Abstract
Mobile genetic elements with the ability to integrate genetic information into chromosomes can cause disease over short periods of time and shape genomes over eons. These elements can be used for functional genomics, gene transfer and human gene therapy. However, their integration-site preferences, which are critically important for these uses, are poorly understood. We analyzed the insertion sites of several transposons and retroviruses to detect patterns of integration that might be useful for prediction of preferred integration sites. Initially we found that a mathematical description of DNA-deformability, called Vstep, could be used to distinguish preferential integration sites for Sleeping Beauty (SB) transposons into a particular 100 bp region of a plasmid [G. Liu, A. M. Geurts, K. Yae, A. R. Srinivassan, S. C. Fahrenkrug, D. A. Largaespada,J. Takeda, K. Horie, W. K. Olson and P. B. Hackett (2005) J. Mol. Biol., 346, 161–173 ]. Based on these findings, we extended our examination of integration of SB transposons into whole plasmids and chromosomal DNA. To accommodate sequences up to 3 Mb for these analyses, we developed an automated method, ProTIS©, that can generate profiles of predicted integration events. However, a similar approach did not reveal any structural pattern of DNA that could be used to predict favored integration sites for other transposons as well as retroviruses and lentiviruses due to a limitation of available data sets. Nonetheless, ProTIS© has the utility for predicting likely SB transposon integration sites in investigator-selected regions of genomes and our general strategy may be useful for other mobile elements once a sufficiently high density of sites in a single region are obtained. ProTIS analysis can be useful for functional genomic, gene transfer and human gene therapy applications using the SB system.
INTRODUCTION
Mobile genetic vectors have been harnessed for genetic studies in model organisms and are being developed as agents for gene-therapy in humans (1–3). For example, the awakening of the Sleeping Beauty (SB) transposon system as a powerful tool for insertional mutagenesis to identify oncogenes (4,5) and other classes of genes (6,7) complements retroviral vectors, which have been used for decades (8). Importantly, understanding the parameters that affect integration of vectors is required to appreciate fully the results of their applications.
Although transposons and some retroviruses integrate in virtually all regions of host genomes, their integration is not random (9–18). Weak consensus sequences have been described surrounding the sites of integration for retroviruses (16,19) and transposable elements (6,20,21). However, the most-favored integration sites do not always conform to these sequences (6). In addition to specific-sequence recognition, DNA structural characteristics, including protein-induced deformability, A-philicity and bendability, have been shown to influence binding of proteins (22). Although these structural characteristics are sequence-dependent, two dissimilar sequences can have similar structural patterns. As a result, distinct preferred integration sites may not match consensus sequences, but rather share similar structural patterns. Unique patterns of these DNA structural characteristics at integration sites have been reported for retroviruses and lentiviruses (19), P-elements (20) and SB transposons (21,23) that may contribute to mechanisms that differentiate potential loci for integration of mobile genetic elements. We previously used a mathematical description of DNA ‘deformability’ called Vstep to identify shared structural patterns among several preferred integration sites for SB transposons into a short 100 bp region of a target plasmid (23). DNA deformation is characterized by a non-uniform twisting of the double helix, alteration in the spacing between the base pairs at the integration site and localized tilting of the target site such that the axis around the insertion site is off center. This initial analysis did not answer the question of whether these parameters can be used to effectively predict integration site preferences into chromosomal DNA in mammalian genomes nor whether other integrating vectors followed similar rules.
Here we describe our strategy of using a small dataset of high-density integrations into a defined region of DNA to formulate rules that govern integration-site preferences in lengths of chromatin of more than 3 Mb. To analyze such long stretches of DNA, we developed an algorithm for rapidly scanning DNA sequences to predict favored sites of integration of mobile elements into mammalian chromosomes. We used SB transposons as a model element to establish a method for finding and testing rules that govern integration-site preferences. We used two datasets from forward-genetic studies to verify the predictions made by our algorithms and then examined potential integration preferences for two other transposons as well as retroviral and lentiviral vectors.
MATERIALS AND METHODS
Algorithm for determining the Vstep profile of SB transposon integration sites
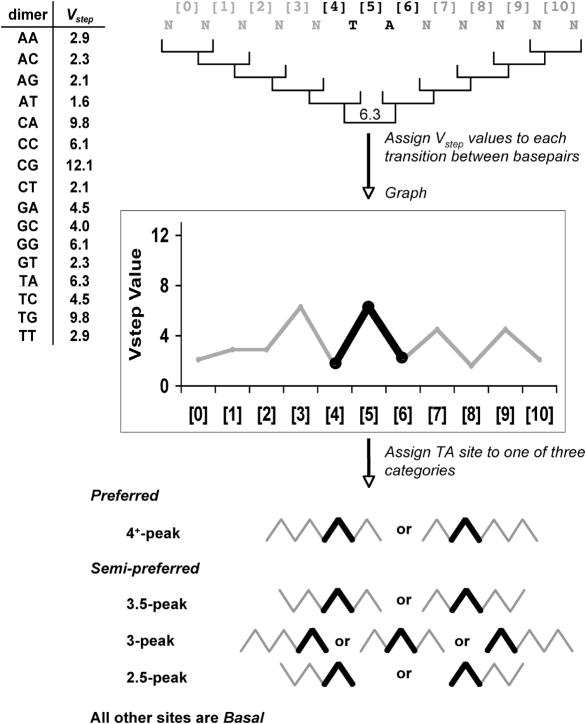
Figure 1 illustrates the steps in establishing Vstep profiles for a given TA site. We developed a Perl script, called ProTIS© (Profiler of Transposon Insertion Sites), to analyze automatically every TA site in an input sequence file (up to 20 Mb tested). For each TA dinucleotide in the input sequence, the script extracts 5 bases on each side of the TA dinucleotide and translates the 12-base sequence into a series of Vstep values for the 11 transitions between consecutive base pairs (referred to as dimer steps) within the sequence (Figure 1). Then, using a series of less-than (<) or greater-than (>) comparisons, the program uses the ordered Vstep values to classify each TA site and its flanking nucleotides into one of five classes (4+-peak, 3.5-peak, 3-peak, 2.5-peak and basal). For each schematic, the TA-peak is shown in bold and is generated from the [4], [5] and [6] Vstep values, where [5] always has a Vstep value of 6.3. For the 4+-peak and 3.5-peak patterns, the light lines represent peaks or half-peaks that must be on at least one or can be on both sides of the central TA-peak. The patterns shown in Figure 1 show mirror images because the transposon can integrate in either direction into the symmetrical TA dinucleotide basepairs. For the 3-peak and 2.5-peak patterns, the peaks flanking the TA-peak can be to the left or the right of the TA-peak. Experimentally, 2.5-, 3- and 3.5-peak pattern target sites exhibited negligible differences in their abilities to attract SB transposons, so they were grouped together. This allowed us to simplify our TA site classification into three groups, preferred sites (4+-peak), semi-preferred sites (3.5/3.2.5-peak) and basal sites. With these definitions, for every bin of a given length of DNA, the total number of each class of TA site per bin is tallied for the input sequence. Using weighted coefficients (shown in bold in the equation below) for each class of TA site from the pFV/Luc data in Table 1, a Total Vstep score can be calculated for each bin using the equation:
The script produces a tab-delineated table output that is then conveniently analyzed and graphed using Microsoft Excel (Microsoft, Redmond, WA).
