Abstract
Missense mutations in the skeletal muscle sodium channel α-subunit gene (SCN4A) are associated with a group of clinically overlapping diseases caused by alterations in the excitability of the sarcolemma. Sodium channel defects may increase excitability and cause myotonic stiffness or may render fibres transiently inexcitable to produce periodic paralysis. A patient with cold-aggravated myotonia did not harbour any of the common SCN4A mutations. We therefore screened all 24 exons by denaturing high-performance liquid chromatography, followed by direct sequencing. Two novel missense changes were found with predicted amino acid substitutions: T323M in the DIS5-S6 loop and F1705I in the intracellular C-terminus. The functional impact of these substitutions was assessed by recording whole-cell Na+ currents from transiently transfected HEK293 cells. T323M currents were indistinguishable from wild-type (WT). Fast inactivation was impaired for F1705I channels, as demonstrated by an 8.6-mV rightwards shift in voltage dependence and a two-fold slowing in the rate of inactivation. Recovery from fast inactivation was not altered, nor was there an increase in the persistent current after a 50- ms depolarization. Activation and slow inactivation were not appreciably affected. These data suggest that T323M is a benign polymorphism, whereas F1705I results in fast inactivation defects, which are often observed for myotonia. This is the first example of a C-terminal mutation in SCN4A associated with human disease. Like the cardiac disorders (long QT syndrome type 3 or Brugada syndrome) and generalized epilepsy with febrile seizures plus (GEFS+) associated with C-terminal mutations in other NaV channels, the primary effect of F1705I was a partial disruption of fast inactivation.
Voltage-gated sodium channels play a vital role in the initiation and propagation of action potentials, which endows the unique characteristic of electric excitability in neurones, skeletal muscle and cardiac cells. Naturally occurring mutations in sodium channel genes have been associated with many human diseases (Lehmann-Horn & Jurkat-Rott, 1999). The first human mutation reported in a voltage-gated ion channel was identified in the tetrodotoxin-sensitive adult skeletal muscle sodium channel gene (SCN4A), encoding the NaV1.4 pore-forming α-subunit. Missense mutations in SCN4A have been associated with dominantly inherited myotonia and periodic paralysis, a group of clinically overlapping diseases caused by alterations in the excitability of the sarcolemma.
Four dominantly inherited skeletal muscle sodium channel disorders (channelopathies) have been delineated in humans on the basis of their clinical phenotype: potassium-aggravated myotonia (PAM, MIM 608390, including myotonia fluctuans, myotonia permanens, acetazolamide-responsive myotonia, painful myotonia); paramyotonia congenita (PC, MIM 168300); hyperkalaemic periodic paralysis (HyperPP, MIM 170500); and hypokalaemic periodic paralysis type 2 (HypoPP2, MIM 170400) (Lehmann-Horn & Jurkat-Rott, 1999). The most common functional abnormality in disease-associated NaV1.4 missense mutations is a gain-of-function defect, due to impaired fast inactivation, or in about 10% of cases due to enhanced activation (Cannon, 2000). Disrupted slow inactivation has been reported for four mutations and predisposes to attacks of depolarization-induced paralysis (Hayward et al. 1999). HypoPP2 is distinct in that the major change is loss-of-function defects due to enhanced inactivation (Jurkat-Rott et al. 2000; Struyk et al. 2000).
Here, we have performed a systematic screen in all 24 exons of the SCN4A gene in a patient with cold-aggravated myotonia but no weakness. Two amino acid substitutions were identified in regions of the channel protein atypical for other SCN4A mutations: T323M in the DIS5-S6 loop and F1705I in the intracellular C-terminus. Functional assessment of mutant channels heterologously expressed in HEK293 cells revealed an impairment of fast inactivation for F1705I channels; whereas the T323M channels behaved like wild-type (WT), suggesting that T323M is a benign polymorphism. In this report we establish that missense mutations in the carboxyl terminus of the skeletal muscle sodium channel can partially disrupt fast inactivation and thereby produce the disease phenotype, as has been previously reported in heart for NaV1.5 mutations associated with long QT syndrome type 3 (Baroudi & Chahine, 2000) and in brain with a NaV1.1 mutation in generalized epilepsy with febrile seizures plus (Spampanato et al. 2004).
Methods
Clinical data
A 49-year-old Caucasian man of Polish heritage complained of frequent stiffness and cramps in the arms and legs. Symptoms had been present for 15 years and were aggravated by cold weather. Muscle stiffness initially impeded his ability to walk upon standing, and he improved over 2–3 min, after which walking was normal. Calf muscles were hypertrophied. Myotonic lid lag, percussion myotonia and grip myotonia were not detected. Muscle strength and tone were normal, except for a slight decrease in the thenar eminence bilaterally, which was worse on the left. The cramps were well controlled with mexiletine although he still complained of stiffness in his neck and back. There was no known family history of muscle stiffness and cramps among his relatives. His electrocardiogram was normal.
Denaturing high-performance liquid chromatography
After obtaining informed consent in accordance with Institutional Review Board protocol 2405 at the Children's National Medical Center, 10 ml whole blood was collected for extraction of genomic DNA (Miller et al. 1988). The patient's DNA sample tested negative for the two most common SCN4A gene mutations (T704M and M1592V), and therefore all 24 exons of the SCN4A gene were screened by denaturing high-performance liquid chromatography (DHPLC). Amplified PCR products were analysed with the Wave DNA Fragment Analysis System (Transgenomic Inc, Omaha, NE, USA). The elution gradient was optimized to allow heteroduplexes and homoduplexes to be eluted between 4 and 6 min.
