Abstract
While non-specific DNA plays a role in target localization for many recombinases, transcription factors and restriction enzymes, the importance of non-specific DNA interactions for transposases has not been investigated. Here, we discuss non-specific DNA-Tn5 Transposase (Tnp) interactions and suggest how they stabilize the Tnp and modulate Tnp localization of the 19 bp Tnp recognition end sequences (ESes). DNA protection assays indicate that full-length Tnp interacts efficiently with supercoiled DNA that does not contain ESes. These interactions significantly prolong the lifetime of Tnp, in vitro. The balance between non-specific DNA bound and free Tnp is affected by DNA topology, yet, intermolecular transfer of active Tnp occurs with both supercoiled and linear non-specific DNA. Experiments with substrates of varying lengths show that Tn5 Tnp can utilize non-specific DNA to facilitate localization of an intramolecular ES over distances less than 464 bp. Finally, synaptic complex formation is inhibited in the presence of increasing concentrations of supercoiled and linear pUC19. These experiments strongly suggest that Tn5 Tnp has a robust non-specific DNA binding activity, that non-specific DNA modulates ES sequence localization within the global DNA, most likely through a direct transfer mechanism, and that non-specific DNA binding may play a role in the cis bias manifested by Tn5 transposition.
INTRODUCTION
Understanding the non-specific DNA binding characteristics of proteins involved in DNA recombination, DNA cleavage and gene regulation is essential for complete comprehension of their reaction mechanisms. The ability to bind DNA non-specifically can affect reaction rates, target recognition and the activity of proteins. While the role of non-specific DNA binding has been well documented for the Lac Repressor and some restriction enzymes (1,2), this important phenomenon has been observed, but not analyzed, in transposition systems.
A review of current literature reveals evidence of non-specific DNA binding by transposases (Tnp), site-specific recombinases and retroviral integrases. Two of the three domains of the bacteriophage Mu Tnp (MuA) independently bind DNA non-specifically. Filter binding experiments with proteolytic fragments of MuA reveal that the N-terminal 26 amino acids of domain III binds supercoiled DNA non-specifically (3) and that domain II can interact with linear non-specific DNA (4). The Tn3 Tnp binds non-specific linear restriction fragments (5,6). Finally, many Tnps, including Tn5 Tnp, can integrate their transposons into random DNA sequences (7–10), a function that depends on non-specific DNA interactions. Remarkably, the importance of these interactions to the stability of Tnp or the transposition mechanism, other than integration, has not been carefully studied.
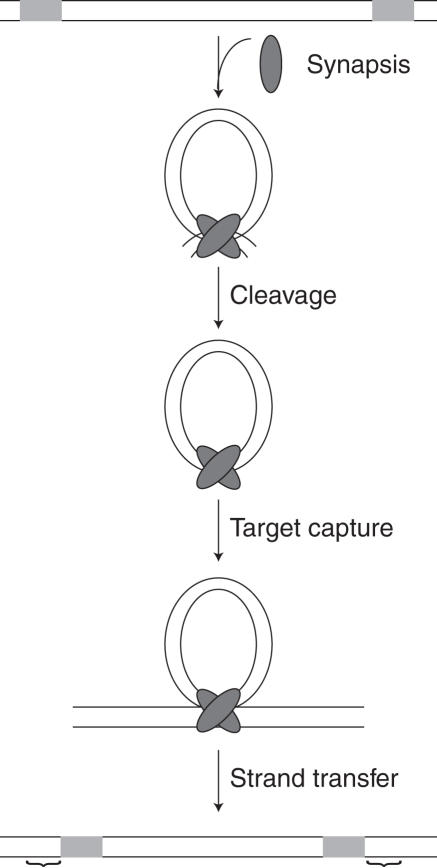
Transposition is the process of moving DNA from one location to another. In its simplest form, this process requires a Tnp, DNA flanked by Tnp recognition end sequences (ESes), target DNA and a divalent metal ion (usually Mg2+) (11). The Tn5 transposon is mobilized using a cut-and-paste mechanism in which Tnp first binds two 19 bp inverted ESes, (12–14) see Figure 1. The two Tnp bound ESes then homodimerize to form a synaptic complex, the nucleoprotein complex required for catalysis (15–17). Following synapsis, a water molecule activated by Mg2+ attacks the phosphodiester backbone of one DNA strand at the junction between the ES and flanking DNA resulting in the generation of a 3′ hydroxyl group (first strand nicking). This 3′ hydroxyl group then attacks the opposite DNA strand creating a hairpin intermediate and releasing the flanking DNA (18). The hairpin is resolved by nucleophilic attack of a second Mg2+-activated water molecule creating a blunt-ended transposon with free 3′ hydroxyl groups (11). Following cleavage, the complex captures a non-specific DNA target and inserts the transposon [for a review see, (19)].
Figure 1.
The Tn5 reaction mechanism. Tn5 Tnp first binds to 19 bp inverted repeat recognition ESes, followed by homodimerization of these bound Tnps to form a synaptic complex. The donor backbone (dbb) DNA is then cleaved from the transposon (Tn) at the dbb/Tn junction. Following cleavage, the synaptic complex can capture a non-specific DNA target and integrate the Tn resulting in a 9 bp duplication (represented by ‘{’). Tn5 Tnp is shown as a gray oval and the 19 bp ES is represented by a light gray box.
In this manuscript, we investigate the effect of Tn5 Tnp non-specific DNA binding on the initial steps of the transposition mechanism leading to ES localization. A previously described hyperactive variant of Tnp containing E54K, M56A and L372P mutations is used in these studies (11,19–22). In this communication, we define non-specific DNA as any DNA that does not contain the Tn5 Tnp ES. First, we use a restriction enzyme blockage assay (REBA) to show that full-length Tn5 Tnp can interact efficiently with many supercoiled pUC19 sites. These interactions allow Tnp to remain active up to 95 times longer than in the absence of non-specific DNA. Tnp bound to both linear and supercoiled pUC19 can transfer to an oligonucleotide having the ES. This conclusion is further supported by single molecule micromanipulations of λ-DNA designed to study intermolecular DNA transfer, which show that Tnp can transfer to competing non-tethered, non-specific DNA or to a DNA having the ES. To investigate intramolecular Tnp movement, in vitro transposition reactions were performed with DNA fragments of different lengths. The reaction rate constants determined on these substrates indicate that DNA covalently linked to the ES can modulate ES localization by Tnp possibly through a direct transfer mechanism. Finally, increasing concentrations of linear or supercoiled pUC19 inhibit synaptic complex formation. These data support a model in which non-specific DNA binding regulates active Tnp concentration in vivo.
MATERIALS AND METHODS
Tn5 Tnps used in this study
A hyperactive Tnp including mutations E54K, M56A and L372P was used for most of the experiments described in this manuscript. The E54K mutation enhances binding to the ES (20). The M56A mutation prevents translation of the inhibitor protein (Inh) and therefore eases Tnp purification. The L372P mutation increases flexibility of Tnp allowing the protein to function in trans (21). This mutant Tnp has been used in many previously published experiments to study Tn5 transposition (15–18). This hyperactive Tnp will be referred to as Tnp throughout the rest of the manuscript. A catalytically inactive mutant having these three mutations plus the D188A mutation is also used in this study (23). The Inh, a protein translated in the same reading frame as Tnp but missing the N-terminal DNA binding domain, was used in non-specific DNA binding experiments. Finally a C-terminally truncated Tnp, Δ369Tnp is missing the last 107 amino acids and includes the E54K and M56A mutations described above (24).
Tn5 Tnp purification
Expression vector pGRTYB35 (18) encoding Tnp (20,25) was transformed into Escherichia coli strain ER2566 (NEB) and plated on TYE (tryptone, yeast extract, NaCl) containing Amp (100 µg/ml) and 0.1% glucose. Following overnight incubation at 37°C, 4 ml of Luria–Bertani media (LB media) (with Amp and glucose) were inoculated with a single colony and the culture was grown at 37°C to OD600 = 0.6. About 2 ml were then pelleted, the media decanted and the cells resuspended in 2 ml of fresh LB. About 100 ml of LB (with Amp and glucose) were inoculated with these 2 ml of culture. Following growth at 37°C to OD600 = 0.6, this culture was placed at 4°C overnight. For the final growth, 20 ml of 4°C culture were pelleted, resuspended as before and used to inoculate 1 l of LB with Amp. The culture was grown to OD600 = 0.25 at 37°C and then transferred to 22°C for growth to OD600 = 0.6. Protein expression was initiated by addition of Isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 1 mM. Induction continued for 2 h before the cells were pelleted in a centrifuge, washed with phosphate-buffered saline (PBS) and placed at −20°C overnight.