Figure 1.
Profiling TA sites using the Vstep algorithm. Sequences of 12 bp (N) with TA sites at positions six and seven were analyzed with respect to the 11 Vstep values ([0]–[10]) for transitions from one base pair to the next (brackets). Profiles are charted and subsequently assigned to one of three categories, preferred, semi-preferred or basal, based upon the graphical pattern. In all profiles there is a ‘TA-peak’ that always exists in such profiles because the T-to-A Vstep value is 6.3 and all steps from N to T and from A to N (N = any base) are always <3.0, as shown on the left side of the figure. The ‘TA peak’ formed by the two lines that connect the three Vstep values for the N-to-T, T-to-A and A-to-N steps are shown in boldface.
Table 1.
SB transposition-site preferences as a function of Vstep profiles
| Vstep Pattern | # Target Sites (% of total) | Sites Hit | Insertions/Site | Preference |
|---|---|---|---|---|
| pFV/Luc: | ||||
| Basal | 299 (61%) | 39 | 0.13 | 1X |
| Semi-Preferred | 154 (31%) | 92 | 0.60 | 5X |
| Preferred | 36 (7%) | 62 | 1.7 | 13X |
| Braf Intron-9: | ||||
| Basal | 209 (60%) | 5 | 0.02 | 1X |
| Semi-Preferred | 105 (19%) | 12a | 0.11 | 6X |
| Preferred | 33 (10%) | 8b | 0.2 | 10X |
| 3.2 Mbp Chromosome 1: | ||||
| Basal | 117 454 (56%) | 5 | 0.00004 | 1X |
| 2.5-peak | 67 070 (32%) | 15 | 0.00022 | 6X |
| Preferred | 23 775 (11%) | 14 | 0.00059 | 15X |
aA total of 11 sites were hit; one was hit twice for a total of 12 hits.
bSix sites were hit; two were hit twice for a total of eight hits.
Analyzing Vstep and A-philicity profiles of insect transposons and retroviruses
An additional script was generated to accept tabulated integration site data from different sources. The Vstep classifier script can accept sequence information accumulated in integration-site studies. The script takes each line of the tabulated data, extracts the pertinent sequence information, assigns both the Vstep and A-philicity values to each dimer step, and generates tab-delineated output files similar to that of ProTIS©. ProTIS©, including further instructions, is available for download on the Hackett lab website http://www.cbs.umn.edu/labs/perry/ as open-source code. Control sequences for piggyBac and P-element analyses were obtained from three separate 1 Mb regions of the Drosophila genome (4.2 BDGP release), chromosome 2L from position 10–11 Mb, chromosome 2L from position 17–18 Mb and chromosome 3L from position 11–12 Mb.
Statistical analysis
To examine the relationship between the ProTIS© prediction, based on the Total Vstep score and known insertion sites, we fit a Poisson regression model. This model takes the number of insertions into each bin as a measurement of ‘insertion activity’ in that region and compares it with the predicted score for that bin made by ProTIS©. To take into account that the incoming transposon does not define a target sequence in terms of 100 bp bins, we fit a lag-1 autocorrelation structure. The autocorrelation structure assumes that neighboring bins are correlated at some estimated level ρ, and that the correlation disappears exponentially with increasing genetic distance, ρd. These combined methods measure the relationship of the insertion activity and Total Vstep scores across the entire target sequence and calculate regression coefficients using generalized estimating equations. The robust standard errors associated with this analysis were used to derive P-values (24). The regression coefficient, in turn, can be used to derive a relative risk value . In this case, correlates an increase in Total Vstep score for any bin, compared with any particular TA site, with the likelihood that a transposon will insert in the bin. For the pFV/Luc integrations, (Results) this fit yielded a regression coefficient and corresponds to a relative risk of . A comparison of the Total Vstep values of bins 17 and 22 in Figure 2a, which have nearly the same number of TA sites, 15 and 16 respectively, but different Total Vstep scores of 72 and 27, respectively, gives a difference of 45. The difference of 45 corresponds to 9.5-fold () increase in the likelihood that an insertion will occur in bin 17 compared to 22. Likewise, fitting an equivalent model for the Braf data (Results) yields a slightly smaller regression coefficient . Interpreting this coefficient means that an increase in the Total Vstep score of 20 raises the probability of an insertion by , while an increase of 40 raises the probability of an insertion by .
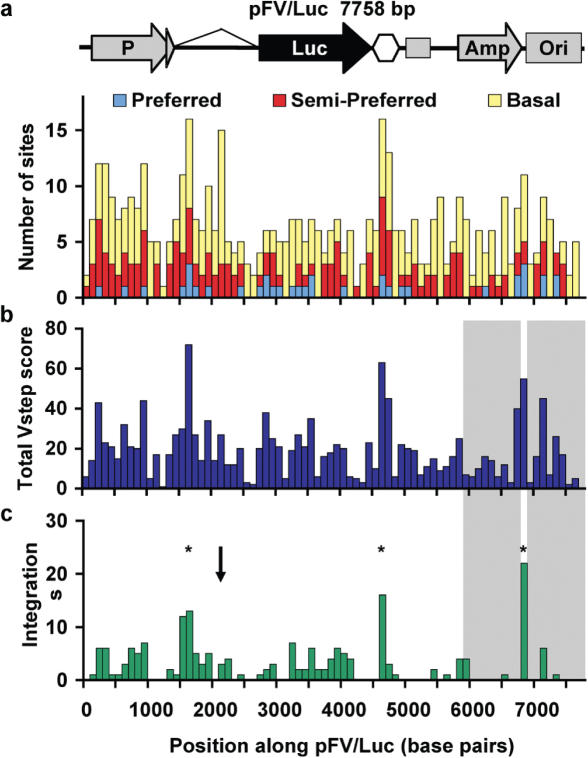
Figure 2.
