 0.05, P < 0.01, and P < 0.001, for one, two, and three asterisks, respectively).
When we divided the patients into subgroups according to neuroleptics received, we noted that mRNA expression tended to differ among the three groups. In general, the expression pattern of the three patient subgroups changed similarly for all genes (Fig. 3). That is, patients treated with typical neuroleptics tended to show increased mRNA levels of all OR genes. In addition, the mRNA levels of patients treated with atypical neuroleptics tended to be somewhat lower than the levels of the untreated patients. This pattern was most clearly seen in the mRNA levels of TF, where the differences among the three neuroleptic subgroups were highly significant (P < 0.001). Although less pronounced, significant differences between the neuroleptic subgroups were also observed for S-MAG, PLP1, SOX10, and PC1 (P = 0.011, 0.002, 0.018, and 0.002, respectively). These differences were confirmed by permuting the subgroup classification among the patients (10,000 permutations, empirical P values for TF, S-MAG, PLP1, SOX10, and PC1 were <0.001, 0.015, <0.001, 0.003, and 0.002, respectively). Moreover, the observed expression differences in OR genes were coordinated, as evident by the response of the PC1 (Fig. 3). We have reported that the expression levels of all QKI splice variants differ among patient subgroups (21). Interestingly, when the differences in total QKI levels are accounted for, the observed expression differences in OR genes are greatly reduced (Table 3).
Fig. 3.
Schizophrenic subgroup differences in expression of OR genes and PC1. Messenger RNA expression levels relative to the expression of reference genes (ACTB, GAPD) for three groups of schizophrenic patients treated with different types of neuroleptics are shown. The zero line indicates the average expression level of all 55 schizophrenic patients. Asterisk indicates significant deviation in mRNA levels between the three groups of patients (P < 0.05, P < 0.01, and P < 0.001 for one, two, and three asterisks, respectively). Mean and standard errors are given.
Discussion
In this work, we show that (i) human QKI coincides with expression levels of several OR genes, and (ii) disturbed expression of myelin-related genes in schizophrenic patients may be explained by alterations in QKI splice-variant expression pattern.
For our experiments, we selected six genes (PLP1, MAG, MBP, TF, SOX10, and CDKN1B) previously shown to be directly or indirectly involved in oligodendrocyte differentiation and maturation. CDKN1B is a cell cycle inhibitor, and SOX10 is a nervous-system-specific transcription factor, both involved in the terminal differentiation of oligodendrocytes (13, 23). The two genes are also related to transcription factor Sp1-dependent activation of the MBP promoter (15). Mature myelinating oligodendrocytes are characterized by the presence of PLP1, MBP, and MAG. PLP1 and MBP are the two most abundant myelin proteins in the CNS, and MAG is a minor but important glycoprotein component of myelin (17). All three genes are exclusively expressed in mature myelinating oligodendrocytes (24). TF is an iron transporter that is predominantly expressed in oligodendrocytes, and that accumulates at the onset of myelination (25).
Normal Function of the Human QKI Gene.
In this study, we show three lines of evidence that suggest that human QKI controls myelination in CNS in a similar way as in mouse, by regulating mRNA expression of genes involved in oligodendrocyte differentiation and maturation.
First, our analysis showed that 78% of the total expression variation of the six OR genes could be explained by an underlying principal component, and because all of the genes loaded similarly on the PC1, this probably reflects interindividual variation in the proportion of oligodendrocytes (immature and mature) in the brain autopsy samples.
Furthermore, the variation in the total mRNA expression of the human QKI gene can explain a large proportion (47%) of the coordinated variation (PC1) of the analyzed genes. Thus, total QKI mRNA levels and the expression of OR genes are tightly correlated in human brain autopsy samples.
Second, we also noted that total QKI mRNA levels could explain 40–55% of the variation of CDKN1B, SOX10, PLP1, and MBP. These results suggest that functional QKI-binding elements should be present within these genes. In fact, mouse mRNA from Cdkn1b and MBP contain Qki-binding elements that are known to physically interact with a region of the QKI protein, present in all QKI splice variants (9, 10). Qki-binding sites for Sox10 and Plp1 have not been characterized previously in mouse or humans. However, we identified conserved putative QKI recognition elements in the 3′ UTR of the human mRNA for MBP, PLP1, and SOX10 and a partially conserved site for CDKN1B. Together, these observations suggest that QKI may have a similar function in the human CNS as it has in mouse, where it regulates oligodendrocyte differentiation and maturation.
