Abstract
We have previously reported a topological model of the electrogenic Na+–HCO3− cotransporter (NBC1) in which the cotransporter spans the plasma membrane 10 times with N- and C-termini localized intracellularly. An analysis of conserved amino acid residues among members of the SLC4 superfamily in both the transmembrane segments (TMs) and intracellular/extracellular loops (ILs/ELs) provided the basis for the mutagenesis approach taken in the present study to determine amino acids involved in NBC1-mediated ion transport. Using large-scale mutagenesis, acidic and basic amino acids putatively involved in ion transport mediated by the predominant variant of NBC1 expressed in the kidney (kNBC1) were mutated to neutral and/or oppositely charged amino acids. All mutant kNBC1 cotransporters were expressed in HEK-293T cells and the Na+-dependent base flux of the mutants was determined using intracellular pH measurements with 2′,7′-bis-(carboxyethyl)-5(6)-carboxyfluorescein (BCECF). Critical glutamate, aspartate, lysine, arginine and histidine residues in ILs/ELs and TMs were detected that were essential for kNBC1-mediated Na+-dependent base transport. In addition, critical phenylalanine, serine, tyrosine, threonine and alanine residues in TMs and ILs/ELs were detected. Furthermore, several amino acid residues in ILs/ELs and TMs were shown to be essential for membrane targeting. The data demonstrate asymmetry of distribution of kNBC1 charged amino acids involved in ion recognition in putative outward-facing and inward-facing conformations. A model summarizing key amino acid residues involved in kNBC1-mediated ion transport is presented.
The electrogenic Na+–HCO3− cotransporter kNBC1 belongs to the SLC4 bicarbonate transporter family (Kurtz et al. 2004, Romero et al. 2004). SLC4 transporters mediate Cl−–HCO3− exchange, Na+–HCO3− cotransport (electrogenic and electroneutral) and Na+-driven Cl−–HCO3− exchange. These transporters share a significant sequence homology especially in their putative membrane domains (Tatishchev et al. 2003). Of the known SLC4 transporters, NBC1 (SLC4A4) and NBC4 (SLC4A5) proteins cotransport sodium and bicarbonate electrogenically. Two human variants of NBC1 have been described, kNBC1 and pNBC1 which differ in their amino terminus (Romero et al. 1997; Burnham et al. 1997; Abuladze et al. 1998, 2000). The electrogenic Na+–HCO3− cotransporter kNBC1 is highly expressed on the basolateral membrane of the renal proximal tubule where it mediates cellular efflux of bicarbonate (Schmitt et al. 1999, Bok et al. 2001). Despite its physiological importance to proximal tubule transepithelial bicarbonate absorption, the transport mechanism of kNBC1-mediated Na+–HCO3− transport is unknown.
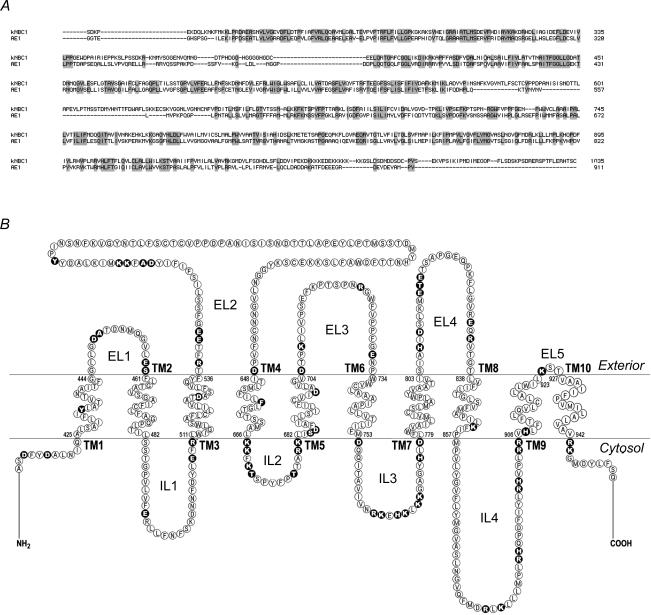
AE1 is the only member of the SLC4 family, whose transport mechanism has been studied in some detail. Despite the fact that AE1 is an anion exchanger and kNBC1 is an electrogenic Na+–HCO3− cotransporter, the two proteins are ∼30% identical at the amino acid level (Fig. 1) and therefore may share specific transport mechanisms (Kurtz et al. 2004). We have previously reported the first topological model of NBC1 (Tatishchev et al. 2003). Whereas both AE1 and kNBC1 have intracellular N- and C-termini (Alper, 2002), in contrast to AE1, which is predicted to have 13 (Fujinaga et al. 1999; Zhu et al. 2003) or 14 (Groves & Tanner, 1999; Kuma et al. 2002) transmembrane segments (TMs), kNBC1 spans the plasma membrane 10 times (Tatishchev et al. 2003). The C-terminal domain of both AE1 (Grinstein et al. 1978) and AE3 (Kopito et al. 1989) appears to be sufficient for mediating Cl−–HCO3− exchange. In addition, co-expression of complementary fragments of AE1 resulted in the generation of a stilbene disulphonate-sensitive uptake of Cl− into Xenopus oocytes (Groves & Tanner, 1995) except the pair of fragments separated within the second exofacial loop (Wang et al. 1997), suggesting that integrity of the second exofacial loop is necessary for transport activity.
Figure 1. Amino acid sequence of human kNBC1.
A, alignment of human kNBC1 and AE1. B, location of kNBC1 amino acids mutated in this study (filled circles).