DNA sequencing
PCR products showing heteroduplexes by DHPLC were re-amplified and purified by QIAquick Gel Extraction kit (QIAGEN, Hilden, Germany). Purified PCR products were then subjected to automated sequence analysis using Dye Terminator and analysed on a CEQ 2000 DNA Analysis System (Beckman Coulter, Fullerton, CA, USA) following the manufacturer's protocols. Data were analysed using Sequencher 3.0 program (Gene Codes Corporation, Ann Arbor, MI, USA).
Site-directed mutagenesis
The QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA) was used to introduce the T323M and F1705I mutations into the human skeletal muscle sodium channel mammalian expression construct pRc/CMV-hSkM1 provided by A. L. George (Vanderbilt University, Nashville, TN, USA). The entire sodium channel coding region was sequenced to confirm the presence of the mutation and to ensure that no other mutations were generated during the site-directed mutagenesis process.
Expression of sodium channels
Human embryonic kidney (HEK293) cells were transiently transfected using the calcium phosphate method (Hayward et al. 1996). In brief, normal WT or mutant (T323M or F1705I) sodium channel α-subunit plasmid (0.6 μg per 35-mm dish) was cotransfected with a human (h) β1-subunit plasmid (McClatchey et al. 1993) at four-fold molar excess over α-subunits, and a CD8 marker (0.1 μg per 35-mm dish). After 24–72 h, HEK293 cells were trypsinized briefly and passaged to 12-mm round glass cover slips for electrophysiological recording. Individual transfection-positive cells were identified by labelling with anti-CD8 antibody cross-linked to microbeads (Jurman et al. 1994) (Dynal, Great Neck, NY, USA).
Whole-cell recording
Na+ currents were measured using conventional whole-cell recording techniques as previously described (Hayward et al. 1996). Recordings were made with an Axopatch 200B amplifier (Axon Instruments, Union City, CA, USA). The output was filtered at 5 kHz and digitally sampled at 40 kHz using an LM900 interface (Dagan, Minneapolis, MN, USA). Data were stored to a Pentium-based (Intel, Santa Clara, CA, USA) computer using a custom-made AxoBasic data acquisition program (Axon Instruments). More than 85% of the series resistance was compensated by the analog circuitry of the amplifier and the leakage conductance was corrected by digital scaling and subtraction of the passive current elicited by a 30-mV depolarization from the holding potential. Only cells with peak currents > 1 nA and < 20 nA upon step depolarization from −120 mV to −10 mV were included. After initially establishing whole-cell access, we often observed a leftward shift in the voltage dependence of gating, an increase in the size of the peak current, and a decrease in the amplitude of persistent Na+ current. To achieve standardized responses across cells, we waited at least 10 min for equilibration after gaining whole-cell access.
Patch electrodes were fabricated from borosilicate capillary tubes with a multi-stage puller (Sutter, Novato, CA, USA). The shank of the pipette was coated with Sylgard and the tip was heat-polished to a final tip resistance (in bath solution) of 0.5–2.0 MΩ. The pipette (internal) solution contained (mm): CsF 105, NaCl 35, EGTA 5 and Cs-Hepes 10; pH 7.4. Fluoride was used in the pipette to prolong seal stability. The bath solution contained (mm): NaCl 140, KCl 4, CaCl2 2, MgCl2 1, glucose 2.5 and Na-Hepes 10; pH 7.4. Tetrodotoxin (Sigma, St Louis, MO, USA) was made in a 3 mm stock and diluted with bath solution to 5 μm before perfusion. Recordings were made at room temperature (24–28°C).
Data analysis
Analysis of raw current traces was performed manually off-line using AxoBasic or Origin (Microcal, Northampton, MA, USA). Conductance was calculated as G(V) =IPEAK(V)/(V−Erev), where the reversal potential (Erev) was measured experimentally for each cell. The voltage dependence of activation was quantified by fitting the conductance measures to a Boltzmann function: G(V) =Gmax/[1 + exp(−(V−V1/2)/k)]; steady-state fast and slow inactivation were fitted to a Boltzmann function with a non-zero pedestal (I0) calculated as I/IPEAK= (1 −I0)/[1 + exp((V−V1/2)/k)]+I0, where V1/2 is the half-maximum voltage and k is the slope factor. Symbols with error bars indicate means ± s.e.m. Statistical significance was determined by the Student's unpaired t test with P-values noted in the text.