Third, we noted that variation in relative expression level of splice-variant QKI-7kb can explain a large fraction (>31%) of additional mRNA variation in three highly coexpressed genes, MAG, PLP1, and TF, which are all directly involved in myelination. Our results are in agreement with findings in mouse, which suggest that a disrupted balance between QKI splice variants is of importance for mRNA expression of myelination-related genes (12). Thus, the balance between splice-variant QKI-7kb and total QKI mRNA levels may be of importance for myelination also in the human brain. For MAG, a QASE has been functionally characterized in mouse (12). Our results indicate that a similar MAG-QASE might exist in humans. Furthermore, we predict that novel QASEs might be found in other myelin-related genes, such as PLP1 and TF.
Potential Involvement of the Human QKI Gene in Schizophrenia.
Recent multiple lines of evidence, including alterations in white matter volumes during adolescence, linkage studies that identify loci containing myelin-related genes, effects of antipsychotic drugs, and mRNA expression studies, support the idea that myelin abnormalities are involved in schizophrenia pathogenesis (17, 19–21, 26, 27).
In this study, we replicated previous findings that showed decrease of myelin-related genes MAG, PLP1, and TF in schizophrenic cases (18–20, 28). Here, as well as in previous studies, expression differences are small. However, in the large sample set investigated, these small differences are strongly significant. Our analysis demonstrated that disturbed expression levels of these genes are strongly associated with a disrupted relative expression of splice-variant QKI-7kb in patients. This suggests that alteration of QKI-7kb might be the primary cause of the down-regulation of myelin-related genes observed in schizophrenia.
It is interesting to note that not all OR genes are down-regulated in schizophrenic patients, because mRNA levels of SOX10 and MBP are not modified and CDKN1B is up-regulated. This suggests that dysmyelination in schizophrenia is caused by a cellular dysfunction rather than being the consequence of a decrease in oligodendrocyte number. It is possible to speculate that the up-regulation of CDKN1B, a cell cycle inhibitor, may be a feedback mechanism caused by the down-regulation of myelin-related genes, to increase oligodendrocyte differentiation.
Typical and atypical neuroleptics are known to bind with different affinities to dopamine and serotonin receptors (29), but the downstream effects of these drugs are not well known. When patients were subdivided according to medication received, we noticed that the expression of all OR genes tended to differ among the three subgroups (atypical neuroleptic, typical neuroleptic, and not treated with neuroleptics before death). However, because this was not a clinical trial, it is possible that the differences in expression pattern of OR genes might be due to one or several unknown confounding factors. Nevertheless, the expression differences in OR genes among the three patient subgroups were coordinated and almost disappeared when the total QKI expression was accounted for. We hypothesize that differences in total QKI among patient subgroups may cause a global change in the number of immature and mature oligodendrocytes.
Taken together, our results point toward QKI being a regulator of oligodendrocyte differentiation and maturation in the human brain, in a similar way as in mouse. We also show that this myelination control is altered in schizophrenia. Future functional analysis of the interaction between QKI and OR genes in, e.g., human cell lines will provide additional clues to the normal control of myelination in the human brain and the functional abnormalities observed in schizophrenia.
Materials and Methods
Case/Control Brain Autopsies.
The Stanley Foundation Brain Consortium (Bethesda) contributed brain autopsies from 30 individuals; 48 samples were provided by Maudsley Brain Bank (Institute of Psychiatry, Department of Neuropathology, London), and 32 came from the Harvard Brain Tissue Resource Center (Massachusetts General Hospital, Boston). The diagnostic criterion used by the Stanley Foundation was DSM-IV, whereas the Maudsley Brain Bank used ICD-10 (30), and the Harvard Resource Center used the Feighner criteria (31). From each brain bank, an equal number of tissue samples from individuals diagnosed with schizophrenia and individuals without psychiatric diagnoses were obtained. Forty-six samples were from females, and 64 were from males. In total, 110 tissue samples from frontal cortex, Brodmann’s area 8 (from the Harvard and Stanley Brain Banks), and the left side of the frontal gyrus (from the Maudsley Brain Bank) were included in the study. The mean time postmortem was 34.3 h for controls (standard error, 22.6) and 34.6 h for schizophrenic cases (standard error, 20.5). Fifty-five tissue samples came from control individuals. Seven of the schizophrenic individuals were treated with atypical neuroleptics, 19 were treated with typical neuroleptics, and 11 were not treated with neuroleptics before death. For the remaining 18 schizophrenic individuals, no information regarding medication was available. The typical neuroleptics group included patients treated with chlorpromazine, promazine, haloperidol, thioridazine, stelazine, trifluroperazine, thiothixene, and sulpiride. The atypical neuroleptics group included patients treated with clozapine and risperidone. This sample set was previously described by Castensson et al. (32). Ethical approval for the use of these samples was received according to Swedish regulations (Ethical Research Committee Ups dnr 99277).