It is generally accepted that AE1 facilitates Cl−–HCO3− exchange by a ping-pong mechanism (Frohlich & Gunn, 1986) wherein the transporter can either be in the inward-facing or the outward-facing state. Human AE1 has an intrinsic functional asymmetry, therefore, at near neutral pH and equal extra- and intracellular concentrations of Cl−, most of the anion exchanger molecules are in the inward-facing state (Knauf et al. 1989). The stilbene inhibitor 4,4′-diisothiocyanatodihydrostilbene-2,2′-disulphonate (DIDS) has been shown to bind preferentially to the outward-facing state (Jennings et al. 1998). In addition, H2DIDS has been shown to covalently bind Lys539 and Lys851 in human AE1 at physiological pH (Okubo et al. 1994), whereas DIDS binds to Lys851 only at pH > 12 (Kang et al. 1992). According to current AE1 topology models, both lysine residues are located in TMs (Fujinaga et al. 1999; Groves & Tanner, 1999; Kuma et al. 2002; Zhu et al. 2003). kNBC1 (Romero et al. 1997; Burnham et al. 1997) and the pancreatic form of NBC1, pNBC1 (Abuladze et al. 1998), as well as all members of the SLC4 family, with the exception of the electroneutral Na+–HCO3− cotransporter NBC3 (Pushkin et al. 1999), are stilbene-inhibitable supporting an idea of possible shared mechanism(s) of ion transport. However against this assumption is the finding that inhibition by stilbenes is not limited to the members of the SLC4 family suggesting that the mechanism of this inhibition is non-specific and involves binding of stilbene to exposed lysine residues leading to steric blocking of the normal ion transport mechanism. For example, various members of the SLC26 anion exchanger family that share no sequence homology with SLC4 transporters, are also stilbene-sensitive (Vincourt et al. 2003; Lohi et al. 2003).
A comparison of the amino acid composition of SLC4 transporters is a useful approach for identifying potential amino acid residues that could play a critical role in the kNBC1-mediated transport mechanism (Kurtz et al. 2004). We utilized this approach as an initial strategy to identify acidic and basic amino acids that were located inside, or in close proximity to, TMs selected based on our NBC1 topology model (Tatishchev et al. 2003). Using large-scale mutagenesis, acidic and basic amino acids putatively involved in ion recognition/transport were mutated to neutral and/or oppositely charged amino acids to determine the effect on kNBC1-mediated function. Furthermore, we modified amino acids in kNBC1 that are homologous to residues in AE1 previously shown to participate in Cl−–HCO3− exchange. Critical amino acids potentially involved in ion transport mediated by kNBC1 were determined.
Methods
Identification of putative amino acid residues involved in ion binding/transport mediated by kNBC1
Alignment of human members of the SLC4 family showed conserved amino acids that are potentially critical residues for kNBC1-mediated ion transport. Using our NBC1 topology model we identified which of these conserved amino acids are acidic or basic and located within or in close proximity to TMs and therefore could be potentially involved in ion binding/transport. In addition, we studied several kNBC1 amino acids homologous to residues in AE1 that have been shown to be involved in DIDS binding (Okubo et al. 1994; Salhany et al. 1995) or important for AE1-mediated ion transport (Müller-Berger et al. 1995a, b; Zhu & Casey, 2004). Amino acid residues studied in this investigation are shown in Fig. 1.
Site-directed mutagenesis
All mutations in full-length wild-type (wt) human kNBC1 were performed using a QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA). Sequences of all constructs were confirmed by bi-directional sequencing using an ABI 310 sequencer (Perkin Elmer, Foster City, CA, USA).
Immunoblotting
SDS-PAGE was performed using 7.5% polyacrylamide ready gels from Bio-Rad (Hercules, CA, USA) to determine the expression of each construct before functional measurements were performed. Proteins separated by SDS-PAGE were electrotransferred onto polyvinyl difluoride (PVDF) membrane (Amersham Biosciences, Piscataway, NJ, USA). Non-specific binding was blocked by incubation for 1 h in Tris-buffered saline (TBS) containing (mm): Tris-HCl 20 (pH 7.5), NaCl 140, and 5% dry milk and 0.05% Tween 20 (Bio-Rad). A previously well-characterised kNBC1-specific antibody (Bok et al. 2001) was used at a dilution of 1 : 1000. Secondary horseradish peroxidase-conjugated goat anti-rabbit antibody (Jackson ImmunoResearch) was used at a dilution of 1 : 20000. Bands were visualized using enhanced chemiluminesence (ECL) kit and ECL hyperfilm (Amersham Biosciences).
Cell surface biotinylation
Cell surface biotinylation experiments were performed as described previously (Li et al. 2000; Gross et al. 2001) with minor modifications to determine the surface expression of each mutant. HEK-293T cells (American Tissue Culture Collection, Manassa, VA, USA) grown on fibronectin-coated coverslips for 24 h after transfection were washed once with ice-cold borate buffer containing (mm): boric acid 10, NaCl 154, KCl 7.2 and CaCl2 1.8 mm; pH 9.0. Each coverslip was subsequently incubated with 1 ml EZ-Link NHS-SS-biotin (Pierce, Rockford, IL, USA) in borate buffer for 15 min at 0°C. Control cells were incubated with borate buffer for 15 min at 0°C. To quench the biotinylation reaction, cells were washed with 0.2 m glycine/25 mm Tris, pH 8.3, and then lysed in a solution containing 10 mm Tris-HCl (pH 7.5), 1% (w/v) deoxycholate, 1% (v/v) Triton X-100, 0.1% SDS, 150 mm NaCl and 1 mm EDTA containing 1 mm phenylmethylsulfonyl fluoride (PMSF), 1 μm leupeptin and 1 μm pepstatin (all protease inhibitors were from Roche, Indianapolis, IN, USA). Following centrifugation at 18 000 g for 10 min, the supernatant was incubated with 0.1 ml streptavidin resin (Pierce) for 1 h at 4°C. The supernatant was removed and analysed by SDS-PAGE and immunoblotting with the kNBC1-specific antibody. No detectable amount of kNBC1 was found in the supernatant in any experiments. The beads were washed with the lysis buffer. Proteins were eluted from the beads with 1 × SDS Laemmli buffer and analysed by SDS-PAGE and immunoblotting.