For cell lysis, pellets were resuspended in 25 ml HEG0.7 buffer [20 mM HEPES (pH 7.5), 1 mM EDTA, 10% glycerol, 0.7 M NaCl] and sonicated at 4°C. Lysed cells were then spun at 10 000 g (Eppendorf F34-6-38) for 45 min at 4°C and the cell lysate was transferred to chitin beads (NEB) pre-equilibrated with HEG0.7 buffer. The lysate-chitin bead mixture shook gently at 4°C for 1 h. Following this incubation, the mixture was transferred to an empty 6 ml polypropylene column (Qiagen) and washed with 25 ml of HEG0.7 buffer. To cleave the protein from the chitin binding domain fusion, 50 mM DTT (Sigma) was included in the final 2 ml of HEG0.7 wash buffer and the column remained undisturbed at 4°C for at least 18 h. Purified Tnp was eluted with 1.25 ml HEG0.7 buffer. Purity was assessed using SDS–PAGE/Coomassie staining and each preparation was quantitated using a standard Bradford assay. All proteins described in this manuscript were purified using this method.
Preparation of linear and supercoiled pUC19
pUC19 was purified from DH5α using a maxiprep kit (Qiagen) and linearized with EcoRI (Promega). The linearized pUC19 was gel purified from uncut plasmid. Both supercoiled and linear pUC19 were extracted twice with phenol–chloroform and ethanol precipitated before use in any non-specific DNA binding experiments.
Single molecule micromanipulation of λ-DNA
All single molecule micromanipulation experiments were performed using the microscope and magnetic tweezers described in (26). Flow cell, DNA tethering, force measurement and calibration were also performed essentially as described in (26).
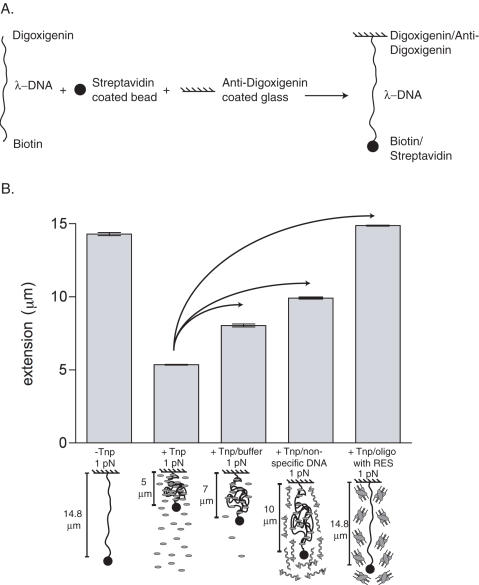
To qualitatively assess stability of a Tnp–λ-DNA complex, a single, 48.5 kb λ-DNA tether was first isolated in the flow cell. A total of 40 nM Tnp was then mixed with DNA binding buffer [20 mM HEPES (pH 7.5), 100 mM NaCl, 1 mM EDTA] in a total volume of 200 µl and immediately injected into the 40 µl flow cell. The λ-DNA tether was held at 1 pN during the injection, but the force was reduced to 0.04 pN (minimum force) following the addition of Tnp. Following Tnp binding to the λ-DNA, competitors were introduced to this system. These included DNA binding buffer, DNA binding buffer plus 1 mg/ml Herring Sperm DNA (Promega) and DNA binding buffer plus 1 µM 60 bp double stranded oligonucleotide having the Tnp ES [sequence described in (15), Integrated DNA Technologies]. Each competitor was added to separate λ-DNA tethers and incubated with the Tnp–λ-DNA complex for 1 h. Following incubation, at least five extension measurements of the λ-DNA were made at 1 pN. These extension measurements were then plotted to visualize the effect of the competitors on the Tnp–λ-DNA complex equilibrium. Each reaction was performed at least three times.
REBA
200 nM to 900 nM Tnp, 200 nM to 900 nM TnpΔ369 or 400 nM to 1.5 µM Inh were incubated with 10 nM super-coiled pUC19 (27 µM nucleotides) in transposition buffer (20 mM HEPES, pH 7.5 and 100 mM potassium glutamate) containing magnesium acetate (MgAc) for 1 h at 37°C. Because MgAc was included, a catalytically incompetent Tnp (DA188 Tnp) was used instead of Tnp in these experiments (27). Following a 1 hour incubation, 20 U of HhaI (NEB), 20 units of Fnu4HI (NEB) and 16 U of Sau3AI (NEB) were added to each reaction and incubation at 37°C continued for an additional three hours. Each reaction was then phenol/chloroform extracted twice and concentrated by ethanol precipitation with glycogen. To analyze the cleavage patterns, the reactions were run on a 9% native polyacrylamide gel followed by brief ethidium bromide staining and visualization on a Typhoon Variable Mode Imager. Traces of each lane were then overlayed to assess the amount of Tnp binding. This experiment was performed at least three times for each Tnp.
Lifetime gel shift assays
To determine the half-life of Tnp, 190 nM Tnp (final concentration) was mixed with transposition buffer and placed immediately at 37°C. At time points 0 through 30 min, aliquots were added to 23 nM fluorescently labeled, 60 bp substrate oligonucleotide. Each time point was then incubated at 37°C for an additional 1.5 hours to allow PEC formation. Each reaction was terminated by adding loading dye and freezing immediately at −20°C. The PECs were separated from unbound DNA by native gel electrophoresis and each gel was visualized on a Typhoon Variable Mode Imager. The percent PEC formation at each time point is defined as the amount of DNA in a PEC divided by the total amount of DNA per lane. These percents were quantitated using Image Quant Total Lab Software, plotted versus time and fit to a one-phase exponential decay equation:
| 1 |
where a = the amplitude, b = the plateau and k equals the observed decrease in complex formation rate constant, kobs,complexformation. The half-life of Tnp equals 0.69/kobs,complexformation.
To determine the half-life of Tnp in the presence of non-specific DNA, 10.4 nM linear or super-coiled pUC19 DNA were included in the initial reaction step. Also, longer time points were taken and the incubation at 37°C following addition of substrate oligonucleotide was extended. Each reaction was performed at least twice.
Single-end substrate in vitro reactions
Substrates for single-end substrate in vitro reactions were constructed as follows. First, the EZ::Tn <KAN-2> transposon (Epicentre) was PCR amplified to add EcoRI restriction sites at each end. Following digestion with EcoRI, EZ::Tn<KAN-2> was cloned into EcoRI digested pUC19 to create pWSR6092. One Tnp recognition end sequence (ES) was then removed from this plasmid by digesting pWSR6092 with XhoI (Promega) and XmaI (NEB) and filling in the sticky ends of the 4096 bp fragment using Klenow fragment (Promega) and dNTPs (Pharmacia). Self-legation of this blunt-ended fragment resulted in pWSR6103. To create linear fragments of differing sizes containing a single Tnp recognition ES, pWSR6103 was digested with PflMI and either XmnI, AatII, NdeI, NarI, BglI or PvuII (all NEB). The restriction enzymes were removed from the reaction using a PCR cleanup kit (Qiagen) and the DNA was quantitated both spectroscopically and by agarose gel electrophoresis.
To determine the cleavage rate constant on single-end substrates, 12.5 nM each DNA substrate was mixed with transposition buffer, 10 mM MgAc, 100 nM Tnp and placed immediately at 37°C. Time points were taken from 0 to 8 h by adding aliquots of the reaction to 1% SDS. Following the last time point, loading dye was added and the time points were placed at 4°C overnight. The reactions containing XmnI, AatII and NdeI single-end substrate fragments were separated using standard 1.5% agarose gels. The reactions containing NarI and BglI single-end substrate fragments were separated using 4% Low-viscosity/low melting NuSieve 3:1 agarose gels (Biowhittaker Molecular Applications) and the PvuII single-end substrate fragments were separated using 6% low-viscosity/low melting NuSieve 3:1 agarose gels. Each gel was stained for 5 minutes with ethidium bromide, destained for 10 min in H2O and then visualized using the Typhoon Variable Mode Imager. The intensity of the substrate and product bands were quantitated using Image Quant Total Lab software. These intensities were normalized to account for size differences and the ratio of normalized product to normalized substrate plus normalized product was calculated. This ratio represents the percent substrate molecules cleaved at each time point. This percent was then plotted versus time and fit to an exponential association equation:
| 2 |
where b = plateau and k = the observed cleavage rate constant, kobs,cleavage. This experiment and analysis were repeated at least three times for each single end substrate. The rate constants corresponding to each experiment were then averaged and plotted versus substrate length to visualize the affect of substrate length on transposition activity.