Real-Time RT-PCR.
Tissue sample preparation, total RNA preparation, and poly(A) RNA purification, as well as reverse transcription reactions, were performed as described (33). In brief, total RNA was prepared by using TRIzol reagent (Life Technologies, Stockholm), mRNA was extracted by using the PolyATtract mRNA isolation system (Promega SDS), and reverse transcription used Superscript II (Life Technologies). The 110 individuals, including 55 patients and 55 controls, were distributed in duplicates on three plates according to a balanced incomplete block design, with respect to diagnosis, sex, and brain bank. Real-time RT-PCR was performed with an ABI PRISM 7000 Sequence Detection System (Applied Biosystems) as follows: 2 min at 50°C and 10 min at 95°C followed by 40 cycles of 15 seconds at 95°C and 1 min at 60°C. Each reaction was carried out in a total volume of 25 μl, consisting of 9.8 μl of TaqMan universal PCR master mix, 0.125 μM probe (Applied Biosystems), a 0.25 μM concentration of each primer (Thermo Electron, Drelelch, Germany), and ≈10–100 ng of cDNA. The primer/probe set for the reference genes (ACTB and GAPD) as well as for the MAG (MAG-tot) and each of its splice variants, MAG-L and MAG-S, was uniquely designed by using primer express (Applied Biosystems). The MAG-tot assay was designed to simultaneously detect both MAG splice variants. Primers for PLP1, MBP, TF, SOX10, and CDKN1B were ordered as “Assay on Demand” and were performed according to the manufacturer’s protocol (Applied Biosystems). The expression data were collected with the abi prism 7000 sds software (Applied Biosystems).
Statistical Analyses.
Before analysis of gene expression levels in the 55 control individuals, systematic variation in target gene expression was removed by fitting a linear model to the observed data (Proc GLM, sas 8.2). The linear model included the categorical factors brain bank, gender, and replicate PCR plate and the covariates age, postmortem time, and reference gene expression (the geometric mean of GAPD and ACTB). The differences between the observed and fitted values (i.e., the residuals) were considered the normalized expression level.
The correlation pattern of normalized expression levels of the OR genes (MAG, MBP, PLP1, TF, SOX10, and CDKN1B) was explored with a principal component analysis (Proc PRINCOMP, sas 8.2), and PC1 was used as an additional derived variable in further statistical analysis.
We then examined whether the variation in the total QKI level can predict the expression variation in OR genes and whether additional splice-variant expression variation in QKI can explain any additional variation in the expression of OR genes. For this analysis, we used a linear regression approach (Proc GLM, sas 8.2). Because the expression levels of the four QKI splice variants were highly correlated to the total QKI level (r2 > 0.81), it was necessary to adjust the expression levels of splice variants (with respect to the total QKI level) before analysis. We did this by regressing expression levels of each splice variant onto the total QKI expression and stored the residuals as the orthogonalized splice-variant expression. These orthogonalized variables are positive when the amount of the splice variant is larger than expected from the total QKI levels and negative when the amount of the splice variant is lower than expected from the total QKI levels. Thus, the orthogonalized splice-variant levels can be interpreted in terms of amounts of the splice variant relative to the total QKI level. To examine to what extent the variation in total QKI expression and QKI splice-variant expression explained variation in the investigated genes, we fitted a regression model, including the total QKI expression and the orthogonalized expression, for one splice variant at a time to the expression levels of the six target genes.
To test whether the expression of the six genes involved in oligodendrocyte differentiation and maturation was disturbed in patients suffering from schizophrenia and/or affected by patient subgroup with respect to neuroleptic treatment, we analyzed the mRNA levels of target genes with an analysis of covariance model (Proc GLM, sas 8.2). The model included the main factors disease status (schizophrenic vs. control) and disease subgroup (atypical neuroleptic, typical neuroleptic, and not treated with neuroleptics before death) nested within disease status. Thus the 18 patients with unknown medication status were included in the comparisons of all patients vs. controls, but they were excluded in the test of patient subgroups. In addition, we included the categorical factors brain bank, gender, and replicate PCR plate and the covariates age, postmortem time, and reference gene expression to account for the effects of these variables on mRNA levels of the target genes. To explore to what extent the disease effect and differences between patient subgroups in mRNA levels of target genes overlapped with the variation explained by total QKI mRNA levels or the relative amount of QKI-7kb (QKI-7kb/QKItotal), models with these additional variables added were compared with the original model described above. We used logarithmic transformed expression data and averaged the mRNA levels obtained from the duplicate samples from each individual before the statistical analysis.