Total Vstep profile of the 7758 bp plasmid pFV/Luc. The sequence is divided into 78 bins of size100 bp. (a) Plot of the number of each type of TA site per bin. The hexagon indicates the Chinook salmon poly(A) addition motif and the following square indicates an M13 origin of replication. (b) Plot of Total Vstep score per bin. (c) Distribution of observed insertion sites [adapted from Liu et al. (23)]. Shaded areas are regions required for selection and thus unlikely to be scored. The asterisks indicate the three most likely regions for integration based on ProTIS analysis and the arrow indicates a region that has a high number of TA sites, but relatively few integrations.
RESULTS
Development of an algorithm for Vstep profiles of transposon-integration sites
Our analysis began with SB transposons, which always integrate into the simple dinucleotide sequence TA (25). The DNA structural parameter Vstep is a measurement of the protein-induced deformability of DNA sequences, gathered from the analysis of DNA molecules bound and unbound by proteins (26). A Vstep value correlates with the level of deformability of the DNA double helix at the transition between two consecutive base pairs in a sequence (dimer steps, Figure 1 top panel) (26). Using an intra-plasmid transposition analysis that examined 100 bp of the 7758 bp pFV/Luc plasmid, we found that potential TA-integration sites could be divided into three groups with a 16-fold range for integration preference based upon Vstep patterns of base pairs flanking the target TA dinucleotide (23).
Based on this very special case, we extended our analysis of target-site selection to refine our ability to predict preferred SB integration sites. Since establishing a Vstep profile for extended regions is extremely tedious, we generated a Perl script that analyzes every TA site in a DNA sequence and assigns a Vstep value to consecutive transitions between base pairs flanking the site. The series of Vstep values corresponding to the dimer steps in the sequence can be graphed to establish a pattern that can be used to distinguish various integration sites. Each TA site is then classified in terms of likelihood of transposon integration based on which of the three categories of Vstep patterns it mimics (Materials and Methods).
Our analysis of the entire 7758 bp plasmid revealed that a 12 bp window, including 5 bp flanking each side of a target TA dinucleotide, was sufficient to distinguish four TA-site Vstep profiles that differed in their integration potentials when compared with a non-preferred, or basal, TA site (Figure 1, bottom panel). To facilitate our analyses, we combined several profiles of TA sites that have similar Vstep patterns into a single category (Figure 1, Semi-preferred), so that any TA site in a target falls into one of three groups—preferred, semi-preferred or basal. As shown in Table 1, these groups vary more than 10-fold in integration preference. We next sought to test whether ‘weighing’ each TA site based on the observed integration frequencies and summing the weighted scores of all TA sites in a given region could be used to predict the likelihood of integration into that region. For this we modified our script to bin the input sequence, tally each class of TA site, sum their relative weights using the preferences in Table 1 as coefficients and generate a ‘Total Vstep score’ for each bin. We called this Perl script ProTIS©.
The distribution of TA sites for the entire pFV/Luc sequence is shown in Figure 2a. The sequence is divided into 100 bp bins and the numbers of each type of TA site within each bin are enumerated. The theoretical plot for the Total Vstep scores for pFV/Luc is shown in Figure 2b and the actual distribution of integrations (23) is shown inFigure 2c. Two regions, Amp and Ori of pFV/Luc, are underrepresented (shaded regions) because insertions into these regions can disrupt the selection method for recovering events. When the entire sequence is divided into 100 bp bins, and the numbers of insertions sites into each bin are treated as events from a Poisson distribution, the experimental data, outside of Amp and Ori, show a statistically significant overlap with the Total Vstep scores plot (P < 0.0001).
As an alternative approach based on the apparent overlap in the distribution of TA dinucleotides in Figure 2a and the integration profile in Figure 2c, we tested whether the TA-dinucleotide distribution alone would be an equally faithful predictor of integration sites. Similar significance of an overlap between the TA distribution and integration pattern was found using the aforementioned statistical method (P < 0.0001). The residual deviance, however, is larger in this model and so the regression fit is inferior to the use of Total Vstep scores when using the number of TAs. The Akaike Information Criterion (AIC) formally compares two model fits based on their likelihoods (27). Fitting the model using a TA tally results in a larger AIC, 328.2, than using the Total Vstep, 312.9. These results suggest that the Total Vstep is the better predictor of insertion sites. Accordingly, using the training set of interplasmid transposition events and the Total Vstep score, we identified a method that could potentially predict the outcomes of applied genetic studies using SB transposons.
Remobilization of transposons into the ninth intron of the mouse Braf gene
The key to identifying preferred sites in chromatin is to examine multiple integrations into a limited genomic region and quantify variations from Poisson statistics. Such data became available from a study in which the SB transposon, T2/Onc, was engineered to elicit gain-of-function mutations and accelerate tumor formation in somatic tissues of mice lacking the p19Arf tumor suppressor (4). The most frequent oncogenic insertion site was intron-9 of the Braf gene. All of the 25 analyzed insertions in intron-9 were oriented toward the 10th exon (Figure 3a), resulting in a transcript encoding the kinase domain of Braf that acts as a dominant oncogene. Of the 347 potential TA-integration sites in the 4069 bp intron, 22 were targets and three sites were hit twice. In this case, the probability of two insertions into a single TA site is 0.07 and the odds of this happening three times are 0.0004, which strongly suggested the existence of preferential insertion sites.
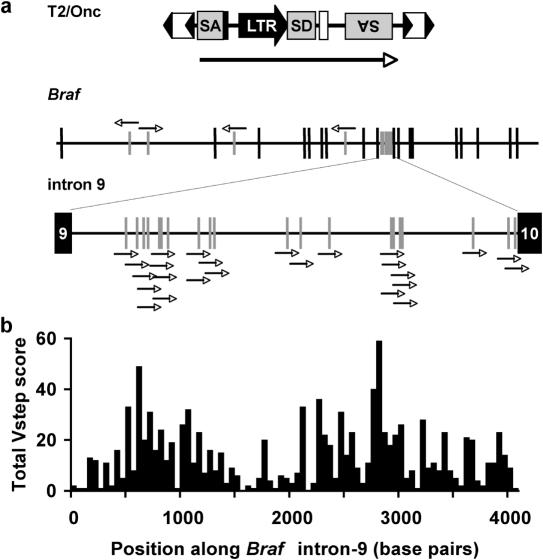
Figure 3.