Immunocytochemistry
All kNBC1 constructs were examined by immunofluorescence microscopy to complement the membrane biotinylation studies in assessing membrane expression. Human embryonic kidney HEK-293T cells (American Type Culture Collection, Manassas, VA, USA) were grown at 37°C, 5% CO2, in Dulbecco's modified Eagle's medium (DMEM)-supplemented with 10% fetal bovine serum, 200 mg l−1l-glutamine and penicillin-streptomycin cocktail (Gemini Bio-Products, Calabasas, CA, USA). Twenty-four hours before transfection, a 90% confluent 10-cm polystyrene plate (Becton Dickinson, Franklin Lakes, NJ, USA) of cells was split 1 : 4 onto a 10-cm polystyrene plate with 12 ml medium that was then immediately divided into a six-well plate (2 ml well−1) containing fibronectin-coated coverslips (Discovery Labware, Bedford, MA, USA). Twenty-four hours later, the six-well plate was transfected with purified plasmids (1 μg μl−1; Qiagen, Santa Clarita, CA, USA) using the standard calcium phosphate method. The transfection medium was removed after 16 h and replaced with fresh medium. After 5 h, the coverslips were rinsed twice with 1 × PBS and then incubated with 1 ml 4% paraformaldehyde for 2 min followed by 1 ml methanol (20°C) for 2 min. The cells were then rinsed twice with 1 × PBS and processed for examination by immunofluorescence microscopy. The kNBC1-specific antibody was applied at 1 : 100 dilution in PBS for 1 h at 20 °C. After several washes in PBS, goat anti-rabbit IgG conjugated with Cy3 (1 : 500 dilution; Jackson ImmunoResearch, West Grove, PA, USA) was applied for 1 h at room temperature. The slides were rinsed in PBS and mounted in Crystal/Mount (Biomeda, Foster City, CA, USA). A PXL charge-coupled device camera (model CH1; Photometrics, USA), coupled to a Nikon Microphot-FXA epifluorescence microscope was used to capture and digitize the fluorescence images for image acquisition.
Na+ -dependent base flux mediated by kNBC1
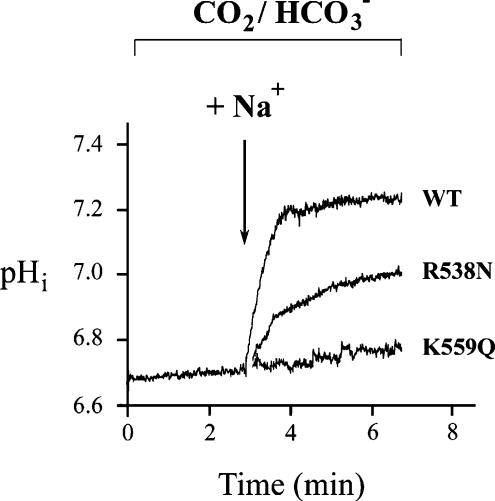
HEK-293T cells were grown on fibronectin-coated coverslips and were transiently transfected using the calcium phosphate precipitation method with a pcDNA3.1 plasmid (Invitrogen, Carlsbad, CA, USA) containing the coding region for kNBC1. Mock-transfected cells were transfected with the vector alone. The plasmids were purified with the Endofree plasmid purification kit (Qiagen) prior to their use. Functional studies were performed 24 h post transfection. Intracellular pH was monitored using the fluorescent probe BCECF (Molecular Probes, Eugene, OR, USA) and a microflourometer coupled to the microscope (Sassani et al. 2002). Data were obtained from ∼20 cells per coverslip. A minimum of five different coverslips were studied for each construct. Calibration of intracellular BCECF was performed at the end of every experiment by monitoring the 500/440-nm fluorescence excitation ratio at various values of intracellular pH (pHi) in the presence of high-K+ nigericin standards. The cells were initially bathed for 25 min in a Na+-free, Cl−-containing Hepes-buffered solution containing (mm): TMACl 140, K2HPO4 2.5, CaCl2 1, MgCl2 1, and glucose 5; pH 7.4. The cells were acutely acidified by exposure to HCO3−-buffered Na+-free, Cl−-containing solution containing (mm): tetramethyl ammonium chloride (TMACl) 115, K2HPO4 2.5, CaCl2 1, MgCl2 1, glucose 5 and TMAHCO3 25; pH 7.4. The cells were then exposed to a HCO3−-buffered Na+- and Cl−-containing solution containing (mm): NaCl 115, K2HPO4 2.5, CaCl2 1, MgCl2 1, glucose 5 and NaHCO3 25; pH 7.4, and the initial rate (initial 15 s) of pHi recovery was calculated. All solutions contained 5′(N-ethyl-N-isopropyl) amiloride (EIPA) (5 μm) to block endogenous Na+–H+ exchange. Intrinsic cell buffer capacity (βi) was measured as previously described (Kurtz et al. 1994). In HCO3−-containing solutions, the total cell buffer capacity (βT) was equal to βi plus the HCO3− buffer capacity calculated as 2.3 ×[HCO3−]i. Base flux through the cotransporter was calculated as dpHi/dt×βT, where dpHi/dt represents the initial rate of change of pHi after the addition of the Na+-containing solution. Typical traces demonstrating the time course of pHi changes in HEK-293T cells expressing wt-kNBC1 or mutant-kNBC1 constructs before and after sodium addition are shown in Fig. 2.