Results
Identification of two novel potential SCN4A mutations
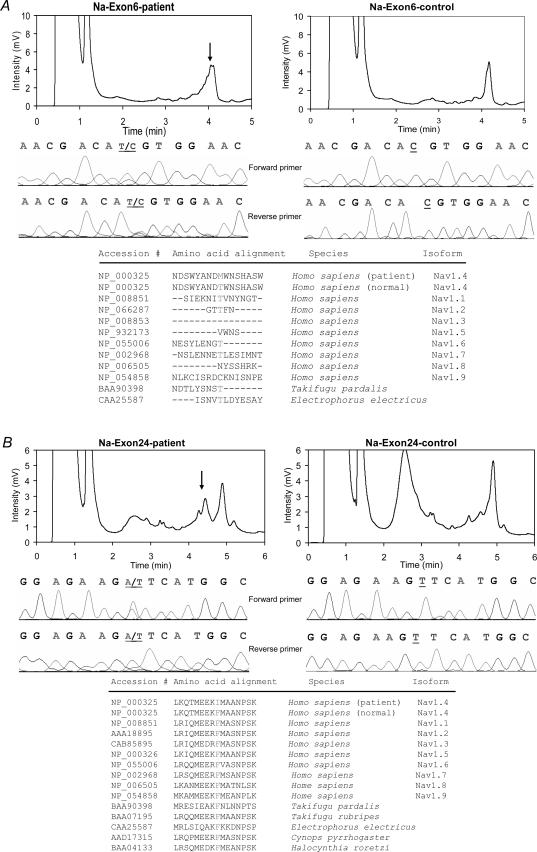
The patient's genomic DNA was screened for sequence changes in all 24 exons of SCN4A by DHPLC analysis. DHPLC heteroduplexes were identified in exons 6 and 24, and DNA sequencing showed heterozygous changes in both coding regions (Fig. 1A and B). The two novel mutations were: a C to A transition at coding sequence 968 in exon 6, which predicts the substitution of methionine for threonine at amino acid 323 (T323M) in an extracellular loop just 5′ to the pore loop in domain I; and a T to A transversion at coding sequence 5113 in exon 24, which results in the substitution of isoleucine for phenylalanine at amino acid 1705 (F1705I) in the C-terminus (Fig. 2). It was not possible to determine whether these mutations co-segregated on the same chromatid, because a single PCR amplimer from genomic DNA could not span such a large distance and no family members were available to test the transmission patterns of these two novel mutations. Therefore, each mutation was compared to normal population controls as a test that either might be a polymorphic variant. Fifty-five control DNA samples from Caucasians of European ancestry were checked by DHPLC chromatographic pattern of exons 6 and 24. None had the same pattern as that of the patient (data not shown).
Figure 1. Identification of two novel potential SCN4A mutations in exons 6 and 24.
A, a heteroduplex occurred at ∼4.1 min due to the T323M mutation in exon 6. B, the exon 24 amplimer had a heteroduplex at ∼4.5 min due to the F1705I mutation. The DHPLC chromatogram of each patient sample shows a heteroduplex pattern (arrow, left panel) compared to control (right panel). DNA sequencing results in both forward and reverse directions reveal single nucleotide transition and transversion in the patient samples (middle panel). The peptide sequences of several sodium channel isoforms were aligned in the region of each mutation using NCBI BLAST. The amino acid of the F1705I mutation is highly conserved in different tissues and species.
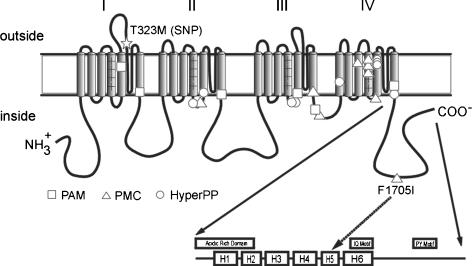
Figure 2. Schematic representation of hNaV1.4 and the location of the human disease mutations.
The canonical membrane-spanning diagram for the sodium channel α-subunit has been modified to illustrate the carboxyl terminus domains proposed by Cormier et al. (2002). Symbols illustrate the location of missense mutations identified in hyperkalaemic periodic paralysis (HyperPP, ○), paramyotonia congenita (PMC, ▵) and potassium-aggravated myotonia (PAM, □). The T323M substitution (*) lies in the extracellular loop connecting transmembrane segments 5 and 6 of domain I. The F1705I substitution (▵) is approximately midway along the carboxyl tail, within the predicted 5th α-helix (H5) (dashed arrow).
The functional consequences of these two putative mutations were ascertained by constructing each mutant in the human NaV1.4 cDNA background (pRc/CMV-hSkM1) and recording whole-cell sodium currents from transiently transfected HEK293 cells. The human Na+ channel β1 subunit was cotransfected to recapitulate the native heterodimeric channel in skeletal muscle.
Activation
Na+ channel activation was characterized by measuring the voltage dependence of the peak Na+ current elicited by 20-ms step depolarizations from a holding potential of −120 mV. Test depolarization from −75 to +80 mV were applied in 5-mV increments (Fig. 3A – C). On average, there was no difference in peak current amplitude for cells expressing T323M (5.0 ± 0.4 nA; n = 24) or F1705I (4.5 ± 0.5 nA; n = 27) compared with WT channels (5.9 ± 0.7 nA; n = 24) (P > 0.05). The Na+ conductance was estimated from the peak current and the measured reversal potential, and its voltage dependence is plotted in Fig. 4A (right). The conductance data were fitted with a Boltzmann function to quantify the midpoint (V1/2) and steepness factor (k) for the voltage dependence of activation (Table 1). There was no difference in the estimated parameter values. WT: V1/2, −27.1 ± 1.2 mV; k, 6.0 ± 0.2 mV; n = 15; T323M: V1/2, −25.8 ± 1.2 mV; k, 5.9 ± 0.2 mV; n = 15; F1705I: V1/2, −27.9 ± 0.7 mV; k, 5.8 ± 0.1 mV; n = 20; P > 0.22 for V1/2, P > 0.17 for k.