Tnp inhibition gel shift assays
To examine Tnp inhibition by supercoiled and linear pUC19 DNA, 190 nM Tnp was mixed with 24 nM fluorescently labeled, 60 bp substrate oligonucleotide and varying concentrations of supercoiled or linear pUC19 (0–64 nM) in transposition buffer and incubated for 24 h at 370 C. PECs were then separated from unbound substrate oligonucleotide by native gel electrophoresis and each gel was visualized using a Typhoon Variable Mode Imager. The quantitated number of PECs (in fluorescence units (FUs)) formed at each concentration of pUC19 were then plotted versus pUC19 concentration.
RESULTS
Tn5 Tnp interacts with supercoiled pUC19
To determine if Tnp could bind supercoiled pUC19, an assay, termed REBA, was developed. In this experiment, increasing concentrations of a catalytically incompetent Tnp were incubated with supercoiled pUC19 plasmid in transposition buffer containing magnesium acetate (MgAc). During this time, Tnp interacted with and bound to the supercoiled pUC19 plasmid. This binding reaction was long enough to ensure that all unbound Tnp was inactive for specific DNA binding activity (see following sections). After the Tnp binding step, a mixture of frequent cutting restriction enzymes HhaI, Sau3AI and Fnu4HI was added to the reaction. These restriction enzymes then cut the supercoiled pUC19 plasmid at their recognition sites. Following further incubation, all proteins were denatured, removed from the DNA and the cleavage fragments were separated on a polyacrylamide gel. If Tnp binds pUC19 non-specifically, some portion of the restriction enzyme sites should be blocked. This can be observed as a change in banding pattern as Tnp concentration increases. It should be noted that even at the highest Tnp concentration (900 nM), blockage of all restriction sites should not be observed because non-specific pUC19 binding sites are not saturated at this Tnp concentration.
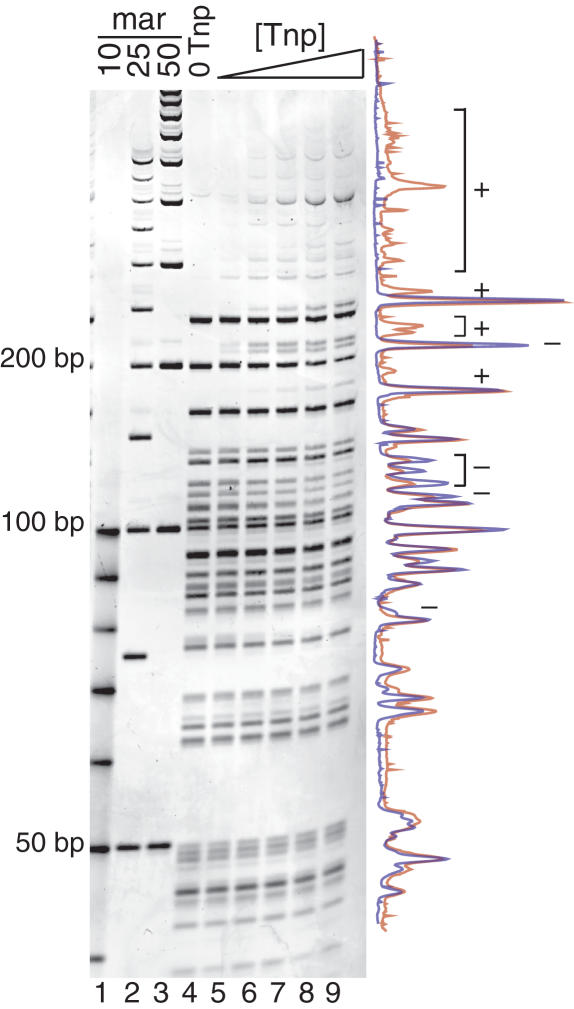
A representative full-length Tnp REBA is seen in Figure 2. When comparing lanes of increasing Tnp concentration (lanes 4 through 9), the intensity of some bands decreases while larger bands appear. This is best seen in overlapping traces of lanes 4 and 9 shown to the right of the gel. DNA fragments caused by blockage of a restriction site are marked with a plus sign (+), while those that decrease in intensity with increased Tnp concentration are marked with a minus sign (−). These data show that more restriction sites are impeded as Tnp concentration increases and indicate that Tnp can interact with supercoiled pUC19 DNA.
Figure 2.
Tn5 Tnp interacts with supercoiled pUC19. REBA of Tnp bound to supercoiled pUC19 is shown in lanes 4–9 and DNA size markers are shown in lanes 1–3. Increasing concentrations of Tnp (lanes 5–9) were incubated with pUC19 followed by addition of frequent cutting restriction enzymes, HhaI, Fnu4HI and Sau3AI. Blockage of restriction sites by Tnp results in the appearance of larger bands and decrease in some smaller bands as Tnp concentration is increased. This is most evident when examining the overlay of the traces from a no Tnp control reaction (blue, lane 4) and 900 µM Tnp lanes (red, lane 9), seen to the right of the gel. Bands increasing in intensity are marked with a plus sign (+). Bands decreasing in intensity are marked with a minus sign (−).
Although the REBA assay clearly shows that Tnp impairs restriction digestion at some sites, not all sites appear equally sensitive. This observation is most easily explained by assuming that non-specific DNA-Tnp binding has some sequence bias. However, analysis of blocked restriction sites did not reveal any sequence similarity or homology beyond the sequence of the restriction site itself. Single molecule experiments with λ-DNA show that Tnp can also interact with linear DNA (see Figure 5B).
Efficient binding to supercoiled pUC19 DNA requires full-length Tnp
To analyze the Tnp structure-function relationship as it applies to pUC19 DNA binding, REBA was performed using two Tnp deletion mutants, TnpΔ369 (Tnp minus 110 C-terminal amino acids) and the Inh (Tnp minus 55 N-terminal amino acids). TnpΔ369 lacks a C-terminal dimerization domain required for synapsis, but retains an N-terminal Tnp recognition ES binding domain (21,24). The Inh can form dimers, but is lacking the N-terminal ES binding domain (28).
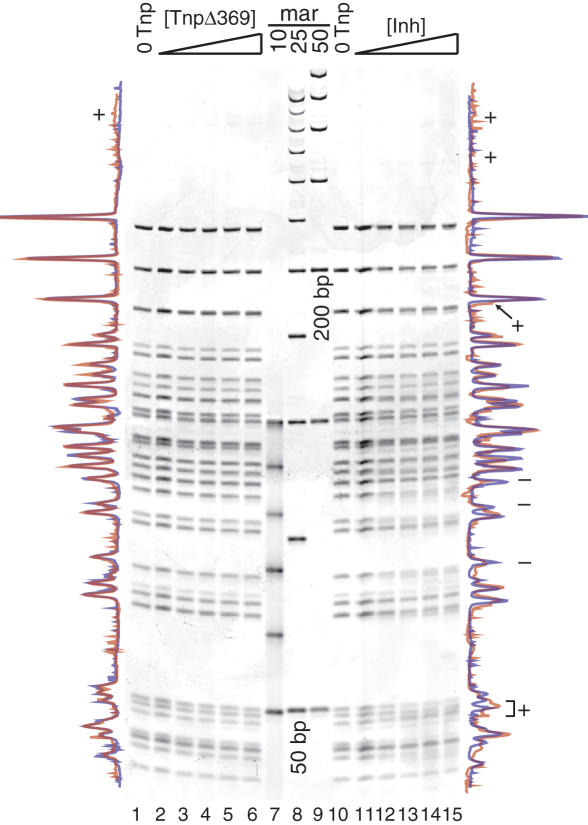
REBA with TnpΔ369 (see Figure 3, lanes 1–6) was performed exactly as for the full-length protein to enable realistic comparison between the two Tnps. Overlapping traces of lanes 1 and 6 are shown to the left of the gel. This Tnp variant does not seem to significantly block any restriction sites and only one faint, higher molecular weight band (marked with +) appears under these reaction conditions.