Vstep analysis of insertion sites of T2/Onc into the mouse Braf gene. (a) Schematic of mapped insertions into Braf (exons shown as tall vertical lines) with an expanded intron-9. Only T2/Onc transposons that integrated in a left-to-right orientation would be identified in the genetic screen. SA, splice-acceptor site, SD, splice-donor site, LTR, retroviral long terminal repeat, double arrowheads, inverted terminal repeats of the integrating transposon. The long arrow represents the direction of transcription from the LTR promoter within T2/Onc. (b) Total Vstep profile of intron-9 in terms of 82 bins of size 50 bp.
Because translation of the N-terminally truncated Braf polypeptide is initiated from an internal start codon in exon-10, we assumed any T2/Onc insertion regardless of location or reading frame in Braf intron-9 would lead to oncogenic selection, and that the uneven distributions of insertions were the result of preferential target site selection. We thus identified these events as a dataset with which to test our method and ran ProTIS© on the intron-9 sequence. The individual Vstep profiles for T2/Onc-targeted sites in intron-9 are shown in Supplementary Figure 1. Table 1 shows the distribution of integrations into the various categories of profiles for the 347 sites. The 33 sites with preferred-site profiles had a 20% hit rate, the 105 predicted semi-preferred sites had an 11% hit rate, and the 209 basal sites showed only a 2% hit rate. These data strongly suggest a 5- to 10-fold preference for integrations at semi-preferred sites and preferred sites in intron-9 compared with basal sites. Figure 3a shows that the distribution of T2/Onc insertions into intron-9matches the plot of Total Vstep scores (Figure 3b). Using the same statistical procedure described for Figure 2, this overlap between the experimental data and the theoretical prediction is highly significant (P < 0.0001).
Vstep-profiling of an extended chromosomal region
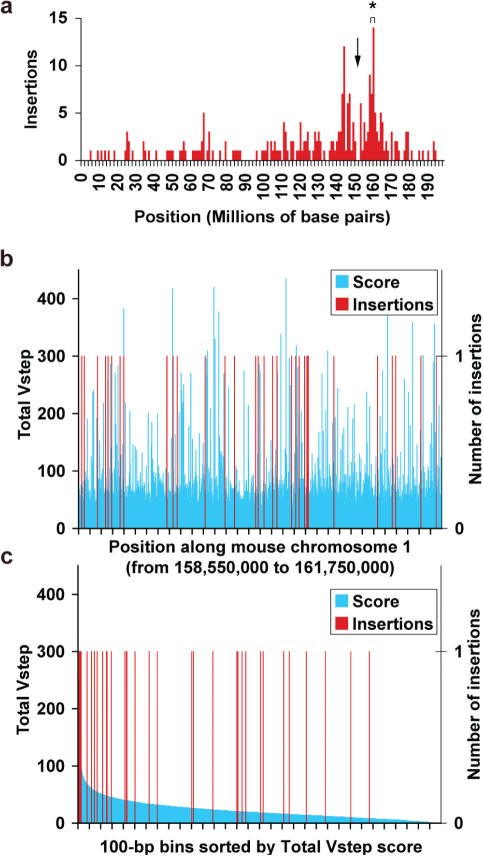
SB transposons resident in a mouse chromosome can be remobilized to new sites, most often within ∼10 Mb of their original locus (6,28–31), providing another source of densely localized transposon integrations. We thus examined 3.2 Mb of mouse chromosome 1 (position 158 550 000–161 750 000 bp according to NCBI m33 build) in which 34 remobilized transposition events were mapped in the vicinity of a transgenic donor concatemer of SB transposons (4). As shown in Figure 4a, this region (asterisk) is ∼15 Mb from the concatemer (arrow). In this region there are 208 299 TA sites corresponding to approximately one TA site per 15 bp. Of these TA sites, ProTIS© predicts 117 454 basal, 67 070 semi-preferred and 23 775 preferred TA sites. The distribution of sites was divided into 32 000 bins of size 100 bp, in terms of either map position (Figure 4b) or Total Vstep score (Figure 4c). The average Total Vstep score per 100 bp bin over the entire region is 23 (range from 0 to 435) and transposons inserted into intervals with an average score of 50 (range from 9 to 250). Thus, the insertions clearly are skewed towards the higher Vstep values (P < 0.0001). Table 1 shows that the distribution of integrations into each Vstep profile category is similar to the integration preferences observed in pFV/Luc and Braf intron-9.
Figure 4.
Transposon insertion sites in 3.2 Mb of mouse chromosome 1. (a) SB integration sites in Chromosome 1, the locations of the concatemer from which the transposons were remobilized (downwards arrow) and the 3.2 Mb region that had the highest density of integrations is marked with an asterisk. Region (b) was divided into 32 000 bins of size 100 bp and the Total Vstep scores for each bin calculated as described in Figure 3. The average Total Vstep value per bin is 23. (b) Blue bars, Total Vstep scores/bin; red bars, insertion sites mapped as a function of position. (c) Insertion sites (red) displayed as a function of Total Vstep score/bin (blue).
Overall, the data from insertions into an active gene (Braf), a region of chromosome 1 comprising ∼0.1% of the mouse genome, and a plasmid are remarkably consistent despite a 1000-fold range in insertion density between pFV/Luc and 3.2 Mb in chromosome 1. These results indicate that ProTIS© and its future derivatives will be valuable predictors of vector integration sites into genomes.