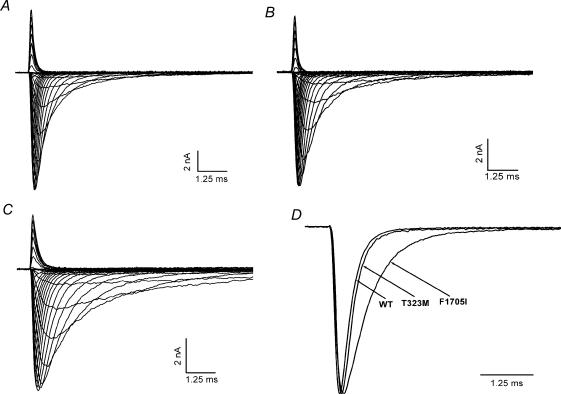
Figure 3. Macroscopic Na+ current decay was slowed for F1705I but not in the T323M channel.
Representative Na+ currents from WT (A), T323M (B) and F1705I (C) channels elicited by a series of 20-ms step depolarizations from a holding potential of −120 mV to between −75 and +80 mV in 5-mV increments. D, Na+ currents at −10 mV were normalized to peak amplitude and superimposed for WT, T323M and F1705I channels to illustrate the slower decay for F1705I (0.65 ms) compared to T323M (0.28 ms) or WT (0.28 ms) channels.
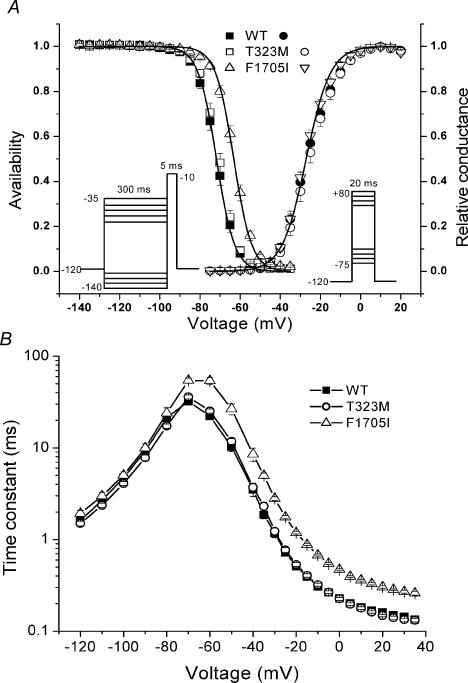
Figure 4. The F1705I mutation disrupted fast inactivation, but not activation.
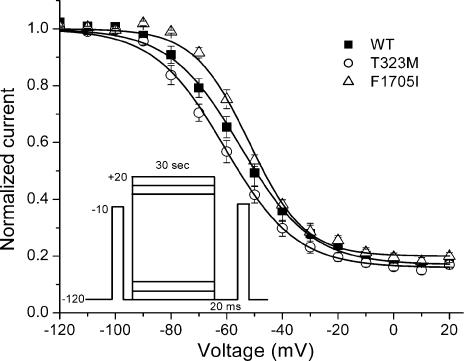
A, activation, as measured by peak Na+ conductance over a range of test potentials (right-hand curves), was not altered by F1705I or T323M. Steady-state fast inactivation was measured as the relative peak current elicited at −10 mV, after a 300-ms conditioning pulse (left-hand curves). The voltage dependence of fast inactivation was shifted in the depolarizing direction (rightwards) by 8.6 mV for F1705I. B, fast inactivation from the open state (test potentials, −35 to +35 mV) and closed states (−70 to −40 mV, measured by a two-pulse protocol see Results) was slowed for F1705I channels. No differences were observed in the rate of recovery from fast inactivation (−120 to −80 mV) between WT and F1705I or T323M.
Table 1.
Gating parameters for NaV1.4 constructs
| Activation | Fast inactivation | Slow inactivation | |||||
|---|---|---|---|---|---|---|---|
| V1/2 (mV) | k (mV) | V1/2 (mV) | k (mV) | V1/2 (mV) | k (mV) | I0 | |
| WT | −27.1 ± 1.2 | 6.0 ± 0.2 | −71.8 ± 0.9 | 4.5 ± 0.1 | −55.0 ± 2.2 | 12.7 ± 0.8 | 0.17 ± 0.019 |
| (15) | (15) | (5) | |||||
| T323M | −25.8 ± 1.2 | 5.9 ± 0.2 | −70.7 ± 0.8 | 4.3 ± 0.1 | −60.4 ± 1.4* | 13.3 ± 0.5 | 0.16 ± 0.008 |
| (15) | (15) | (7) | |||||
| F1705I | −27.9 ± 0.7 | 5.8 ± 0.1 | − 63.2 ± 0.8** | 4.5 ± 0.1 | − 51.9 ± 0.9 | 9.9 ± 0.6* | 0.20 ± 0.011 |
| (20) | (5) | ||||||
Values are means +s.e.m. with number of experiments in parenthesis
Significantly different from WT, P < 0.05;
significantly different from WT, P < 0.001.