Figure 3.
Efficient Tnp binding to supercoiled pUC19 DNA requires full-length Tnp. REBA (see Figure 2) was performed for two Tnp deletion mutants, TnpΔ369 and Inh. Lanes 2–6 show increasing concentrations of TnpΔ369, lanes 7–9 show DNA size markers and lanes 11–15 show increasing concentrations of Inh. Traces for the no protein controls (lanes 1 and 10) and highest protein concentration (lanes 6 and 15) are shown to the left of the gel for TnpΔ369 and to the right for the Inh protein. Colors and symbols are as in Figure 2.
REBA was also performed with Inh (see Figure 3, lanes 10–15), but both a wider protein concentration range and a higher maximum protein concentration were used in this experiment. Examination of the overlapping traces of lanes 10 and 15 reveals a small amount of restriction site blockage by Inh. Bands with greater intensity at higher concentrations of the Inh are marked with a plus sign (+) while bands decreasing in intensity as Inh concentrations increase are labeled with a minus sign (−).
Comparison of REBA for these two proteins to REBA of full-length Tnp (Figure 2) shows that removal of either the N-terminal (Inh) or C-terminal (TnpΔ369) Tnp domains causes a reduction in pUC19 DNA binding. These data imply that the full-length Tnp is required for efficient binding to supercoiled pUC19 DNA.
Binding to non-specific DNA increases the half-life of Tnp
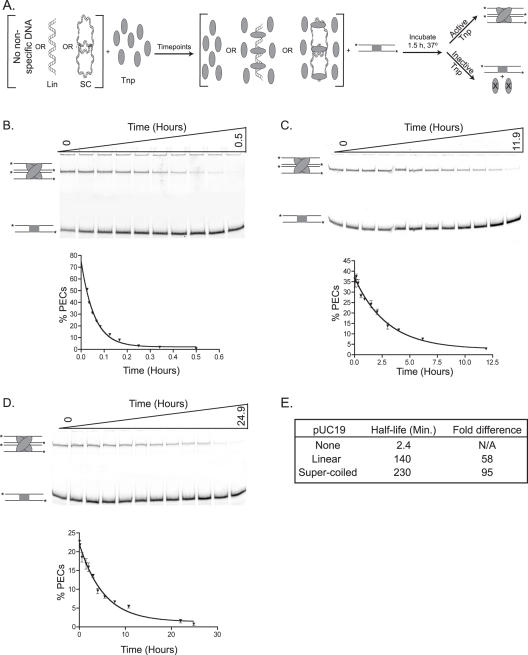
To begin defining characteristics of Tnp pUC19 DNA binding, the lifetime of active Tnp in the presence of linear, supercoiled or no pUC19 DNA was determined. Tnp was mixed with transposition buffer having no pUC19 DNA (control), linear or supercoiled pUC19. At time points, aliquots of each reaction were removed and added to fluorescently labeled, double stranded oligonucleotide having the transposon ES. Each time point was then incubated for an additional 1.5 h. During this time, active protein could interact with the ES and form a paired ends complex (PEC). A PEC containing two ES containing oligonucleotides is analogous to a synaptic complex on a transposon with two inverted repeat ESes. See Figure 4A for a schematic of this reaction. Following conclusion of the assay, each time point was run on a native polyacrylamide gel where the PECs are shifted from unbound DNA. If the lifetime of Tnp is extended by interaction with pUC19 DNA, then the ability of Tnp to form PECs should be maintained for a longer period of time under these conditions as compared to the control reaction lacking non-specific DNA.
Figure 4.
Non-specific DNA binding extends the lifetime of Tnp. (A) A schematic of reactions to determine Tnp half-life in the presence of non-specific DNA is shown. Tnp was incubated in the absence of non-specific DNA (control) or in the presence of linear or supercoiled pUC19. Aliquots were removed at various times and a ES containing oligonucleotide was added to each time point. The aliquots were then incubated for an additional 90 min during which active Tnp could form a PEC. Tnp is represented as a gray oval, linear and supercoiled non-specific DNA are labeled. The ES containing oligonucleotide is shown as two parallel lines containing a gray box, the *'s represent the fluorescent label at each 5′ end. Inactive Tnp is marked with an ‘X.’ The PEC contains two ES containing oligonucleotides and two molecules of Tnp. (B) The lifetime of Tnp in the absence of non-specific DNA (control) was assessed. PECs were separated from unbound oligonucleotide using polyacrylamide gel electrophoresis. The PECs and unbound DNA are labeled as in (A). This experiment was performed twice and the mean percentage of DNA in PECs was determined for each time point. These mean percentages were plotted versus time and the data were fit to a one-phase exponential equation. The error bars associated with each point show the standard error. (C) The lifetime of Tnp in the presence of linear pUC19 was assessed. This experiment was performed as in (B) except the time course was extended to 11.9 h. (D) The lifetime of Tnp in the presence of supercoiled pUC19 was assessed. This experiment was performed as in (B) except the time course was extended to 24.9 h. (E) The lifetime of Tnp is quantitatively expressed as the half-life, or the time at which Tnp activity is half maximal. The half-life of Tnp under each of the previous conditions was determined from the exponential fit of each dataset (see Materials and Methods). This table shows the half-life of Tnp under each reaction condition and the fold extension in half-life due to the presence of non-specific DNA.
Figure 4 shows the results of these experiments. Figure 4B is the control and the reactions including linear and supercoiled pUC19 are shown in Figure 4C and D, respectively. The lifetime of Tnp under each condition is quantitatively expressed as the half-life. The plots used to determine the Tnp half-life are shown with their corresponding experiments in Figure 4 and the calculated half-life of Tnp under each reaction condition is shown in Figure 4E.
The half-life of Tnp in buffer without DNA is only 2.4 min. Adding linear pUC19 extends the half-life of Tnp to 140 minutes, while inclusion of supercoiled pUC19 increases the half-life to 230 min. These dramatic extensions of Tnp lifetime show that non-specific DNA binding is important for Tnp activity and exemplifies differences in Tnp binding affinity for linear and supercoiled non-specific DNA.
Opening of Tnp-condensed DNA is strongly enhanced by DNA fragments in solution
To confirm that Tnp can interact with other linear non-specific DNAs and to further investigate transfer between non-specific DNA and competitor DNAs, the stability of a Tnp λ-DNA complex was assessed using single DNA molecule micromanipulations. These experiments allow Tnp–DNA interactions to be directly observed on single non-specific DNA molecules (C. D. Adams, manuscript in preparation). First, λ-DNA molecules (total length 48.5 kb or 16.4 µm) having a 2.8 µm-diameter paramagnetic particle at one end were attached to a glass surface in a flow cell using the opposite end (see Figure 5A for substrate preparation details). Single λ-DNA molecules could then be manipulated with ‘magnetic tweezers’ (a localized magnetic field gradient generated using small permanent magnet poles). First, a single λ-DNA molecule was located and its extension was measured at a few different forces. Note that at 0.04 pN the extension of naked λ-DNA is about 6 µm, while at 1 pN, its extension is about 14.5 µm (see Figure 5B, first bar). Following this, force was reduced to 0.04 pN and Tnp was introduced into the flow cell to form the Tnp-λ-DNA complex. DNA condensation was monitored as a drop in extension below the 6 µm extension for bare DNA, which occurs over a few-minute period. We note that no condensation was ever observed for DNA held by a 1 pN force (which stretches out naked DNA), indicating a necessity for the random-coil fluctuations and DNA self-collision that occurs at low forces. Following condensation at 0.04 pN, force was increased to 1.0 pN and λ-DNA extension was observed to be only 5 µm (Figure 5B, second bar). 1.0 pN was used because this force is high enough to observe the Tnp's mode of non-specific DNA binding, but not high enough to pull the Tnp-λ-DNA complex apart. In separate experiments, the Tnp-λ-DNA complex was then incubated with either DNA binding buffer, herring sperm DNA or doubled stranded oligonucleotide having the transposon ES. Following a 1 h incubation, while force was held at 1 pN, the extension of the λ-DNA was measured and the resulting extensions were plotted versus the reaction condition to compare the effect of these competitors on the Tnp-λ-DNA complex (Figure 5B, third, fourth and fifth bars). Washing the Tnp-λ-DNA complex with DNA binding buffer caused the λ-DNA extension to increase by only 2 µm from 5 µm to 7 µm (see Figure 5B, compare bar 2 to bar 3); note that this is still far shorter than the naked DNA length for this force of 14.5 µm, indicating that much of the Tnp bound to DNA is immobile. When a new Tnp-λ-DNA complex was washed with DNA binding buffer containing non-specific DNA, the λ-DNA extension increased ∼5 µm from 5 µm to 10 µm (see Figure 5B, compare bar 2 to bar 4). Finally, when a third separate Tnp-λ-DNA complex was washed with DNA binding buffer containing an oligonucleotide having the transposon ES, the λ-DNA extension increased from 5 µm to its fully extended form at ∼14.5 µm (see Figure 5B, compare bar 2 to bar 5).