Application of the ProTIS© method to a genomic target of therapeutic vectors
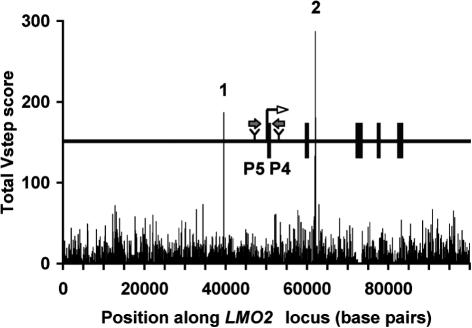
The randomness of integration sites is an area under discussion with regard to vectors for gene therapy, including SB-based vectors (2,3,32). However, the potential of severe adverse effects following random integration has been a concern (33,34). In particular, two cases of acute lymphoblastic leukemia in children followed transfer of the IL-2 γc gene in retrovirus-based vectors. Each apparently resulted from an insertional activation of the LMO2 oncogene followed by selective outgrowth of the treated cells (35,36). Because SB transposons are being developed as gene therapy vectors (3) and LMO2, out of more than 291 identified cancer genes (37), is associated with all of the severe adverse events incurred in the IL-2 γc trials, we examined the LMO2 locus using ProTIS© as a model for how the program can be applied to any genetic locus of interest. Figure 5 shows the plot of Total Vstep scores of 100 kb of genomic sequence containing the LMO2 gene, with 50 kb of upstream sequence, and the relative positions of the two activating retroviruses (P4, P5). ProTIS© predicts two sequences with prominent Vstep scores, labeled 1 and 2, that derive from a simple tandem repeat, (TCTA)n, and a 165 bp sequence that is replete with tandem (TA)n repeats, respectively. SB apparently has a 10-fold preference to land in microsatellite repeat regions containing TA dinucleotides (18), which is consistent with our findings that preferred sites such as (TA)n repeats have a 13-fold predicted preference using ProTIS© profiling. The ProTIS© plot of the LMO2 locus suggests that SB vectors would target regions 1 and 2, which are more than 10 kb from the transcriptional initiation site, and three times the distances of the activating proviruses, P4 and P5. Similar analyses can be done for any gene of interest.
Figure 5.
Total Vstep Profile for the human LMO2 gene plotted as 100 bp bins. The map of 100 kb of the LMO2 locus is shown above the center of the Vstep profile. Rectangles, exons; block arrows, sites of two activating retroviral insertions [P4 and P5, Ref. (36)]. Spikes 1 and 2 in the Total Vstep profile correspond to short tandem repeats of (TCTA)n and (TA)n, respectively.
Profiling other transposable elements and retroviruses
Although SB was the first DNA-based transposable element developed to deliver DNA sequences into mammalian genomes, lepidopteran piggyBac transposons and Drosophila P-elements are powerful germline-transformation tools in insects (38,39). Although both of these vectors have significantly strong preferences for transcriptional units, we hypothesized that they might exhibit target-site selection patterns related to DNA structure that would further define sites of integration within genes. Accordingly, we examined the integration-site sequence-tags deposited in GenBank from multiple investigations. The single largest deposit of integration sites was generated by Exelixis and comprises over 18 000 piggyBac and 6500 P-element insertions (20,40). We refined the piggyBac data to 11 791 integrations that could be identified by the TTAA sequence recognized by piggyBac transposase and 5070 P-element integrations into validated genomic sequences. For both transposons we used the same procedure to identify preferential integration sites as we did for SB integrations: (i) find insertion hotspots, (ii) develop rules based on these sequences and (iii) test the rules against a much larger set of integrations. In contrast with what we found for SB transposons, there was no consistent Vstep pattern shared amongst either the piggyBac or P-element integration sites (Supplementary Figures 2 and 3).
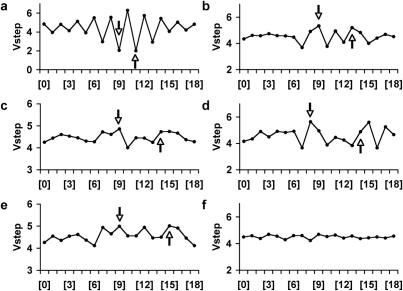
Retroviruses have been utilized in genetic screens and for germline and somatic transgenesis in vertebrates for decades. Weak consensus sequences are found at the integration sites of several retroviruses (16,19), based upon the examination of relatively few integration sites scattered across a target genome. Using curated data kindly provided by Drs Xiaolin Wu and Alex Holman, we examined 695 murine leukemia virus (19), 1371 human immunodeficiency virus-1(10,13), 148 simian immunodeficiency virus (13) and 551 avian sarcoma-leukosis virus (13,14) integration sites for Vstep patterns that would aid in predicting integration preferences (Figure 6). As with P-elements, we found symmetric patterns that overlap with the base pairs involved in the target site duplication for most family members. Importantly, these patterns are based on the same compilations used to identify unique, weak consensus sequences for the various viruses (16,19) and cannot be used to generate algorithms alone. The indicated patterns shown in Figure 6 suggest that Vstep rules for identifying preferential integration sites might exist, but adequately dense sources of in vivo integration sites for these vectors, along with the identification of hotspots, are still required to generate appropriate algorithms.
Figure 6.
Vstep analysis of insertion sites of proviruses and transposons. The arrows in the profiles indicate the boundaries of the TSD sequence that occurs with the staggered cuts made by the various integrase enzymes. (a) Average Vstep profiles for 573 SB transposon integrations. (b) Average Vstep profiles for murine leukemia virus. (c) Average Vstep profiles for human immunodeficiency virus. (d) Average Vstep profiles for simian immunodeficiency virus. (e) Average Vstep profiles for avian sarcoma/leucosis virus. (f) Average Vstep profiles for 1006 random DNA 20mer sequences.
DISCUSSION
The observation that hotspots for SB transposon integration do not always match the published consensus sequence from different studies (23) led us to investigate other properties of sequences surrounding target sites. The data presented in this report confirm our hypothesis that SB transposase recognizes distinct structural features in DNA sequences, regardless of primary DNA sequence, that can be described by the Vstep DNA-deformation parameter. Preferential TA-integration sites can be identified by specific Vstep profiles of the DNA sequences flanking a TA site, regardless of whether the target sequence is a 100 bp segment of a plasmid, an entire plasmid, a portion of an actively transcribed gene or bulk chromatin (Table 1). This method is more accurate than a simple distribution of TA sites in a target sequence. As transposon insertions approach saturation of a target genome, the ProTIS© algorithm will provide a closer approximation, in part, because some simple repeat sequences containing TA dinucleotides have a greater ability to attract SB transposons than other repeats containing TA dinucleotides. For instance, a 100 bp target consisting of the repeat (TATC)25 translates into a Total Vstep score of 325, whereas a 100 bp sequence consisting of the repeat (TACT)25 has a Total Vstep score of only 25. Each sequence represents an equal number of TA target sites and the same base composition, but when compared, translate into a 13-fold difference in the number of integrations that would be observed. Nevertheless, genomes are vast and non-uniform in terms of structure, protein associations, methylation, compaction, etc. Thus, it would not be surprising that in some cases predictions made by ProTIS will fail.