Figure 2. Functional characterization of wt-kNBC1 and mutant-kNBC1.
Constructs were expressed in HEK-293T cells.
Statistics
Results are reported as mean ± s.e.m. Dunnett's t test was used when more than one experimental group was compared. A value of P < 0.05 was considered statistically significant.
Results
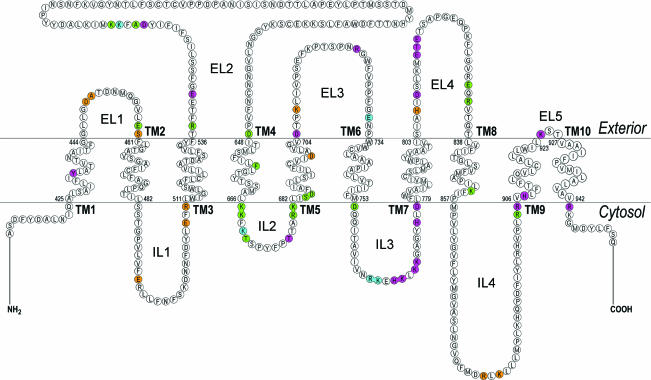
The data corresponding to specific amino acid substitutions are shown in Figs 2–5 and Tables 1–3. Substitution of acidic/basic amino acids with neutral or oppositely charged amino acids in internal (ILs) and external loops (ELs) flanking TMs or located in very close proximity to TMs significantly decreased kNBC1-mediated function or blocked membrane targeting of the cotransporter (E459Q, S460L, E508N, R510D, R538D, D647R, K667E, K668N, R680E, K681N, D705N, K708Q, D754R, H776N, D778N, H807N, R833Q, R904E and K924N).
Figure 5. Amino acid mutations, which had a severe (< 25% of wt-kNBC1 function,  ), moderate (25–60% of wt-kNBC1 function,
), moderate (25–60% of wt-kNBC1 function,  ) and a small (> 60% of wt-kNBC1 function,
) and a small (> 60% of wt-kNBC1 function,  ) effect on kNBC1 function.
) effect on kNBC1 function.
Designation of TMs, ELs and ILs is same as in Fig. 4. The residues that are required for plasma membrane expression are shown in brown.
Table 1.
Effect of mutating residues in the intracellular loops, N- and C-termini on kNBC1 function
| Plasmid | pHi minimum | Total buffer capacity (mm pH−1) | Na+-dependent base flux (mm min−1) | Na+-dependent base flux (% of wt) |
|---|---|---|---|---|
| wt-kNBC1 | 6.68 ± 0.19 | 38.1 ± 2.08 | 22.5 ± 1.84 | 100 ± 8.21 |
| Vector | 6.70 ± 0.06 | 39.4 ± 5.69 | 0 ± 0.05* | 0* |
| D416N | 6.65 ± 0.02 | 35.5 ± 2.21 | 19.5 ± 1.38 | 86.7 ± 6.15 |
| D419N | 6.65 ± 0.03 | 34.5 ± 2.66 | 24.4 ± 1.69 | 108 ± 7.52 |
| K667R | 6.69 ± 0.19 | 39.0 ± 8.62 | 23.8 ± 3.61 | 106 ± 16.0 |
| K667E | 6.65 ± 0.28 | 34.6 ± 2.49 | 4.16 ± 0.32* | 18.5 ± 1.40* |
| K667N | 6.67 ± 0.02 | 34.1 ± 2.53 | 8.27 ± 1.04* | 36.8 ± 4.64* |
| K667Q | 6.67 ± 0.07 | 35.1 ± 5.95 | 5.79 ± 0.48* | 25.8 ± 2.14* |
| K668N | 6.71 ± 0.02 | 36.1 ± 1.82 | 6.82 ± 0.62* | 30.4 ± 2.78* |
| 667KK-667NN | 6.71 ± 0.03 | 36.7 ± 2.47 | 2.27 ± 0.19* | 10.1 ± 0.86* |
| 667KK-667QQ | 6.70 ± 0.02 | 34.1 ± 1.53 | 0 ± 0.04* | 0* |
| K670Q | 6.69 ± 0.03 | 38.7 ± 3.10 | 16.0 ± 2.30* | 71.1 ± 10.0* |
| T671N | 6.54 ± 0.02 | 35.8 ± 1.72 | 3.10 ± 0.19* | 13.8 ± 0.87* |
| T677G | 6.74 ± 0.04 | 40.4 ± 2.98 | 12.9 ± 1.52* | 57.5 ± 6.76* |
| R680Q | 6.65 ± 0.02 | 36.8 ± 1.34 | 21.0 ± 1.97 | 93.0 ± 8.74 |
| R680E | 6.62 ± 0.02 | 34.3 ± 2.56 | 5.35 ± 0.30* | 23.8 ± 1.33* |
| K681N | 6.72 ± 0.05 | 35.5 ± 0.04 | 9.96 ± 0.64* | 44.3 ± 2.82* |
| 680RK-680NN | 6.69 ± 0.02 | 35.1 ± 1.44 | 4.94 ± 0.48* | 22.0 ± 2.12* |
| D754E | 6.51 ± 0.02 | 35.4 ± 1.99 | 5.26 ± 0.43* | 23.3 ± 1.90* |
| D754N | 6.69 ± 0.04 | 37.8 ± 5.92 | 1.94 ± 0.16* | 8.64 ± 0.74* |
| D754R | 6.66 ± 0.07 | 35.0 ± 7.37 | 0 ± 0.27* | 0* |