Figure 5.
Tnp can dissociate from λ-DNA in the absence of the Tnp ES. (A) To prepare DNA for single molecule micromanipulation experiments, digoxigenin-labeled oligonucleotide was ligated to the λ-DNA molecules using the cosR site and biotin was added using the cosL site. A magnetic strepavidin coated bead was attached to the λ-DNA via interaction with the biotin and the λ-DNA molecules were then coupled to an anti-digoxigenin coated glass slide to create the final substrate. (B) To investigate the stability of a linear non-specific DNA–Tnp complex, single molecule experiments were performed. Reaction components are defined as in Figure 4A. For each experiment, a single λ-DNA molecule was isolated and allowed to fully condense with Tnp at 0.04 pN. The force was increased to 1.0 pN and then three individual experiments were performed (represented by arrows). First, the affect of non-specific DNA binding buffer alone was investigated. Next, the affect of non-specific DNA on the Tnp–λ-DNA complex was assessed. Finally, the Tnp–λ-DNA complex was challenged with a double stranded oligonucleotide having the transposon ES. At least five extension measurements were made following the 1 h incubation at 1 pN. The average λ-DNA extensions are shown as gray bars above schematics of Tnp–λ-DNA binding behavior under each condition. The error bars represent one standard deviation from the mean.
These data show that addition of DNA binding buffer alone causes some lengthening of the λ-DNA, in the absence of any transposon ES site. This indicates that some of the DNA condensation by Tnp has a finite lifetime, decaying either by dissociation of some Tnp from the λ-DNA, or possibly via release of either protein-protein or DNA-protein interactions without loss of Tnp from DNA. On the other hand, much of the Tnp-bound DNA is stable even in protein-free buffer. The amount and rate of opening of the Tnp–DNA complex are strongly enhanced by non-specific DNA fragments in solution, and even more strongly by ES–containing DNA fragments in solution. This enhancement by DNA fragments in solution indicates that Tnp can transfer, intermolecularly, from non-specific DNA to both a different non-specific DNA molecule and a DNA molecule containing the ES, with a preference for the DNA molecule containing the ES. Intermolecular transfer to the ES is greatly favored as virtually all Tnp dissociates from the λ-DNA under these conditions.
Single ES substrates of differing lengths have variable observed cleavage rate constants
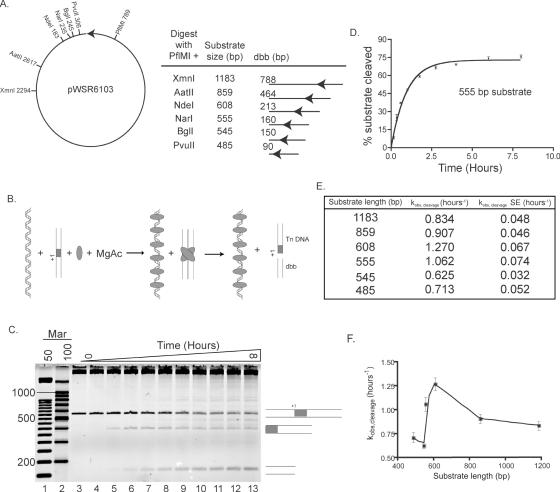
To investigate the intramolecular mechanism of transposon ES localization, DNA cleavage reactions were performed. If covalently linked non-specific DNA facilitates ES localization, then the cleavage rate constant should increase as substrate length increases. Linear DNA Tnp substrates each having one ES, 395 bp of transposon (Tn) and 90 bp to 788 bp of donor backbone (dbb) DNA (see Figure 6A), were incubated with Tnp in transposition buffer plus MgAc. Each reaction contained the same total amount of non-specific DNA (see Figure 6B). Time points were taken and reaction products were separated on agarose gels (see Figure 6C for a representative gel). To determine the observed cleavage rate constant (kobs, cleavage), the percentage of total cleavage events was calculated for each time point. These percentages were plotted versus time and the results were fit to an exponential curve to determine kobs,cleavage for each substrate (see Figure 6D for an example). kobs,cleavage was then plotted versus substrate length to better assess the affect of DNA length on kobs,cleavage (see Figure 6E and F).
Figure 6.
Single ES substrates of differing lengths are cleaved with variable rate constants. (A) A partial restriction map of the plasmid (pWSR6103) used to create substrates for in vitro transposition reactions is shown. The Tnp ES is represented as a black arrow. This plasmid was digested with PflMI and either PvuII, BglI, NarI, NdeI, AatII or XmnI to create substrates varying in size from 485 to 1183 bp. Each restriction fragment contained 395 bp of transposon (Tn) DNA and varying lengths of donor backbone (dbb) DNA as shown. The location of the transposon ES in each substrate is marked with a black arrow. (B) A schematic of the in vitro transposition reactions with single-ended substrates is shown. Each substrate DNA was incubated (together with non-specific DNA remaining from the restriction digest) with Tnp and MgAc at 37°C. Time points were taken from 0 to 8 h. Following PEC formation, the substrate was cleaved into two products, the dbb and Tn DNA. In this figure, the single ended substrate DNA is shown as two parallel lines containing a transposon ES (gray box). The cleavage site is marked with +1. The non-specific DNA remaining from the restriction digest is shown as linear double stranded DNA. Both product DNAs are appropriately labeled and other reaction components are described as in Figure 4. (C) Each time point was run on an appropriate agarose gel to separate the full-length, unreacted substrate from the dbb and Tn DNA products. In this representative gel of the 555 bp substrate, time points are shown in lanes 3–13 and DNA size markers are shown in lanes 1 and 2. The substrate, dbb and Tn DNAs are represented as in (B). (D) The percentage of substrates cleaved was determined for each time point as described in the Materials and Methods. The mean percentage cleaved at each time point was calculated from at least three independent experiments and was then plotted (together with error bars representing the standard error) versus time and the data were fit to a one-phase exponential equation. The plot shown here represents data for the 555 bp substrate. In vitro transposition reactions and analysis were performed in this fashion for each of the six single end substrates. (E) kobs,cleavage and the standard error (SE) of this value were calculated from the fits described in (D). These are shown for each of the six substrates tested. (F) To better visualize the effect of substrate length on kobs,cleavage, kobs,cleavage was plotted versus substrate length for each substrate. The error bars represent the standard error of kobs,cleavage for each substrate.
As the dbb length increases from 90 bp to 213 bp, kobs,cleavage increases from 0.713 h−1 to 1.270 hours−1 with a standard error (SE) of less than 10% for each group of reactions. The cleavage rate constant, kobs,cleavage, is largest for the substrate having 213 bp of dbb DNA. For the two substrates with dbb DNA longer than 213 bp (464 bp and 788 bp), kobs,cleavage decreases to 0.907 h−1 and 0.834 hours−1, respectively. The significance of the DNA length dependency on the cleavage rate constant will be explored in the discussion.