Fast inactivation
The voltage dependence and kinetics of Na+ channel fast inactivation were characterized by recording whole-cell currents. Figure 3A–C shows whole-cell Na+ currents for WT, T323M and F1705I, respectively, elicited by a series of 20-ms step depolarization from −75 to +80 mV in 5-mV increments. The decay of the transient Na+ current was slower for F1705I than WT channels, whereas the decay rate for T323M was indistinguishable from WT. To illustrate this difference more clearly, currents elicited at −10 mV were normalized to the peak value and traces were superimposed for WT, T323M and F1705I (Fig. 3D). In this example, the current decayed 2.3 times more slowly for F1705I (0.65 ms) than T323M (0.28 ms) and WT (0.28 ms).
The voltage dependence of steady-state fast inactivation (availability) was measured as the peak current elicited following a 300-ms conditioning prepulse from −140 to −35 mV in 5-mV increments. Figure 4A left shows the voltage dependence of the steady-state availability for WT, T323M and F1705I. The data for F1705I were shifted rightward (depolarized) compared with WT, whereas T323M showed no difference from WT. To quantify this difference, the data were fitted with a Boltzmann function (Table 1). For F1705I channels, the average for the midpoint of the curve (V1/2) was shifted +8.6 mV (P < 10−8) while no difference in the slope factor (k) was identified. T323M showed no difference from WT (WT: V1/2, −71.8 ± 0.9 mV; k, 4.5 ± 0.1 mV, n = 15; T323M: V1/2, −70.7 ± 0.8 mV; k, 4.3 ± 0.1 mV, n = 15; F1705I: V1/2, −63.2 ± 0.8 mV; k, 4.5 ± 0.1 mV, n = 20).
The kinetics of fast inactivation were characterized by quantifying the time course of entry to and recovery from inactivation. Entry at depolarized potentials (−35 to +35 mV) was measured by fitting the decay of sodium currents measured from macroscopic current decay with a single exponential. A defect in fast inactivation for the F1705I mutant was discernible at voltages above −35 mV as a two- to three-fold slower time constant, whereas no difference was observed between T323M and WT channels. Over an intermediate range of voltages, −70 to −40 mV, the time course of fast inactivation was measured using a two-pulse protocol. First, a conditioning pulse depolarization was applied for 1–500 ms. Inactivation was measured as a decrease in the peak Na+ current elicited by the second pulse to −10 mV. The relative Na+ current as a function of conditioning pulse duration was fitted with a single exponential, and the time constants were plotted against the conditioning pulse voltage in Fig. 4B. From −70 to −40 mV, the time constant for inactivation from closed states was two- to three-fold slower for F1705I than for WT (n = 11–14; P < 0.004), whereas T323M showed no difference from WT.
We measured the time course of recovery from the fast-inactivated state at a more hyperpolarized range of voltages (−120 to −80 mV) using a two-pulse protocol. A 30-ms conditioning pulse to −10 mV was applied to fully fast-inactivate the channel. The membrane was then hyperpolarized for 0.05–2000 ms, and then recovery was monitored by measuring the relative peak Na+ current elicited by a second pulse to −10 mV. The recovery of the peak amplitude was fitted with a single exponential and the time constants are plotted as a function of the recovery potential in Fig. 4B. Over the voltage range studied, recovery of the peak currents of both T323M and F1705I channels were no different from WT channel. The depolarized shift in the V1/2 of availability and the slower rate of entry are all indicative of disrupted fast inactivation for F1705I channels.
Slow inactivation
The voltage dependence of slow inactivation is shown for WT, T323M and F1705I mutant channels in Fig. 5. Steady-state slow inactivation was measured using a 30-s conditioning pulse from −120 to +20 mV followed by a 20-ms gap at −120 mV to allow recovery from fast inactivation, before a −10-mV test pulse (inset). The voltage for half inactivation (V1/2) was shifted by −5 mV for T323M mutants (P = 0.028), whereas the depolarized shift of +5 mV for F1705I was not statistically different from WT (P = 0.11). The voltage dependence of slow inactivation was steeper for F1705I (lower k-value, P = 0.01), but the maximal extent of slow inactivation (I0) did not differ significantly for either mutant compared to WT (Table 1).
Figure 5. Voltage dependence of slow inactivation was similar for WT, T323M and F1705I channels.
Steady-state slow inactivation was measured as the relative Na+ current after a 30-s conditioning pulse and an intervening 20-ms recovery at −120 mV to remove fast inactivation (inset). Fitting the data with a Boltzmann function did not reveal a difference in the voltage for half inactivation or the maximal extent of slow inactivation for mutants compared to WT (see text).