| 764RK-764NN | 6.67 ± 0.14 | 38.4 ± 5.66 | 14.2 ± 1.47* | 63.2 ± 6.53* |
| 767HK-767NN | 6.68 ± 0.06 | 39.4 ± 5.73 | 21.3 ± 3.87 | 94.7 ± 17.2 |
| 770KK-770NN | 6.67 ± 0.26 | 37.8 ± 2.19 | 9.29 ± 0.73* | 41.3 ± 3.25* |
| 767HKLKK- 767NNLNN | 6.68 ± 0.03 | 38.1 ± 2.82 | 8.97 ± 0.90* | 39.9 ± 4.00* |
| H776D | 6.70 ± 0.03 | 35.1 ± 2.90 | 18.0 ± 1.10* | 80.0 ± 4.91* |
| H776N | 6.75 ± 0.02 | 40.5 ± 3.76 | 12.2 ± 0.86* | 54.1 ± 5.83* |
| H776S | 6.75 ± 0.03 | 38.3 ± 3.14 | 9.56 ± 0.75* | 42.5 ± 3.33* |
| H776R | 6.70 ± 0.03 | 38.1 ± 2.82 | 8.97 ± 0.90* | 39.9 ± 4.00* |
| D778N | 6.64 ± 0.24 | 35.2 ± 2.41 | 5.77 ± 0.24* | 25.6 ± 1.07* |
| 890KH-890NN | 6.72 ± 0.03 | 38.4 ± 3.07 | 25.0 ± 1.48 | 111 ± 6.60 |
| 899RH-899NN | 6.75 ± 0.04 | 38.6 ± 4.32 | 25.1 ± 1.79 | 112 ± 7.97 |
| R904E | 6.59 ± 0.02 | 35.3 ± 4.23 | 5.45 ± 0.27* | 24.2 ± 1.47* |
| R905E | 6.60 ± 0.02 | 34.2 ± 0.40 | 10.8 ± 1.02* | 48.1 ± 4.55* |
| 904RR-904QQ | 6.74 ± 0.02 | 37.4 ± 2.44 | 19.1 ± 2.85* | 85.0 ± 12.7 |
| R943N | 6.68 ± 0.04 | 37.2 ± 3.63 | 8.74 ± 1.01* | 38.9 ± 4.60* |
| R943E | 6.56 ± 0.07 | 37.0 ± 9.70 | 12.9 ± 1.47* | 57.3 ± 6.53* |
P < 0.05 versus wt-kNBC1.
Table 3.
Effect of mutating residues in the extracellular loops on kNBC1 function
| Plasmid | pHi minimum | Total buffer capacity (mm pH−1) | Na+-dependent base flux (mm min−1) | Na+-dependent base flux (% of wt) |
|---|---|---|---|---|
| E459Q | 6.67 ± 0.04 | 34.5 ± 2.94 | 2.02 ± 0.12* | 8.99 ± 0.55* |
| R538N | 6.73 ± 0.04 | 37.8 ± 4.19 | 11.5 ± 0.92* | 51.1 ± 4.19* |
| R538Q | 6.66 ± 0.02 | 34.1 ± 1.75 | 4.79 ± 0.36* | 21.3 ± 1.61* |
| E541Q | 6.78 ± 0.04 | 38.2 ± 2.65 | 19.3 ± 2.26 | 85.7 ± 10.10 |
| E542Q | 6.63 ± 0.02 | 38.8 ± 1.45 | 7.55 ± 0.08* | 33.6 ± 0.36* |
| D555N | 6.49 ± 0.04 | 38.8 ± 3.61 | 12.8 ± 1.04* | 56.9 ± 4.62* |
| A556T | 6.61 ± 0.04 | 34.6 ± 3.35 | 1.68 ± 0.14* | 7.48 ± 0.64* |
| K558Q | 6.54 ± 0.03 | 35.4 ± 2.81 | 12.0 ± 0.96* | 53.4 ± 4.27* |
| K559Q | 6.70 ± 0.03 | 39.8 ± 3.25 | 0.97 ± 0.05* | 4.30 ± 0.22* |
| 558KK-558QQ | 6.77 ± 0.03 | 41.0 ± 2.98 | 3.28 ± 0.24* | 14.6 ± 1.06* |
| Y567H | 6.61 ± 0.05 | 35.6 ± 1.81 | 20.6 ± 1.88 | 91.8 ± 8.34 |
| D647R | 6.62 ± 0.02 | 34.7 ± 0.42 | 2.43 ± 0.21* | 10.8 ± 0.95* |
| D705N | 6.68 ± 0.19 | 35.6 ± 1.58 | 8.82 ± 0.99* | 39.2 ± 4.40* |
| R722Q | 6.65 ± 0.04 | 38.2 ± 1.72 | 10.8 ± 1.30* | 48.1 ± 5.78* |
| E731N | 6.77 ± 0.02 | 42.0 ± 2.28 | 16.4 ± 1.48* | 72.9 ± 6.56* |
| D809N | 6.66 ± 0.02 | 35.2 ± 1.82 | 11.4 ± 0.75* | 50.7 ± 3.30* |
| K812T | 6.76 ± 0.03 | 36.4 ± 3.48 | 20.4 ± 3.11 | 90.6 ± 13.9 |
| 814ETE-814MSK | 6.68 ± 0.19 | 37.0 ± 3.07 | 7.18 ± 0.20* | 31.9 ± 1.86* |
| G828E | 6.65 ± 0.07 | 36.0 ± 3.45 | 25.8 ± 1.84 | 114.7 ± 8.18 |
| E831Q | 6.68 ± 0.02 | 35.0 ± 2.23 | 7.22 ± 0.82* | 32.1 ± 3.63* |
| 831EQR-831QQQ | 6.63 ± 0.03 | 35.6 ± 1.85 | 0.71 ± 0.04* | 3.17 ± 0.18* |
| K924Q | 6.78 ± 0.02 | 41.6 ± 2.28 | 8.11 ± 0.44* | 36.1 ± 1.96* |
P < 0.05 versus wt-kNBC1.