Increasing concentrations of pUC19 DNA inhibit PEC formation
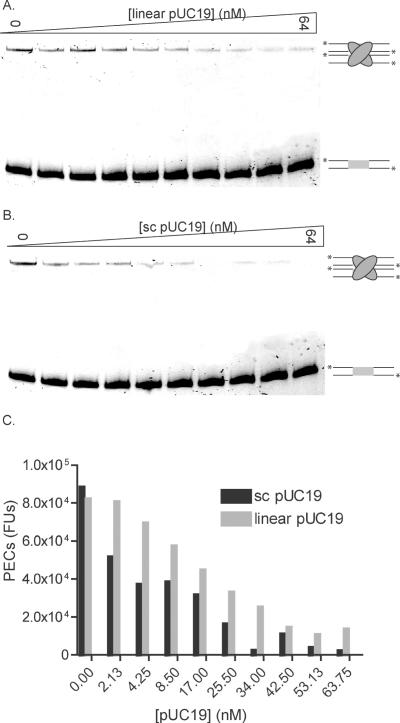
To determine if inactivation of Tnp could occur from interaction with linear or supercoiled non-specific DNA, Tnp was incubated with both increasing concentrations of competing linear or supercoiled pUC19 and oligonucleotides containing the ES. Each reaction was incubated for 24 h to ensure maximal PEC formation followed by native gel electrophoresis. Figure 7A shows the affect of linear pUC19 while Figure 7B shows the affect of supercoiled pUC19. The number of PECs formed was determined and plotted versus pUC19 concentration to assess the affect of including non-specific DNA in a PEC reaction. See Figure 7C.
Figure 7.
Increasing concentrations of pUC19 DNA inhibit PEC formation. (A) To determine if linear pUC19 DNA could inhibit PEC formation, Tnp was incubated with 0 to 64 nM linear pUC19 and fluorescently labeled oligonucleotide containing the ES in transposition buffer for 24 h. Reactions were then separated on a native polyacrylamide gel where PECs are shifted relative to unbound substrate oligonucleotide. Both PECs and unbound substrate are labeled as in Figure 4A. (B) To determine if supercoiled pUC19 DNA could inhibit PEC formation, Tnp was incubated with 0 to 64 nM supercoiled pUC19 as in (A). Both PECs and unbound substrate are labeled as in Figure 4A. (C) The number of PECs formed at each concentration of linear and supercoiled pUC19 [in Fluorescence Units (FUs)] were quantitated and plotted versus DNA concentration. Gray bars represent PECs formed in the presence of linear pUC19 while black bars represent PECs formed in the presence of supercoiled pUC19.
Increasing concentrations of both linear and supercoiled pUC19 cause inhibition of PEC formation with the supercoiled pUC19 causing greater inhibition. These results imply that the role of Tnp non-specific DNA binding includes regulation of the total amount of active Tnp available for transposition.
DISCUSSION
Full-length Tnp interacts with non-ES containing DNA
Experiments described here clearly demonstrate that Tn5 Tnp can interact with supercoiled non-specific DNA. Specifically, the REBA assay (Figure 2) shows blockage of supercoiled pUC19 restriction sites by Tnp, causing changes in the restriction digest pattern as the Tnp concentration is increased. Single molecule micromanipulations show not only non-specific DNA binding but also condensation of linear λ-DNA upon addition of Tnp (Figure 5B). Several factors indicate that the primary mode of DNA binding and condensation upon Tnp binding is via Tnp-mediated looping of the DNA (C. D. Adams, manuscript in preparation). This mode of non-specific DNA binding is a subject of current investigation. In short, both the low force of 0.04 pN required for this condensation, and the large amount of shortening of the apparent DNA length (via the extension observed at 1 pN force, of 14.5 µm before Tnp incubation, to only 5 µm after Tnp incubation) both are indicative of the DNA–Tnp complex containing Tnp-synapsed DNA loops.
Interestingly, Tnp deletion mutants lacking either the N-terminal 55 amino acids (Inh) or the C-terminal 106 amino acids (TnpΔ369) cannot bind non-specific DNA efficiently under these reaction conditions (see Figure 3). These results are surprising because both mutants retain a region of basic amino acids hypothesized to be important for capture of a non-specific target DNA by Tnps in PECs (22). Since Tn5 Tnp can strand transfer into any 9 bp target DNA sequence, we hypothesized that this domain might also be required for pre-synapsis non-specific DNA binding. Also, because TnpΔ369 has an N-terminal domain required for Tnp interaction with the recognition ES, we thought this protein might be able to interact with non-specific DNA. Because neither the target capture nor the ES DNA binding domains are sufficient for the non-specific DNA binding observed with the REBA assay, we conclude that domains present in the full-length Tnp, but not in these deletion mutants, are required for efficient non-specific DNA binding.
The effect of DNA topology on non-specific DNA binding
The half-life of active Tnp in the absence of DNA is 2.4 min. Adding supercoiled pUC19 increases the half-life 95-fold while adding linear pUC19 increases the half-life 58-fold (see Figure 4E). These data show that the increased Tnp lifetime must be due to non-specific DNA binding. While it is unclear exactly what causes Tnp to lose activity, moderate salt concentration (less than ∼300 mM monovalent salt ions) in the absence of DNA is clearly detrimental to the protein.
The effect of DNA topology on Tnp non-specific binding can be inferred from both Figures 4 and 7. Figure 4 shows that the total number of PECs formed in the presence of supercoiled pUC19 (24%) is less than with linear pUC19 (35%, compare the zero time points from Figure 4D and C).Also, the lifetime of Tnp is longer when incubated with supercoiled pUC19 (95-fold) than when Tnp is incubated with linear pUC19 (58-fold, see Figure 4E). While both linear and supercoiled pUC19 inhibit PEC formation (see Figure 7), the linear pUC19 has a less inhibitory effect (compare Figure 7A to B, see Figure 7C). These experiments indicate that Tn5 Tnp can interact with both linear and supercoiled non-specific DNA, but the protein binds more tightly to supercoiled DNA.
Intermolecular transfer mechanism
The previously discussed half-life data show that Tnp can transfer between DNA molecules. To investigate the balance between Tnp bound to λ-DNA and free Tnp, single molecule micromanipulations were performed (see Figure 5). If a λ-DNA molecule fully condensed with Tnp is incubated in DNA binding buffer for 1 h while subjected to a 1 pN force, the λ-DNA extension increases from 5 to 7 µm, indicating that much of the Tnp–DNA complex is stable in protein-free buffer. In contrast, if a λ-DNA molecule fully condensed with Tnp is incubated with herring sperm DNA, the λ-DNA extension increases from 5 to 10 µm. If a λ-DNA molecule fully condensed with Tnp is incubated with an oligonucleotide having the Tnp ES, λ-DNA extension increases from 5 to 14.5 µm (the bare DNA extension for that force). Thus, presence of non-specific DNA in solution greatly enhances opening of Tnp-condensed DNA; ES-containing DNA fragments have an even stronger complex-destabilizing effect.
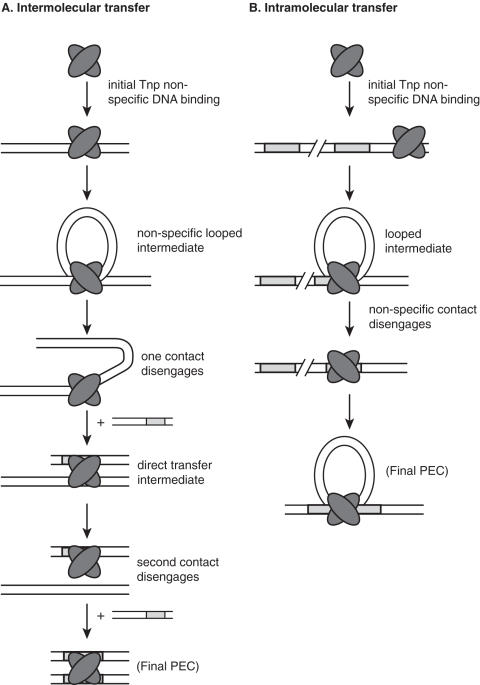
These data can be explained with the following intermolecular transfer model. While it is possible that some Tnp may spontaneously dissociate after non-specifically binding DNA (e.g. possibly explaining the partial opening observed for a single λ-DNA during a long buffer wash), the stability of most of the Tnp–DNA complex indicates that most of the Tnp bound to DNA is essentially immobile. However, we observed DNA fragments in solution to strongly stimulate the opening of the complex. This suggests that Tnp can move from one non-specific DNA site to another via a direct (or intersegment) transfer mechanism. Direct transfer is a type of DNA target localization mechanism in which a protein finds a specific DNA target while simultaneously bound to non-specific DNA (29). This type of transfer mechanism can often be excluded because most DNA binding proteins cannot interact with two DNA molecules concurrently. However, because Tn5 Tnp condensation of non-specific DNA is blocked by a point mutation in the Tnp dimerization domain (C. D. Adams, manuscript in preparation), and thus likely involves a Tnp dimer, Tn5 Tnp will have the capacity to undergo this type of DNA transfer mechanism. If the Tnp–λ-DNA complex is challenged with a preferred DNA sequence (such as the transposon ES), then we hypothesize that the Tnp can also move to this DNA by direct transfer. But, in this case, Tnp binds the ES sequence far more tightly and does not dissociate. Therefore, all Tnp is transferred from the λ-DNA to an oligonucleotide with the Tnp ES and the λ-DNA is fully extended. This model is presented schematically in Figure 8A.