QKI-Binding Motif Identification.
The core Qki265-binding site matrix described by Galarneau and Richard (9) was used to search RefSeq cDNA sequences for MBP (NM_001025081), MAG (NM_080600), PLP1 (NM_000533), SOX10 (NM_006941), TF (NM_001063), and CDKN1B (NM_004064), using the transcription factor-binding site perl modules (34). The presence of QKI half sites (AYYA) (9) in the vicinity the QKI core-binding sites was identified by using the fuzznuc program from the emboss suite (35). Orthologous human and mouse sequences were aligned by using clustalw (36).
Acknowledgments
We thank the patients who participated in this study and their families. This work was supported by a strategic grant from the Swedish University of Agricultural Sciences; the Swedish Research Council; and the School of Biology, Uppsala University, Uppsala, Sweden. Postmortem brain samples were donated by the Stanley Foundation Brain Consortium, Bethesda; by the Maudsley Brain Bank, Institute of Psychiatry, Department of Neuropathology, London; and by the Harvard Brain Tissue Resource Center, Massachusetts General Hospital, Boston.
Abbreviations
- QRE
Qki response element
- OR
oligodendrocyte related
- PC1
principal component 1
- QASE
quaking alternative splicing element
- MBP
myelin basic protein
- TF
transferrin.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Ebersole T. A., Chen Q., Justice M. J., Artzt K. Nat. Genet. 1996;12:260–265. doi: 10.1038/ng0396-260. [DOI] [PubMed] [Google Scholar]
- 2.Jones A. R., Schedl T. Genes Dev. 1995;9:1491–1504. doi: 10.1101/gad.9.12.1491. [DOI] [PubMed] [Google Scholar]
- 3.Lin Q., Taylor S. J., Shalloway D. J. Biol. Chem. 1997;272:27274–27280. doi: 10.1074/jbc.272.43.27274. [DOI] [PubMed] [Google Scholar]
- 4.Vernet C., Artzt K. Trends Genet. 1997;13:479–484. doi: 10.1016/s0168-9525(97)01269-9. [DOI] [PubMed] [Google Scholar]
- 5.Chen T., Richard S. Mol. Cell. Biol. 1998;18:4863–4871. doi: 10.1128/mcb.18.8.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sidman R. L., Dickie M. M., Appel S. H. Science. 1964;144:309–311. doi: 10.1126/science.144.3616.309. [DOI] [PubMed] [Google Scholar]
- 7.Hardy R. J., Loushin C. L., Friedrich V. L., Jr., Chen Q., Ebersole T. A., Lazzarini R. A., Artzt K. J. Neurosci. 1996;16:7941–7949. doi: 10.1523/JNEUROSCI.16-24-07941.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen T., Damaj B. B., Herrera C., Lasko P., Richard S. Mol. Cell. Biol. 1997;17:5707–5718. doi: 10.1128/mcb.17.10.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galarneau A., Richard S. Nat. Struct. Mol. Biol. 2005;12:691–698. doi: 10.1038/nsmb963. [DOI] [PubMed] [Google Scholar]
- 10.Larocque D., Galarneau A., Liu H. N., Scott M., Almazan G., Richard S. Nat. Neurosci. 2005;8:27–33. doi: 10.1038/nn1359. [DOI] [PubMed] [Google Scholar]
- 11.Larocque D., Pilotte J., Chen T., Cloutier F., Massie B., Pedraza L., Couture R., Lasko P., Almazan G., Richard S. Neuron. 2002;36:815–829. doi: 10.1016/s0896-6273(02)01055-3. [DOI] [PubMed] [Google Scholar]
- 12.Wu J. I., Reed R. B., Grabowski P. J., Artzt K. Proc. Natl. Acad. Sci. USA. 2002;99:4233–4238. doi: 10.1073/pnas.072090399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Casaccia-Bonnefil P., Tikoo R., Kiyokawa H., Friedrich V., Jr., Chao M. V., Koff A. Genes Dev. 1997;11:2335–2346. doi: 10.1101/gad.11.18.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miskimins R., Srinivasan R., Marin-Husstege M., Miskimins W. K., Casaccia-Bonnefil P. J. Neurosci. Res. 2002;67:100–105. doi: 10.1002/jnr.10080. [DOI] [PubMed] [Google Scholar]