In general, mutations of acidic/basic amino acids located in loop segments more distant to TMs (D555N, K558Q, Y567H, K670Q, R722Q and E731N, and in addition 764RK to 764NN and 770KK to 770NN motifs) had a smaller effect on kNBC1 function. It is important to note that specific mutant constructs that were an exception to this tendency included E542Q, K559Q, 558KK to 558QQ, 767HKLKK to 767NNLNN and 831EQR to 831QQQ, wherein kNBC1-mediated transport was significantly decreased. D449N, E492Q, 764RKEHKLKK to 764NNENNLNN, and 881RLK to 881NLN prevented plasma membrane targeting of the cotransporter.
Certain amino acids in kNBC1 homologous to residues in AE1 that were previously shown to participate in anion exchanger function provided an additional rationale for mutational analysis. kNBC1-Lys559 (AE1-Lys539) and kNBC1-Lys924 (AE1-Lys851) located in EL1 and EL5, respectively (Fig. 1B) have been shown to participate in AE1-mediated transport and also covalently bind DIDS (Okubo et al. 1994; Müller-Berger et al. 1995b). Moreover, these two amino acids are conserved in all members of the SLC4 family. Importantly, in the K559Q mutant, kNBC1 function was completely inhibited, and decreased by ∼65% in the K924Q mutant (Table 3). Substitution of residues located in close proximity to Lys559 (D555N and A556T) also significantly decreased kNBC1 function, supporting the findings of Müller-Berger et al. (1995b) that the AE1 homologue of kNBC1-D555 (AE1-D535) participates in anion exchanger function. The specificity of Lys559 for kNBC1 transport function was further supported by mutating neighbouring Lys558 to glutamine. As shown in Table 3, unlike the K559Q mutant, the function of the K558Q mutant was only partially inhibited.
Various charged and uncharged residues in kNBC1 were specific for Na+-dependent members of the SLC4 family and differ in their respective positions in AE transporters. The effect of substitution of these residues to homologous residues in AE proteins was also examined. In addition to basic amino acids located in IL2, two threonine residues, Thr671 and Thr677, when mutated to asparagine and glycine located in homologous positions in AE1 demonstrated significantly decreased transport as a percentage of control (∼14% and ∼58%, respectively; Fig. 5; Table 1). However, substitution of residues specific for Na+-dependent SLC4 members with residues present at homologous sites in AE proteins was also found in certain instances to affect membrane targeting. Specifically, when Ala450 and Ser460 were mutated to their homologous residues present in AE proteins (lysine and leucine, respectively), plasma membrane expression was blocked (Fig. 5).
The TMs have a significantly smaller number of charged amino acids than ILs and ELs of kNBC1. Specifically, there are only five charged amino acids within kNBC1 TMs (Fig. 1B). Asp685, Asp699 and His907 are conserved in the SLC4 family whereas Asp528 and Lys854 are not conserved. Three of these residues, Asp685, Lys854 and His907 located in TM5, TM8 and TM9, respectively, were important for kNBC1 function (Table 2). Specifically, the D685R, D685N, K854E and H907D mutants had approximately 29, 23, 14 and 40% of normal function, respectively. In addition, H907R and H907N mutants were not targeted to the plasma membrane. Furthermore, we were not able to determine the effect of conserved Asp699 in TM5 on cotransporter function because both a D699K and D699N mutant also had impaired plasma membrane targeting (Fig. 5). Substitution of non-conserved Asp528 (TM3) to arginine did not affect cotransporter function.
Table 2.
Effect of mutating residues in transmembrane segments on kNBC1 function
| Plasmid | pHi minimum | Total buffer capacity (mm pH−1) | Na+-dependent base flux (mm min−1) | Na+-dependent base flux (% of wt) |
|---|---|---|---|---|
| Y433F | 6.58 ± 0.05 | 34.2 ± 1.69 | 12.1 ± 1.73* | 54.0 ± 7.70* |
| 433Y797Y-433F797F | 6.60 ± 0.04 | 34.0 ± 0.68 | 12.7 ± 1.03* | 56.4 ± 4.58* |
| D528N | 6.64 ± 0.03 | 36.0 ± 1.67 | 20.0 ± 2.50 | 89.0 ± 11.1 |
| D528R | 6.56 ± 0.02 | 38.9 ± 1.50 | 25.6 ± 1.49 | 114.3 ± 6.63 |
| F656M | 6.64 ± 0.03 | 37.0 ± 2.72 | 5.27 ± 0.33* | 23.4 ± 1.46* |
| S684G | 6.63 ± 0.02 | 34.9 ± 1.56 | 2.20 ± 0.18* | 9.79 ± 0.80* |
| D685N | 6.71 ± 0.02 | 37.5 ± 2.06 | 5.24 ± 0.32* | 23.3 ± 1.44* |
| D685R | 6.58 ± 0.02 | 34.4 ± 0.74 | 6.45 ± 0.43* | 28.7 ± 1.93* |
| K854E | 6.61 ± 0.02 | 34.7 ± 1.65 | 3.15 ± 0.16* | 14.0 ± 1.21* |
| H907D | 6.71 ± 0.02 | 37.2 ± 1.70 | 8.97 ± 0.79* | 39.9 ± 3.50* |
P < 0.05 versus wt-kNBC1.