Figure 8.
Models of intermolecular and intramolecular ES localization by Tn5 Tnp. (A) A model for intermolecular ES localization is shown. We propose that Tnp first binds a non-specific DNA molecule followed by simultaneous interaction with another non-specific DNA site to form a looped intermediate. Disengagement of one Tnp-non-specific contact allows simultaneous interaction with the ES to form a direct transfer intermediate. Subsequent release of Tnp from the non-specific DNA results in transfer of Tnp to the ES. Tnp is represented as a dimer because dimerization is required for condensation of λ-DNA via looping (C. D. Adams, manuscript in preparation). Therefore, we hypothesize that dimerization is also necessary for intermolecular transfer. It should be noted that Tnp can presumably also transfer intermolecularly via complete dissociation from non-specific DNA and reassociation with the ES. Tnp and the ES are depicted as in Figure 1. (B) A model for intramolecular ES localization is shown. This model is similar to the intermolecular transfer model except, in this case, the ES is located on the same DNA molecule as the non-specific sequence where Tnp is bound. Therefore, when Tnp interacts with the ES and non-specific DNA simultaneously, a looped intermediate is formed with optimal loop sizes being between 200 and 300 bp. While Tnp dimerization is required for condensation of λ-DNA via looping, it is unclear at which ES localization step dimerization is necessary. Therefore, the looped intermediate may be formed by interaction of one non-specific DNA bound Tnp and one ES bound Tnp (data not shown). Tnp and the ES are depicted as in Figure 1.
Intramolecular transfer mechanism
To determine if non-specific DNA covalently linked to the ES could facilitate ES localization by Tnp, the rate of catalysis on single-ended Tnp ES containing DNA substrates was investigated (2) (see Figure 6). While transposons are usually defined as DNA flanked by inverted repeats, cleavage at the ES-donor backbone (dbb) DNA junction occurs efficiently when the two ESes on different DNA molecules form a PEC (30). Six DNA substrates with lengths ranging from 485 to 1183 bp, each with 395 bp of transposon DNA, were obtained by restriction digest of a plasmid with six different enzymes. The Tnp mediated cleavage rate constant was determined for each substrate using an in vitro transposition time course and the rate constants were compared to determine if the non-specific DNA could facilitate Tnp ES localization. It should be noted that each reaction contained the substrate DNA together with all remaining plasmid DNA from that digest. This ensures that the total amount of DNA and, therefore, the total amount of Tnp bound non-specifically, were equal in all reactions. Because the total amount of active Tnp was equal in each reaction, differences in rate constants can only be due to length variation of the DNA fragment containing the Tnp ES.
These experiments show that cleavage rate constants increase (with one exception) as the dbb DNA length increases from 90 to 213 bp, but then decrease when the dbb DNA is between 464 and 788 bp. These data indicate that Tnp might use the non-specific DNA to facilitate ES localization, but only with substrates containing less than 464 bp of dbb DNA. If Tnp only bound the ES through a direct mechanism, then the probability of ES localization (and therefore the cleavage rate constant) would be equal for all DNA molecules containing the ES. But, if Tnp can use the non-specific DNA to localize the ES, then the rate of ES localization will increase as the non-specific binding target gets longer due to a higher probability of a longer DNA capturing Tnp.
Given the observation in single molecule micromanipulation experiments that Tnp mediates non-specific DNA looping (C. D. Adams, manuscript in preparation), we propose that Tnp locates the ES after initial non-specific DNA binding through a direct transfer mechanism that involves looping of the non-specific DNA between the initial non-specific DNA Tnp binding site and the ES. This model is explored in Figure 8B. The optimal size for this loop is between 213 and 464 bp. Tnps that bind non-specific DNA within 160 bp of the ES have a slower rate of ES localization because loop formation is inefficient for DNAs shorter than this length. This model is supported by the observation that the optimal length for DNA between transposon ends in vivo is ∼400 bp when the transposon is contained on a plasmid (31). Finally, we have shown that full length Tnp is required for interaction with non-specific DNA. This requirement is consistent with a direct transfer mechanism in which a Tnp dimer is involved and one Tnp monomer binds non-specifically while the second monomer interacts with the ES.
The reduction in ES localization observed for substrates with longer dbbs could have multiple explanations. First, Tnps bound more than one persistence length from the ES would be more likely to undergo competitive sampling of non-specific sites and possible dissociation/inactivation before locating the ES. This would result in a decrease in the observed catalytic rate constant. Alternatively, if the loop size is too large [longer than 1.5 to 3 persistence lengths (213 to 464 bp)], the non-specific DNA bound Tnp cannot appropriately target the ES resulting in intramolecular transfer to another non-specific site instead of ES localization. This would also result in the observed decrease in rate constant for substrates with longer dbbs. It should be noted that the decrease in observed rate constant for substrates with longer dbbs is consistent with lower in vivo transposition frequencies for transposons with ends separated by greater than 400 bp (31).
An in vivo, wild-type (wt) Tnp model for non-specific DNA binding
While certain aspects of intramolecular and intermolecular ES localization by Tnp remain unclear, the data suggest that non-specific DNA binding is an important part of the in vivo Tn5 transposition mechanism. Although a hyperactive Tnp was used in these in vitro studies, the behavior exhibited by this Tnp is likely relevant to the wt protein. Previous studies indicate that an inhibitory interaction between the N- and C-termini of the protein partially explains the low activity and cis bias of the wt Tnp, although, there must be some relief of this inhibition by a conformational change to explain the low level occurrence of transposition in trans (25). The L372P mutation is proposed to act by enhancing the probability of this conformational change and thus results in a more trans active protein (25). Therefore, this mutation only biases the Tnp towards one of two wt conformations and should not affect the non-specific DNA binding characteristics of the wt protein. While the E54K mutation enhances binding to the ESes, the wt Tnp still binds ESes preferentially to non-specific DNA (or transposition would never occur at specific sites). Therefore, while there may be a quantitative difference between an E54K containing Tnp and the wt, all qualitative conclusions stated in this manuscript are valid for the wt Tnp. Therefore, it is certain that wt Tnp manifests non-specific DNA binding properties and, we believe, similar direct transfer properties as the hyperactive Tnp. For both wt and hyperactive Tnp, the qualitative results should be the same regardless of whether the hyperactive ME ES or the wt OE ES is used because the focus of these experiments is how Tnp finds a preferred sequence in the midst of non-specific DNA. Because the affinity of Tnp for all ES containing DNA is substantially higher than for non-specific DNA, the results obtained from these experiments are valid regardless of the ES sequence.
It is also of interest to consider how the non-specific DNA binding would impact our understanding of the cis preference of Tn5 Tnp. Our previous model suggested that the inhibitory interaction between the N- and C-termini would lead to a cis bias because the N-terminus of Tnp could bind to the ES before complete translation of the C-terminus (21). We now extend this model to include co-translational binding of Tnp to non-specific DNA. If wt Tnp initially interacts with a non-specific DNA site near the ES (as might occur in the cis conformation), then, following Tnp dimerization, direct transfer could lead to productive formation of synaptic complexes. However, if the non-specific DNA were located some distance from the ES, Tnp would likely be impaired in ES localization as suggested from the data in Figure 6. In addition, Tnp that eluded the N-terminal, C-terminal inhibitory interaction, would still need to find the ES. This trans Tnp would likely bind non-specific DNA quite some distance from the ES and this binding would impair ES localization.
In this manuscript we have characterized non-specific DNA binding activity of Tn5 Tnp using biochemical and biophysical techniques. While non-specific DNA binding has been observed in other transposition systems, its relevance to any transposition mechanism, other than integration, has never been investigated. Non-specific DNA binding complicates our current transposition models, but also illuminates the intricacies of Tn5 transposition initiation. Hopefully, a better understanding of Tn5 transposition will facilitate greater knowledge of all transposition, integration and site-specific recombination systems.