The application of SB to forward-genetic studies (4,5) has opened possibilities for the identification of novel genes that influence the formation of various tumor types. Repeated observation of transposon-induced mutations in the same gene in several different tumor samples identifies that gene as a candidate cancer gene. ProTIS© will be a valuable tool in this field, helping geneticists to distinguish between those events that are truly biologically significant common sites of integration from those events that are biased to be repeatedly tagged because of an abundance of preferred integration sites.
SB transposase has catalytic properties that are shared by other DDE-type recombinases, including retroviral integrases (41). Consequently, we reasoned that these other enzymes might also have integration site preferences that are based on local DNA structure. However, even though thousands of integration sites have been recorded for various viral vectors, there are no reports of regions of chromatin that harbor densities of integrations that result in multiple integrations into a single site, a requirement for defining Vstep-based rules for preferential integration sites. Thus, quantitative measurements that generate rules for prediction of structure-based preferential integration sites for piggyBac and P-element transposons as well as for retroviruses are not possible using this approach with currently available datasets. Although sequence-based assays for examining some retroviral integration patterns in defined targets have been developed (42–44), hundreds of integration sites for any vector will likely have to be generated to generate rules for predicting preferred sites. Many factors have been shown to influence the integration of retroviral and lentiviral integration including preferences to integrate into transcription units, gene expression profiles of the target cell genome, nucleosome packing of chromatin, sequence motifs such as CpG islands (45) and growth arrest of cultured cells in the case of HIV integration (46). Our understanding of the contributions of these factors is insufficient for prediction of retroviral integration sites. Perhaps local DNA structure, as we have shown for SB transposons, plays yet an additional role in defining preferential sequences for integration. For example, it may provide a mechanism by which HIV prefers to avoid integration into or near CpG islands because the structure of dimer steps in the CpG sequence is not favorable to integration. Validation of this hypothesis requires a substantial dataset of numerous integrations into a small, defined target sequence to identify specific Vstep patterns common to the most preferred insertion sites. Otherwise, Vstep analyses provide essentially the same information as a consensus sequence.
Our examination of SB transposon integrations in ∼0.1% of the euchromatic genome (Table 1) suggest that of the ∼200 million TA sites in the mouse genome, ∼10% (20 million) will be preferred sites that would account for 55% of transposon insertions, whereas 120 million (60%) basal TA sites would attract only 5% of transposon insertions. We expect the same results in humans. Thus, although SB transposons can integrate into practically any TA site, within a given region about half will go to only 10% of the available sites. This information is important for evaluating SB transposons for both insertional mutagenesis and as a vector for gene therapy.
Our analysis of integration sites is applicable to understanding the biology of other transposons whose consensus preferences are already known. For example, the Tc1 transposon in Caenorhabditis elegans that integrates into TA sites has a consensus sequence GA(G/T)(A/G)TA(T/C)(G/C)T (47,48). One hotspot, TGGTGTATGTCT, was hit 51 times in 166 mapped insertions (49). Vstep analyses of the consensus and hot spot match the most preferred category for SB transposition. In contrast, the integration consensus sequence for a related C.elegans transposon, Tc3, does not match that of Tc1 and the Vstep profile of both its consensus and most preferred integration site, ACTAATATTATG, are distinctly different from Tc1 and SB (49,50). Specifically, there is extra spacing in the most preferred Tc3 profile on both sides of the TA peak compared with the profiles for Tc1 and SB (Supplementary Figure 4). Likewise, some of the hottest sites for Drosophila Himar1 integration (51) also match the Vstep profiles of SB and Tc1.
Repetitive (mobile) elements play a significant role in genome evolution (52–54). For instance, the most prominent differences in the human and chimpanzee genomes are rates of transposable element insertions and new insertions of novel retroviral elements (55). Until now, parameters governing the integration of transposons and proviruses have been ignored. By identifying preferences for the different classes of repetitive elements, it should be possible to determine the role(s) of natural selection on newly introduced elements by comparing their observed distributions compared with the theoretical expectations. Because viral elements comprise a significant proportion of mammalian genomes, further work in identifying the rules for their integration preferences will be of interest to those studying evolution as well as those interested introducing new genetic sequences into genomes for functional genomic studies and therapeutic purposes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
The authors thank the Arnold and Mabel Beckman Foundation for supporting their work and all members of the Beckman Center for Transposon Research for a long history of contributions of ideas and results. The authors are especially grateful to Drs Michael Simmons, Reuben Harris, William A. Weiss (UCSF) and Elena Aronovich for careful reading of the manuscript, Dr Keiko Akagi (NCI) for mapping the T2/Onc integrations, Michael H. L Lee for technical assistance and Joe Carlson (Berkeley Drosophila Genome Project) for sending P-element datasets. The authors were also supported by NIH grants 1PO1 HD32652-07 and R43 HL076908-01 (P.B.H.) and DA014764 (D.A.L.) and T32 HD007480 (A.M.G.), NIGMS GM008568 (C.S.H.) and NCI T32CA09138-28 (L.S.C. and C.M.C.) and F32CA10619-01 (L.S.C.). Funding to pay the Open Access publication charges for this article was provided by NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ivics Z., Izsvak Z. Transposable elements for transgenesis and insertional mutagenesis in vertebrates: a contemporary review of experimental strategies. Methods Mol. Biol. 2004;260:255–276. doi: 10.1385/1-59259-755-6:255. [DOI] [PubMed] [Google Scholar]