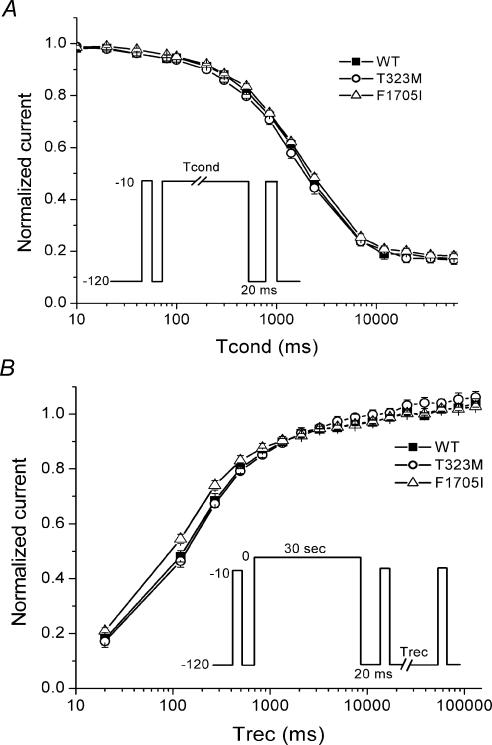
The kinetics of slow inactivation were characterized by measuring the time course of entry to and recovery from inactivation. Entry to slow inactivation was monitored by using a two-pulse protocol. A conditioning pulse at −10 mV with varying duration from 10 ms to 60 s was applied to inactivate Na+ channels. A 20-ms gap at −120 mV was then applied to remove fast inactivation, and finally a brief depolarization to −10 mV was used to activate available Na+ channels. Recovery from slow inactivation was determined by measuring the peak Na+ current at various times after a 30-s conditioning pulse to 0 mV. The kinetics of entry (Fig. 6A) and recovery (Fig. 6B) from slow inactivation were not different for T323M or F1705I channels compared to WT.
Figure 6. Kinetics of slow inactivation were not altered in T323M and F1705I channels.
A, the time course of entry to slow inactivation at −10 mV was indistinguishable between WT (▪), T323M (○) and F1705I (▵). B, recovery from slow inactivation was monitored after a 30-s conditioning pulse at 0 mV, by applying very brief −10-mV test depolarizations during recovery at −120 mV (inset). Neither T323M (○) nor F1705I (▵) channels showed any alteration in recovery from slow inactivation compared to WT (▪) channels.
Persistent Na+ current
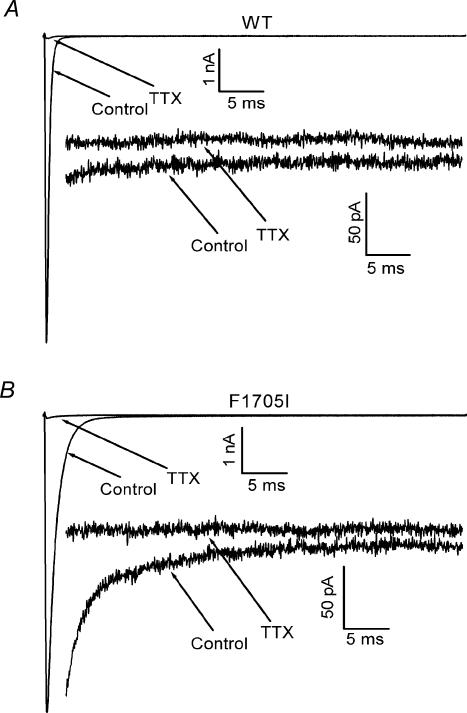
Disease-associated mutations may also disrupt the completeness of fast inactivation, which is detected as an anomalous Na+ current that persists for several tens of milliseconds after the test depolarization (Cannon et al. 1991). Because even a small persistent Na+ current can alter the excitability of the cell, a tetrodotoxin-subtraction protocol was used to detect the presence of Na+ currents of the order of 0.2% of the initial inward peak transient. The current elicited by a 50-ms step depolarization from −120 mV to −10 mV was measured in control bath solution and in the presence of a saturating 5 μm concentration of tetrodotoxin (Fig. 7). The difference between the two responses is a sensitive measure of the small persistent current conducted only through Na+ channels, without relying on the assumption of a linear leakage current subtraction as in the P/N subtraction protocol. The amplitude of the time-averaged steady-state current during the last 5 ms of a 50-ms pulse was normalized to the initial peak amplitude of the transient current. The small persistent Na+ current observed for mutant channels was no different from that observed for WT channels (F1705I, 0.22 ± 0.05%, n = 5; WT, 0.15 ± 0.02%, n = 5; P > 0.09).
Figure 7. The F1705I mutation does not increase the persistent Na+ current.
A tetrodotoxin-subtraction technique was used to detect small persistent Na+ currents. The current elicited by a 50-ms step depolarization from −120 mV to −10 mV was recorded at normal and high amplifier gains to measure the peak and persistent current, respectively. The protocol was repeated after application of a saturating concentration of tetrodotoxin (5 μm) to isolate the background current, which was subtracted from the control response to isolate the tetrodotoxin-sensitive Na+ current. Representative traces in low (1 nA scale) and high (50 pA scale) gain are shown for WT (A) and F1705I (B) channels. Peak transient currents were comparable for these two cells, so the separation of the persistent currents in the high-gain traces directly show the comparable amplitude of persistent current for WT and F1705I channels.