In addition to acidic/basic amino acids located in TM3, TM5, TM8 and TM9 we analysed several non-charged amino acids in these and other TMs as candidate residues forming an ion-binding site. The involvement of non-charged amino acids in substrate binding has been shown for several transporters including lactose permease of E. coli (DeFelice, 2004). Two amino acids Phe656 (TM4) and Ser684 (TM5) that are conserved in Na+-dependent members of the SLC4 family were replaced with methionine and glycine, respectively, present in homologous positions in AEs. These amino acids were substituted with residues present in homologous positions in AE proteins. Both the F656M and S684G mutants demonstrated significantly decreased kNBC1-mediated transport (Table 2) indicating that these amino acids are critical for kNBC1 function. In addition, substitution of Tyr433 (TM1) which is conserved in the SLC4 family to phenylalanine decreased kNBC1 function by ∼50%. The latter finding complements the data of Dinour et al. (2004) who characterized a naturally occurring NBC1 S427L mutation also located in TM1. Finally, unlike Tyr433 in TM1, non-conserved Tyr797 in TM7 was not critical as its substitution with phenylalanine did not affect the function of the cotransporter.
Both N- and C-termini of kNBC1 are located in the cytoplasm and therefore could be directly or indirectly involved in cotransporter-mediated ion transport. It was previously shown (Igarashi et al. 1999) that the R298S mutant in the N-terminus of human kNBC1 has ∼55% of normal function. The C-terminus of human NBC1 is involved in binding of carbonic anhydrase II (Pushkin et al. 2004), which is important for the cotransporter function (Gross et al. 2001, 2002, 2003; Gross & Kurtz, 2002). We performed additional experiments to evaluate the involvement of acidic/basic amino acids in both termini flanking or located in close proximity to TM1 and TM10. Mutation of Arg943 in the C-terminus of kNBC1 flanking TM10 to glutamate and asparagine decreased the cotransporter function to ∼57% and ∼39% of normal, respectively (Table 1). Asp416 and Asp419 in the N-terminus of kNBC1 when mutated to asparagine did not affect kNBC1-mediated transport.
Discussion
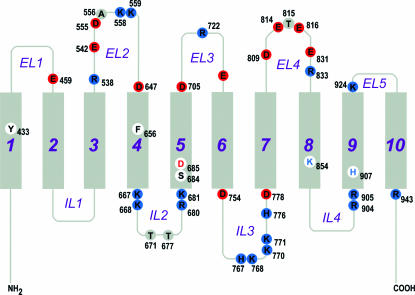
This report represents the first large-scale mutagenesis study of kNBC1 (as well as of any Na+-dependent HCO3− transporter) where critical amino acid residues involved in the ion transport process have been successfully characterized. Prior to the current study, the only data available to provide potential mechanistic information regarding kNBC1 cotransporter function were based on mutations in the human SLC4A4 gene resulting in proximal renal tubular acidosis and ocular abnormalities (Igarashi et al. 1999; Dinour et al. 2004). Based on our NBC1 topology model we were able to categorize all critical amino acid residues spatially into two groups (Figs 4 and 5). Group 1 residues were amino acids located within TMs, and therefore potentially involved in ion translocation/binding. Group 2 residues were amino acids located outside TMs, and therefore potentially involved in ion selectivity. The data in this study provided strong evidence for a model detailing: (1) amino acid residues in ELs and ILs potentially involved in an ion selectivity filter (ISF) for Na+ and HCO3− in inward-facing and outward-facing conformations; and (2) amino acids in TMs involved in ion translocation/binding site (ITS). In this transport model, ions interact initially with residues in the ISF and subsequently with the ITS. A similar ISF has been previously described in AE1 that acts as a substrate channel leading from the aqueous medium to a separate transport site located in TMs (Falke & Chan, 1986b). The model complements a kinetic analysis of kNBC1-mediated transport in mouse proximal tubule cells (Gross & Hopfer, 1998).
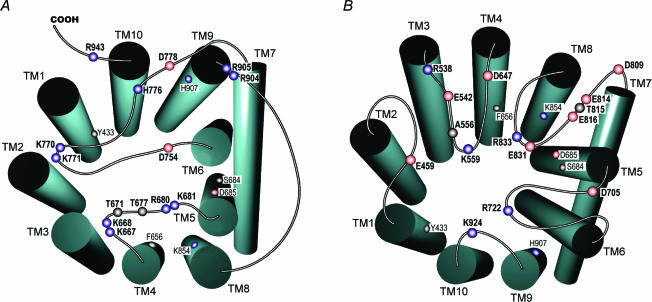
Figure 4. Critical amino acid residues in kNBC1 that determine its functional properties.
Grey rectangles represent transmembrane segments (TMs). Extracellular loops are designated as EL1–EL5, and intracellular loops are designated as IL1–IL4. Acidic, basic and neutral amino acid residues are shown in red, blue and grey, respectively.