Acknowledgments
The authors would like to thank Dr Ruth Saeker and Dr M. Thomas Record for helpful discussions and insight and Dr Brandon Ason and Dr Ambrose R. Kidd III for critical review of the manuscript. The authors would also like to thank Dr Dunja Skoko for assistance with the single DNA molecule experiments. This work was supported by NIH grant GM50692 to W.S.R. (M.S. and C.D.A) and by NSF grants DMR-0203963 and PHY-0445565, and by an award from the Focused Giving Program of Johnson & Johnson Corporation to J.F.M. (C.D.A.). Funding to pay the Open Access publication charges for this article was provided by NIGMS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Stanford N.P., Szczelkun M.D., Marko J.F., Halford S.E. One- and three-dimensional pathways for proteins to reach specific sites. EMBO J. 2000;19:65476–66557. doi: 10.1093/emboj/19.23.6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ehbrecht H.-J., Pingoud A., Urbanke C., Maass G., Gualerzi C. Linear diffusion of restriction endonucleases on DNA. J. Biol. Chem. 1985;260:6160–6166. [PubMed] [Google Scholar]
- 3.Wu Z., Chaconas G. A novel DNA binding and nuclease activity in domain III of Mu transposase: evidence for a catalytic region involved in donor cleavage. EMBO J. 1995;14:3835–3843. doi: 10.1002/j.1460-2075.1995.tb00053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakayama C., Teplow D.B., Harshey R.M. Structural domains in phage Mu transposase: identification of the site-specific DNA binding domain. Proc. Natl Acad. Sci. USA. 1987;84:1809–1813. doi: 10.1073/pnas.84.7.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wishart W.L., Broach J.R., Ohtsubo E. ATP-dependent specific binding of Tn3 transposase to Tn3 inverted repeats. Nature. 1985;314:556–558. doi: 10.1038/314556a0. [DOI] [PubMed] [Google Scholar]
- 6.Ichikawa H., Ikeda K., Wishart W.L., Ohtsubo E. Specific binding of transposase to terminal inverted repeats of transposable element Tn3. Proc. Natl Acad. Sci. USA. 1987;84:8220–8224. doi: 10.1073/pnas.84.23.8220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goryshin I.Y., Miller J.A., Kil Y.V., Lanzov V.A., Reznikoff W.S. Tn5/IS50 target recognition. Proc. Natl Acad. Sci. USA. 1998;95:10716–10721. doi: 10.1073/pnas.95.18.10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yanagihara K., Mizuuchi K. Mismatch-targeted transposition of Mu: a new strategy to map genetic polymorphism. Proc. Natl Acad. Sci. USA. 2002;99:11317–11321. doi: 10.1073/pnas.132403399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halling S.M., Kleckner N. A symmetrical six-base-pair target site sequence determines Tn10 insertion specificity. Cell. 1982;28:155–163. doi: 10.1016/0092-8674(82)90385-3. [DOI] [PubMed] [Google Scholar]
- 10.Peters J.E., Craig N.L. Tn7 recognizes transposition target structures associated with DNA replication using the DNA-binding protein TnsE. Genes Dev. 2001;15:737–747. doi: 10.1101/gad.870201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goryshin I.Y., Reznikoff W.S. Tn5 in vitro transposition. J. Biol. Chem. 1998;273:7367–7374. doi: 10.1074/jbc.273.13.7367. [DOI] [PubMed] [Google Scholar]
- 12.Johnson R.C., Reznikoff W.S. DNA sequences at the ends of transposon Tn5 required for transposition. Nature. 1983;304:280–282. doi: 10.1038/304280a0. [DOI] [PubMed] [Google Scholar]
- 13.Johnson R.C., Reznikoff W.S. Copy number control of Tn5 transposition. Genetics. 1984;107:9–18. doi: 10.1093/genetics/107.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson R.C., Reznikoff W.S. Role of the IS50 R proteins in the promotion and control of Tn5 transposition. J. Mol. Biol. 1984;177:645–661. doi: 10.1016/0022-2836(84)90042-1. [DOI] [PubMed] [Google Scholar]
- 15.Bhasin A., Goryshin I.Y., Steiniger-White M., York D., Reznikoff W.S. Characterization of a Tn5 pre-cleavage synaptic complex. J. Mol. Biol. 2000;302:49–63. doi: 10.1006/jmbi.2000.4048. [DOI] [PubMed] [Google Scholar]
- 16.Naumann T.A., Reznikoff W.S. Trans catalysis in Tn5 transposition. Proc. Natl Acad. Sci. USA. 2000;97:8944–8949. doi: 10.1073/pnas.160107997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steiniger-White M., Reznikoff W.S. The C-terminal alpha helix of Tn5 transposase is required for synaptic complex formation. J. Biol. Chem. 2000;275:23127–23133. doi: 10.1074/jbc.M003411200. [DOI] [PubMed] [Google Scholar]
- 18.Bhasin A., Goryshin I.Y., Reznikoff W.S. Hairpin formation in Tn5 transposition. J. Biol. Chem. 1999;274:37021–37029. doi: 10.1074/jbc.274.52.37021. [DOI] [PubMed] [Google Scholar]
- 19.Reznikoff W.S. Tn5 as a model for understanding DNA transposition. Mol. Microbiol. 2003;47:1199–1206. doi: 10.1046/j.1365-2958.2003.03382.x. [DOI] [PubMed] [Google Scholar]
- 20.Zhou M., Reznikoff W.S. Tn5 transposase mutants that alter DNA binding specificity. J. Mol. Biol. 1997;271:362–373. doi: 10.1006/jmbi.1997.1188. [DOI] [PubMed] [Google Scholar]
- 21.Weinreich M.D., Mahnke-Braam L., Reznikoff W.S. A functional analysis of the Tn5 transposase. Identification of domains required for DNA binding and multimerization. J. Mol. Biol. 1994;241:166–177. doi: 10.1006/jmbi.1994.1486. [DOI] [PubMed] [Google Scholar]
- 22.Davies D.R., Goryshin I.Y., Reznikoff W.S., Rayment I. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science. 2000;289:77–85. doi: 10.1126/science.289.5476.77. [DOI] [PubMed] [Google Scholar]
- 23.Naumann T.A., Reznikoff W.S. Tn5 transposase with an altered specificity for transposon ends. J. Bacteriol. 2002;184:233–240. doi: 10.1128/JB.184.1.233-240.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.York D., Reznikoff W.S. DNA binding and phasing analyses of Tn5 transposase and a monomeric variant. Nucleic Acids Res. 1997;25:2153–2160. doi: 10.1093/nar/25.11.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weinreich M.D., Gasch A., Reznikoff W.S. Evidence that the cis preference of the Tn5 transposase is caused by nonproductive multimerization. Genes Dev. 1994;8:2363–2374. doi: 10.1101/gad.8.19.2363. [DOI] [PubMed] [Google Scholar]
- 26.Skoko D., Wong B., Johnson R.C., Marko J.F. Micromechanical analysis of the binding of DNA-bending proteins HMGB1, NHP6A, and HU reveals their ability to form highly stable DNA-protein complexes. Biochem. 2004;43:13867–13874. doi: 10.1021/bi048428o. [DOI] [PubMed] [Google Scholar]
- 27.Naumann T.A., Reznikoff W.S. Tn5 transposase active site mutants. J. Biol. Chem. 2002;277:17623–17629. doi: 10.1074/jbc.M200742200. [DOI] [PubMed] [Google Scholar]
- 28.Braam L.A., Goryshin I.Y., Reznikoff W.S. A mechanism for Tn5 inhibition. carboxyl-terminal dimerization. J. Biol. Chem. 1999;274:86–92. doi: 10.1074/jbc.274.1.86. [DOI] [PubMed] [Google Scholar]
- 29.Halford S.E., Marko J.F. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 2004;32:3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ason B., Reznikoff W.S. Mutational analysis of the base flipping event found in Tn5 transposition. J. Biol. Chem. 2002;277:11284–11291. doi: 10.1074/jbc.M111119200. [DOI] [PubMed] [Google Scholar]
- 31.Goryshin I.Y., Kil Y.V., Reznikoff W.S. DNA length, bending, and twisting constraints on IS50 transposition. Proc. Natl Acad. Sci. USA. 1994;91:10834–10838. doi: 10.1073/pnas.91.23.10834. [DOI] [PMC free article] [PubMed] [Google Scholar]