Discussion
We have identified two novel amino acid substitutions in a cold-aggravated myotonia patient through molecular genetic studies, and have characterized the functional defects of mutant channels through whole-cell patch-clamp analysis. Neither the T323M nor the F1705I mutations identified here were present in 55 control DNA samples (110 chromosomes), arguing that the changes were not common polymorphisms. The degree of conservation at these residues across channel isoforms and species was examined as a further indicator of their potential significance. The F1705 residue is absolutely conserved in all α-subunits of the human NaV family (NaV1.1–NaV1.9), fish and marine invertebrates (sea squirt). The expression studies herein confirm that F1705I is not a benign polymorphism, as the mutant channel has a gain-of-function defect with disrupted fast inactivation. However, the T323M is not conserved, even in the closely related NaV1.5 cardiac Na+ channel, and is more likely to be a benign polymorphism. Indeed, the electrophysiological properties of the T323M channel are indistinguishable from WT, except for a −5 mV shift in the voltage dependence of slow inactivation. This modest enhancement of slow inactivation is a change in the wrong direction to cause susceptibility to myotonia. Moreover, we have previously established that even more pronounced enhancement of slow inactivation (−10 mV shift and two-fold reduction of I0) does not suppress myotonia caused by gain-of-function defects in fast gating (Takahashi & Cannon, 1999). Consequently, we conclude that T323M is a rare benign polymorphism. There are rare instances in the cardiac Na+ channel, NaV1.5, wherein the presence of a polymorphism, which does not alter function alone, modifies the functional consequence of a disease-causing missense mutation (Ye et al. 2003). This possibility remains untested in the proband herein because it was not feasible to determine whether T323M and F1705I are on the same allele. We propose that F1705I is a new missense mutation that causes cold-aggravated myotonia without attacks of weakness. Paradoxical myotonia (i.e. worsening with repeated muscular activity) was not a feature in this patient. Nevertheless, the cold sensitivity and lack of spontaneously fluctuating susceptibility to myotonia suggest this phenotype is a variant of paramyotonia congenita. The F1705I substitution reported herein is also the first example of a mutation in the C-terminus of NaV1.4, out of over 30 reported mutations associated with myotonia or periodic paralysis.
The functional defects observed for F1705I channels expressed in HEK293 cells are consistent with those reported for other NaV1.4 mutants associated with myotonia. The common feature of these mutations is disrupted fast inactivation, which was also the primary change for F1705I. The slower rate of inactivation from open state (Figs 3D and 4B, −35 to +35 mV range) and closed states (Fig. 4B, −70 to −40 mV range), and the 8.6-mV depolarized shift in steady-state availability (Fig. 4A), are all indicators of impaired fast inactivation. Unlike some other myotonia-associated mutations (e.g. T1313M in the III–IV linker (Hayward et al. 1996) or substitutions in DIVS4 (Yang et al. 1994)), recovery from fast inactivation was not hastened in F1705I channels. Computer simulations in prior studies have demonstrated that fast-inactivation defects of the magnitude observed for F1705I are sufficient to produce trains of repetitive discharges in response to a brief stimulus (Green et al. 1998), which is the electromyographic hallmark of myotonia. Finally, slow inactivation was not impaired by F1705I, which is consistent with the prevailing view that slow-inactivation defects predispose to attacks of paralysis, not myotonia (Cannon, 2000).
The C-terminus of voltage-gated Na+ channels has recently been implicated in fast inactivation. Chimeric exchange of C-termini between rat NaV1.2 and rNaV1.5, as well as hNaV1.4 and hNaV1.5 have demonstrated that the proximal half (5′-end) of the C-terminus determines the phenotype of the fast-inactivation kinetics (Deschenes et al. 2001; Mantegazza et al. 2001). In addition, many missense mutations in NaV1.5 associated with the long QT syndrome type 3 are located in the mid-portion of the C-terminus, in the acidic rich domain near H1 or in the H5 and H6 α-helical segments (Fig. 2). Biochemical studies of NaV1.5 have revealed a potential interaction of the C-terminal domain with the III–IV loop to form a molecular complex that subserves inactivation; mutations in either portion destabilized the closed gate and increased persistent current (Motoike et al. 2004). A C-terminal mutation in the neuronal NaV1.1 channel causes generalized epilepsy with febrile seizures plus, and this D1866Y substitution impairs fast inactivation by disrupting interaction of the C-terminus and the cytoplasmic tail of the β1 subunit (Spampanato et al. 2004). The F1705I mutation studied herein lies just seven residues downstream from a 40-amino acid peptide segment centred about D1866 (from NaV1.1), that was shown to interact with the β1 subunit. This observation raises the possibility that a similar mechanism may underlie the fast-inactivation defect for F1705I. Another consideration might be disruption of Ca2+ regulation of NaV1.4 by F1705I (yet to be tested rigorously), although this site is not within either the IQ motif for calmodulin binding (30 residues downstream) or a putative paired EF hand motif (42 residues upstream). Taken together, these other studies suggest a coordinated interaction of the III–IV cytoplasmic loop, the 5′-end of the carboxyl terminus, and the cytoplasmic tail of the β1 subunit are required for fast inactivation. The report herein of a similar effect for a C-terminal mutation of NaV1.4 channels of skeletal muscle strengthens the notion that this is a conserved feature of voltage-gated Na+ channels.
Acknowledgments
The authors thank Jim Giron for assistance with molecular diagnosis, and Joseph Devaney for assistance with DHPLC operation and analysis. This work was supported by grants from the National Institutes of Health (E.P.H and S.C.C.).