- 15.Wei Q., Miskimins W. K., Miskimins R. J. Neurosci. Res. 2004;78:796–802. doi: 10.1002/jnr.20342. [DOI] [PubMed] [Google Scholar]
- 16.Li Z. Z., Kondo T., Murata T., Ebersole T. A., Nishi T., Tada K., Ushio Y., Yamamura K., Abe K. Jpn. J. Cancer Res. 2002;93:167–177. doi: 10.1111/j.1349-7006.2002.tb01255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stewart D. G., Davis K. L. Int. Rev. Neurobiol. 2004;59:381–424. doi: 10.1016/S0074-7742(04)59015-3. [DOI] [PubMed] [Google Scholar]
- 18.Aston C., Jiang L., Sokolov B. P. J. Neurosci. Res. 2004;77:858–866. doi: 10.1002/jnr.20208. [DOI] [PubMed] [Google Scholar]
- 19.Hakak Y., Walker J. R., Li C., Wong W. H., Davis K. L., Buxbaum J. D., Haroutunian V., Fienberg A. A. Proc. Natl. Acad. Sci. USA. 2001;98:4746–4751. doi: 10.1073/pnas.081071198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tkachev D., Mimmack M. L., Ryan M. M., Wayland M., Freeman T., Jones P. B., Starkey M., Webster M. J., Yolken R. H., Bahn S. Lancet. 2003;362:798–805. doi: 10.1016/S0140-6736(03)14289-4. [DOI] [PubMed] [Google Scholar]
- 21.Aberg K., Saetre P., Lindholm E., Ekholm B., Pettersson U., Adolfsson R., Jazin E. Am J. Med. Genet. 2006;141:84–90. doi: 10.1002/ajmg.b.30243. [DOI] [PubMed] [Google Scholar]
- 22.Karolchik D., Baertsch R., Diekhans M., Furey T. S., Hinrichs A., Lu Y. T., Roskin K. M., Schwartz M., Sugnet C. W., Thomas D. J., et al. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stolt C. C., Rehberg S., Ader M., Lommes P., Riethmacher D., Schachner M., Bartsch U., Wegner M. Genes Dev. 2002;16:165–170. doi: 10.1101/gad.215802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takahashi Y. Prog. Neurobiol. 1992;38:523–569. doi: 10.1016/0301-0082(92)90041-c. [DOI] [PubMed] [Google Scholar]
- 25.Connor J. R., Menzies S. L. Glia. 1996;17:83–93. doi: 10.1002/(SICI)1098-1136(199606)17:2<83::AID-GLIA1>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 26.Maier M., Ron M. A. Schizophr. Res. 1996;22:5–17. doi: 10.1016/0920-9964(96)00044-8. [DOI] [PubMed] [Google Scholar]
- 27.Bartzokis G. Neuropsychopharmacology. 2002;27:672–683. doi: 10.1016/S0893-133X(02)00364-0. [DOI] [PubMed] [Google Scholar]
- 28.Aston C., Jiang L., Sokolov B. P. Mol. Psychiatry. 2005;10:309–322. doi: 10.1038/sj.mp.4001565. [DOI] [PubMed] [Google Scholar]
- 29.Kapur S., Remington G. Annu. Rev. Med. 2001;52:503–517. doi: 10.1146/annurev.med.52.1.503. [DOI] [PubMed] [Google Scholar]
- 30.World Health Organization. The ICD-IO Classification of Mental and Behavioural Disorders. Geneva: W.H.O.; 1992. [Google Scholar]
- 31.Feighner J. P., Robins E., Guze S. B., Woodruff R. A., Jr., Winokur G., Muñoz R. Arch. Gen. Psychiatry. 1972;26:57–63. doi: 10.1001/archpsyc.1972.01750190059011. [DOI] [PubMed] [Google Scholar]
- 32.Castensson A., Aberg K., McCarthy S., Saetre P., Andersson B., Jazin E. Am J. Med. Genet. 2005;134:84–89. doi: 10.1002/ajmg.b.30151. [DOI] [PubMed] [Google Scholar]
- 33.Castensson A., Emilsson L., Preece P., Jazin E. E. Genome Res. 2000;10:1219–1229. doi: 10.1101/gr.10.8.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lenhard B., Wasserman W. W. Bioinformatics. 2002;18:1135–1136. doi: 10.1093/bioinformatics/18.8.1135. [DOI] [PubMed] [Google Scholar]
- 35.Rice P., Longden I., Bleasby A. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 36.Thompson J. D., Higgins D. G., Gibson T. J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]