- 2.Izsvak Z., Ivics Z. Sleeping Beauty transposition: biology and applications for molecular therapy. Mol. Ther. 2004;9:147–156. doi: 10.1016/j.ymthe.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 3.Hackett P.B., Ekker S.C., Largaespada D.A., McIvor R.S. Sleeping Beauty transposon-mediated gene therapy for prolonged expression. Adv. Genet. 2005;54:189–232. doi: 10.1016/S0065-2660(05)54009-4. [DOI] [PubMed] [Google Scholar]
- 4.Collier L.S., Carlson C.M., Ravimohan S., Dupuy A.J., Largaespada D.A. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature. 2005;436:272–276. doi: 10.1038/nature03681. [DOI] [PubMed] [Google Scholar]
- 5.Dupuy A.J., Akagi K., Largaespada D.A., Copeland N.G., Jenkins N.A. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436:221–226. doi: 10.1038/nature03691. [DOI] [PubMed] [Google Scholar]
- 6.Carlson C.M., Dupuy A.J., Fritz S., Roberg-Perez K.J., Fletcher C.F., Largaespada D.A. Transposon mutagenesis of the mouse germline. Genetics. 2003;165:243–256. doi: 10.1093/genetics/165.1.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keng V.W., Yae K., Hayakawa T., Mizuno S., Uno Y., Yusa K., Kokubu C., Kinoshita T., Akagi K., Jenkins N.A., et al. Region-specific saturation germline mutagenesis in mice using the Sleeping Beauty transposon system. Nat. Methods. 2005;2:763–769. doi: 10.1038/nmeth795. [DOI] [PubMed] [Google Scholar]
- 8.Mikkers H., Berns A. Retroviral insertional mutagenesis: tagging cancer pathways. Adv. Cancer Res. 2003;88:53–99. doi: 10.1016/s0065-230x(03)88304-5. [DOI] [PubMed] [Google Scholar]
- 9.Zhang P., Spradling A.C. Insertional mutagenesis of Drosophila heterochromatin with single P elements. Proc. Natl Acad. Sci. USA. 1994;91:3539–3543. doi: 10.1073/pnas.91.9.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schroder A.R.W., Shinn P., Chen H., Berry C., Ecker J.R., Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 11.Wu X., Li Y., Crise B., Burgess S.M. Transcription start regions in human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 12.Nakai H., Montini E., Fuess S., Storm T.A., Grompe M., Kay M.A. AAV serotype 2 vectors preferentially integrate into active genes in mice. Nature Genet. 2003;34:297–302. doi: 10.1038/ng1179. [DOI] [PubMed] [Google Scholar]
- 13.Mitchell R.S., Beitzel B.F., Schroder A.R., Shinn P., Chen H., Berry C.C., Ecker J.R., Bushman F.D. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLOS. 2004;2:1127–1136. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Narezkina A., Taganov K.D., Litwin S., Stoyanova R., Hayashi J., Seeger C., Skalka A.M., Katz R.A. Genome-wide analyses of avian sarcoma virus integration sites. J. Virol. 2004;78:11656–11663. doi: 10.1128/JVI.78.21.11656-11663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maxfield L.F., Fraize C.D., Coffin J.M. Relationship between retroviral DNA-integration-site selection and host cell transcription. Proc. Natl Acad. Sci. USA. 2005;102:1436–1441. doi: 10.1073/pnas.0409204102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holman A.G., Coffin J.M. Symmetrical base preferences surrounding HIV-1, avian sarcoma/leukosis virus, and murine leukemia virus integration sites. Proc. Natl Acad. Sci. USA. 2005;102:6103–6107. doi: 10.1073/pnas.0501646102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hematti P., Hong B.K., Ferguson C., Adler R., Hanawa H., Sellers S., Holt I.E., Eckfeldt C.E., Sharma Y., Schmidt M., et al. Distinct genomic integration of MLV and SIV vectors in primate hematopoietic stem and progenitor cells. PLoS Biol. 2004;2:e423. doi: 10.1371/journal.pbio.0020423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yant S.R., Wu X., Huang Y., Garrison B., Burgess S.M., Kay M.A. High-resolution genome-wide mapping of transposon integration in mammals. Mol. Cell. Biol. 2005;25:2085–2094. doi: 10.1128/MCB.25.6.2085-2094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu X., Li Y., Crise B., Burgess S.M., Munroe D.J. Weak palindromic consensus sequences are a common feature found at the integration target sites of many retroviruses. J. Virol. 2005;79:5211–5214. doi: 10.1128/JVI.79.8.5211-5214.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liao G.C., Rehm E.J., Rubin G.M. Insertion site preferences of the P transposable element in Drosophila melanogaster. Proc. Natl Acad. Sci. USA. 2000;97:3347–3351. doi: 10.1073/pnas.050017397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vigdal T.J., Kaufman C.D., Izsvak Z., Voytas D.F., Ivics Z. Common physical properties of DNA affecting target site selection of Sleeping Beauty and other Tc1/mariner transposable elements. J. Mol. Biol. 2002;323:411–452. doi: 10.1016/s0022-2836(02)00991-9. [DOI] [PubMed] [Google Scholar]
- 22.Olson W.K., Zhurkin V.B. Modeling DNA deformations. Curr. Opin. Struct. Biol. 2000;10:286–297. doi: 10.1016/s0959-440x(00)00086-5. [DOI] [PubMed] [Google Scholar]
- 23.Liu G., Geurts A.M., Yae K., Srinivassan A.R., Fahrenkrug S.C., Largaespada D.A., Takeda J., Horie K., Olson W.K., Hackett P.B. Target-site preference for Sleeping Beauty transposons. J. Mol. Biol. 2005;346:161–173. doi: 10.1016/j.jmb.2004.09.086. [DOI] [PubMed] [Google Scholar]
- 24.Zeger S.L., Liang K.Y. Longitudinal data analysis for discrete and continuous outcomes. Biometrics. 1986;42:121–130. [PubMed] [Google Scholar]
- 25.Plasterk R.H.A., Izsvák Z., Ivics Z. Resident aliens: the Tc1/mariner superfamily of transposable elements. Trends Genet. 1999;15:326–332. doi: 10.1016/s0168-9525(99)01777-1. [DOI] [PubMed] [Google Scholar]
- 26.Olson W.K., Gorin A.A., Lu X.J., Hock L.M., Zhurkin V.B. DNA sequence-dependent deformability deduced from protein-DNA crystal complexes. Proc. Nat. Acad. Sci. USA. 1998;95:11163–11168. doi: 10.1073/pnas.95.19.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Akaike H. Prediction and entropy. In: Atkinson A.C., Fienberg S.E., editors. A Celebration of Statistics. New York, NY: Springer; 1985. pp. 1–24. [Google Scholar]