References
- Baroudi G, Chahine M. Biophysical phenotypes of SCN5A mutations causing long QT and Brugada syndromes. FEBS Lett. 2000;487:224–228. doi: 10.1016/s0014-5793(00)02360-7. [DOI] [PubMed] [Google Scholar]
- Cannon SC. Spectrum of sodium channel disturbances in the nondystrophic myotonias and periodic paralyses. Kidney Int. 2000;57:772–779. doi: 10.1046/j.1523-1755.2000.00914.x. [DOI] [PubMed] [Google Scholar]
- Cannon SC, Brown RH, Jr, Corey DP. A sodium channel defect in hyperkalemic periodic paralysis: potassium-induced failure of inactivation. Neuron. 1991;6:619–626. doi: 10.1016/0896-6273(91)90064-7. [DOI] [PubMed] [Google Scholar]
- Cormier JW, Rivolta I, Tateyama M, Yang AS, Kass RS. Secondary structure of the human cardiac Na+ channel C terminus: evidence for a role of helical structures in modulation of channel inactivation. J Biol Chem. 2002;277:9233–9241. doi: 10.1074/jbc.M110204200. [DOI] [PubMed] [Google Scholar]
- Deschenes I, Trottier E, Chahine M. Implication of the C-terminal region of the α-subunit of voltage-gated sodium channels in fast inactivation. J Membr Biol. 2001;183:103–114. doi: 10.1007/s00232-001-0058-5. 10.1007/s00232-001-0058-5. [DOI] [PubMed] [Google Scholar]
- Green DS, George AL, Jr, Cannon SC. Human sodium channel gating defects caused by missense mutations in S6 segments associated with myotonia: S804F and V1293I. J Physiol. 1998;510:685–694. doi: 10.1111/j.1469-7793.1998.685bj.x. 10.1111/j.1469-7793.1998.685bj.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward LJ, Brown RH, Jr, Cannon SC. Inactivation defects caused by myotonia-associated mutations in the sodium channel III–IV linker. J Gen Physiol. 1996;107:559–576. doi: 10.1085/jgp.107.5.559. 10.1085/jgp.107.5.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward LJ, Sandoval GM, Cannon SC. Defective slow inactivation of sodium channels contributes to familial periodic paralysis. Neurology. 1999;52:1447–1453. doi: 10.1212/wnl.52.7.1447. [DOI] [PubMed] [Google Scholar]
- Jurkat-Rott K, Mitrovic N, Hang C, Kouzmekine A, Iaizzo P, Herzog J, et al. Voltage-sensor sodium channel mutations cause hypokalemic periodic paralysis type 2 by enhanced inactivation and reduced current. Proc Natl Acad Sci U S A. 2000;97:9549–9554. doi: 10.1073/pnas.97.17.9549. 10.1073/pnas.97.17.9549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurman ME, Boland LM, Lin Y, Yellen G. Visual identification of individual transfected cells for electrophysiology using antibody-coated beads. Biotechniques. 1994;17:874–881. [PubMed] [Google Scholar]
- Lehmann-Horn F, Jurkat-Rott K. Voltage-gated ion channels and hereditary disease. Physiol Rev. 1999;79:1317–1372. doi: 10.1152/physrev.1999.79.4.1317. [DOI] [PubMed] [Google Scholar]
- McClatchey AI, Cannon SC, Slaugenhaupt SA, Gusella JF. The cloning and expression of a sodium channel β1-subunit cDNA from human brain. Hum Mol Genet. 1993;2:745–749. doi: 10.1093/hmg/2.6.745. [DOI] [PubMed] [Google Scholar]
- Mantegazza M, Yu FH, Catterall WA, Scheuer T. Role of the C-terminal domain in inactivation of brain and cardiac sodium channels. Proc Natl Acad Sci U S A. 2001;98:15348–15353. doi: 10.1073/pnas.211563298. 10.1073/pnas.211563298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motoike HK, Liu H, Glaaser IW, Yang AS, Tateyama M, Kass RS. The Na+ channel inactivation gate is a molecular complex: a novel role of the COOH-terminal domain. J Gen Physiol. 2004;123:155–165. doi: 10.1085/jgp.200308929. 10.1085/jgp.200308929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spampanato J, Kearney JA, de Haan G, McEwen DP, Escayg A, Aradi I, et al. A novel epilepsy mutation in the sodium channel SCN1A identifies a cytoplasmic domain for beta subunit interaction. J Neurosci. 2004;24:10022–10034. doi: 10.1523/JNEUROSCI.2034-04.2004. 10.1523/JNEUROSCI.2034-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struyk AF, Scoggan KA, Bulman DE, Cannon SC. The human skeletal muscle Na channel mutation R669H associated with hypokalemic periodic paralysis enhances slow inactivation. J Neurosci. 2000;23:8610–8617. doi: 10.1523/JNEUROSCI.20-23-08610.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi MP, Cannon SC. Enhanced slow inactivation by V445M: a sodium channel mutation associated with myotonia. Biophys J. 1999;76:861–868. doi: 10.1016/S0006-3495(99)77249-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Ji S, Zhou M, Ptacek LJ, Barchi RL, Horn R, George AL. Sodium channel mutations in paramyotonia congenita exhibit similar biophysical phenotypes in vitro. Proc Natl Acad Sci U S A. 1994;91:12785–12789. doi: 10.1073/pnas.91.26.12785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye B, Valdivia CR, Ackerman MJ, Makielski JC. A common human SCN5A polymorphism modifies expression of an arrhythmia causing mutation. Physiol Genomics. 2003;12:187–193. doi: 10.1152/physiolgenomics.00117.2002. [DOI] [PubMed] [Google Scholar]