- 28.Dupuy A.J., Fritz S., Largaespada D.A. Transposition and gene disruption using a mutagenic transposon vector in the male germline of the mouse. Genesis. 2001;30:82–88. doi: 10.1002/gene.1037. [DOI] [PubMed] [Google Scholar]
- 29.Dupuy A.J., Clark K.J., Carlson C.M., Fritz S., Davidson A.E., Markley K.M., Finley K., Fletcher C.F., Ekker S.C., Hackett P.B., et al. Mammalian germ-line transgenesis by transposition. Proc. Natl Acad. Sci. USA. 2002;99:4495–4499. doi: 10.1073/pnas.062630599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horie K., Kuroiwa A., Ikawa M., Okabe M., Kondoh G., Matsuda Y., Takeda J. Efficient chromosomal transposition of a Tc1/mariner- like transposon Sleeping Beauty in mice. Proc. Nat. Acad. Sci. USA. 2001;98:9191–9196. doi: 10.1073/pnas.161071798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horie K., Yusa K., Yae K., Odajima J., Fischer S.E., Keng V.W., Hayakawa T., Mizuno S., Kondoh G., Ijiri T., et al. Characterization of Sleeping Beauty transposition and its application to genetic screening in mice. Mol. Cell. Biol. 2003;23:9189–9207. doi: 10.1128/MCB.23.24.9189-9207.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Essner J.J., McIvor R.S., Hackett P.B. Awakening of gene therapy with Sleeping Beauty transposons. Curr. Opin. Pharmacol. 2005;5 doi: 10.1016/j.coph.2005.04.015. (in press) [DOI] [PubMed] [Google Scholar]
- 33.Yi Y., Hahm S.H., Lee K.H. Retroviral gene therapy: safety issues and possible solutions. Curr. Gene Therap. 2005;5:25–35. doi: 10.2174/1566523052997514. [DOI] [PubMed] [Google Scholar]
- 34.Baum C., von Kalle C., Staal F.J., Li Z., Fehse B., Schmidt M., Weerkamp F., Karlsson S., Wagemaker G., Williams D.A. Chance or necessity? Insertional mutagenesis in gene therapy and its consequences. Mol. Therap. 2004;9:5–13. doi: 10.1016/j.ymthe.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 35.Dave U.P., Jenkins N.A., Copeland N.G. Gene therapy insertional mutagenesis insights. Science. 2004;303:33. doi: 10.1126/science.1091667. [DOI] [PubMed] [Google Scholar]
- 36.Hacein-Bey-Abina S., Von Kalle C., Schmidt M., McCormack M.P., Wulffraat N., Leboulch P., et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 37.Futreal P.A., Coin L., Marshall M., Down T., Hubbard T., Wooster R., Rahman N., Stratton M.R. A census of human cancer genes. Nature Rev. Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ryder E., Russell S. Transposable elements as tools for genomics and genetics in Drosophila. Brief Funct. Genomic Proteomic. 2003;2:57–71. doi: 10.1093/bfgp/2.1.57. [DOI] [PubMed] [Google Scholar]
- 39.Handler A.M. Use of the piggyBac transposon for germ-line transformation of insects. Insect Biochem. Mol. Biol. 2002;32:1211–1220. doi: 10.1016/s0965-1748(02)00084-x. [DOI] [PubMed] [Google Scholar]
- 40.Thibault S.T., Singer M.A., Miyazaki W.Y., Milash B., Dompe N.A., Singh C.M., Buchholz R., Demsky M., Fawcett R., Francis-Lang H.L., et al. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nature Genet. 2004;36:283–287. doi: 10.1038/ng1314. [DOI] [PubMed] [Google Scholar]
- 41.Craig N.L. Target site selection in transposition. Annu Rev. Biochem. 1997;66:437–474. doi: 10.1146/annurev.biochem.66.1.437. [DOI] [PubMed] [Google Scholar]
- 42.Pryciak P.M., Muller H.P., Varmus H.E. Simian virus 40 minichromosomes as targets for retroviral integration in vivo. Proc Natl Acad Sci U S A. 1992;89:9237–9241. doi: 10.1073/pnas.89.19.9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pryciak P.M., Sil A., Varmus H.E. Retroviral integration into minichromosomes in vitro. Embo J. 1992;11:291–303. doi: 10.1002/j.1460-2075.1992.tb05052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fitzgerald M.L., Grandgenett D.P. Retroviral integration: in vitro host site selection by avian integrase. J. Virol. 1994;68:4314–4321. doi: 10.1128/jvi.68.7.4314-4321.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bushman F., Lewinski M., Ciuffi A., Barr S., Leipzig J., Hannenhalli S., Hoffmann C. Genome-wide analysis of retroviral DNA integration. Nat. Rev. Microbiol. 2005;3:848–858. doi: 10.1038/nrmicro1263. [DOI] [PubMed] [Google Scholar]
- 46.Ciuffi A., Mitchell R.S., Hoffmann C., Leipzig J., Shinn P., Ecker J.R., Bushman F.D. Integration site selection by HIV-based vectors in dividing and growth-arrested IMR-90 lung fibroblasts. Mol. Ther. 2006;13:366–373. doi: 10.1016/j.ymthe.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 47.Eide D., Anderson P. Insertion and excision of Caenorhabditis elegans transposable element Tc1. Mol. Cell biol. 1988;8:737–746. doi: 10.1128/mcb.8.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mori I., Benian G.M., Moerman D.G., Waterston R.H. Transposable element Tc1 of Caenorhabditis elegans recognizes specific target sequences for integration. Proc. Natl Acad. Sci. USA. 1985;85:861–864. doi: 10.1073/pnas.85.3.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Luenen H.G.A.M., Plasterk R.H.A. Target site choice of the related transposable elements Tc1 and Tc3 of Caenorhabditis elegans. Nucleic Acids Res. 1994;22:262–269. doi: 10.1093/nar/22.3.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Preclin V., Martin E., Segalat L. Target sequences of Tc1, Tc3 and Tc5 transposons of Caenorhabditis elegans. Genet. Res. 2003;82:85–88. doi: 10.1017/s0016672303006359. [DOI] [PubMed] [Google Scholar]
- 51.Lampe D.J., Grant T.E., Robertson H. Factors affecting transposition of the Himar1 mariner transposon in vitro. Genetics. 2006;149:179–187. doi: 10.1093/genetics/149.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Britten R.J. Coding sequences of functioning human genes derived entirely from mobile element sequences. Proc. Natl Acad. Sci. USA. 2004;101:16825–16830. doi: 10.1073/pnas.0406985101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Charlesworth B., Sniegowski P., Stephan W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature. 1994;371:215–220. doi: 10.1038/371215a0. [DOI] [PubMed] [Google Scholar]
- 54.Shapiro J.A., von Sternberg R. Why repetitive DNA is essential to genome function. Biol. Rev. Camb. Philos. Soc. 2005;80:227–250. doi: 10.1017/s1464793104006657. [DOI] [PubMed] [Google Scholar]
- 55.Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]