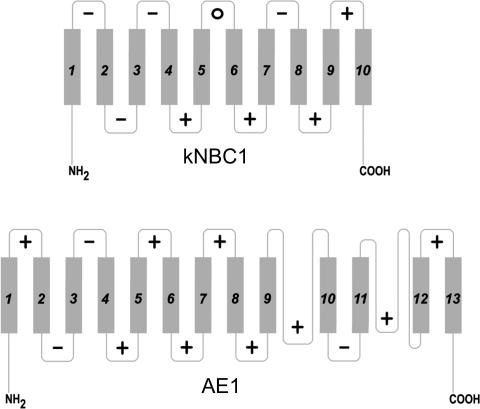
Given that AE1 mediates exchange of negatively charged chloride and bicarbonate ions and kNBC1 cotransports negatively charged bicarbonate and positively charged sodium, one would expect a priori that the ISF in AE1 would be composed mostly of positively charged residues, whereas in kNBC1 the ISF would also contain negatively charged amino acids that interact with Na+. It is interesting that a global analysis of the net charge (pI) of ELs and ILs in AE1 and kNBC1 based on current topological models yielded fundamentally different results (Fig. 6). Specifically, the majority of human AE1 ELs and ILs are positively charged at physiological pH. In contrast, three of five ELs in kNBC1 (EL1, EL2 and EL4) have an acidic pI, whereas only EL5 has basic pI (Fig. 6). EL5 consisted of only three amino acids 924KST and its positive charge was provided by Lys924, an analogue of human AE1-Lys851 shown to interact with DIDS (Okubo et al. 1994; Salhany et al. 1995). Three of four kNBC1 ILs (IL2, IL3 and IL4) have an alkaline pI and IL1 has acidic pI. The asymmetric distribution of specific negatively and positively charged residues in the ELS and ILS of kNBC1 could play an important role in optimizing the interaction of the cotransporter with Na+ and HCO3− in the inward-facing and outward-facing conformation.
Figure 6. Isoelectric point (pI) of ELs and ILs of human kNBC1 and AE1.
–, pI below 7; +, pI above 7; 0, pI ∼7.
Several basic lysine and arginine residues in ILs 2–4 shown in this study to be important for kNBC1-mediated transport, are apparently involved in the ISF in the inward-facing conformation (Fig. 7A). His776 whose AE1 homologue has been hypothesized to be involved in the anion exchange mechanism (Müller-Berger et al. 1995b) was also shown in this study to be extremely important for kNBC1-mediated transport. kNBC1-His776 may play a role of proton donor as its substitution with arginine decreased function by ∼60%, whereas in the H776D mutant kNBC1 function was only decreased by ∼20%. In addition, Asp754 and Asp778 flanking TM6 and TM7, respectively, and Thr671 and Thr677 located in IL2 are also involved in the ISF. Asp778 is conserved in the SLC4 family and glutamate is located in the position homologous to Asp754 in AE transporters. AE1-Glu681 homologous to kNBC1-Asp754 has been implicated in the AE transport mechanism (Müller-Berger et al. 1995b). When mouse AE1-Glu699 (human AE1-Glu681) was mutated to glutamine, the exchanger lost Cl−–Cl− exchange transport activity and demonstrated electrogenic SO42−–Cl− exchange in Xenopus oocytes (Chernova et al. 1995).
Figure 7. Amino acids in ILs and in ELs involved in ISF of kNBC1.
A, amino acids in ILs involved in ISF in the inward-facing conformation of kNBC1. B, amino acids in ELs involved in ISF in the outward-facing conformation of kNBC1. Amino acids involved in the ITS are shown with a smaller font in A and B.
Several amino acids located in EL1–5 are apparently a part of the ISF in the kNBC1 outward-facing conformation (Fig. 7B). Asp647 and Asp705 flanking TM4 and TM5, respectively, are the only acidic amino acids flanking TMs extracellularly. Our results demonstrated that the local negative charge at Asp647 and Asp705 is required for the cotransporter to function normally. It is important that Asp647 is not conserved in all members of the SLC4 family and is notably absent from the anion exchangers AE1–3. Of interest, the analogous residue is present in AE4 suggesting that AE4 is Na+-dependent as has been reported in preliminary studies by Parker et al. (2002). Acidic Asp647 flanking the junction of EL2 and TM4, is potentially involved in the interaction of external Na+ with kNBC1. Asp705 is conserved in electrogenic Na+–HCO3− cotransporters suggesting that this residue is involved in the electrogenicity of NBC1- and NBC4-mediated transport. Acidic Glu459 in EL1 is conserved among members of the SLC4 family. Substitution of this residue with glutamine significantly decreased kNBC1 function (Table 3). In the topology model of AE1 (Zhu et al. 2003), the analogous residue Glu439 is located within the second TM unlike kNBC1. There are currently no studies addressing whether AE1-Glu439 plays a role in AE1-mediated anion exchange. Differential topographic localization of kNBC1-Glu459 compared with AE1 suggests that this residue is required for Na+ recognition in the outward-facing conformation. In addition, Arg538, Glu542, Asp555, Asp705, Asp809, Glu814, Glu816, Glu831 and Arg833 could also be involved in the ISF in the outward-facing conformation.
Although Asp555 is not conserved in all SLC4 members, acidic charge conservation at this site may play an important transport role as suggested by the involvement of homologous Glu535 in the human AE1 transport (Müller-Berger et al. 1995b). The importance of this site is highlighted by the fact that adjacent residue, Ala556, when mutated to threonine inhibited kNBC1 function to a significantly greater degree (Table 3). Asp555 is located in close proximity to Lys559, which is very important for kNBC1 function (Table 3). It is also important that the homologous residue in AE1, Lys539, covalently binds DIDS (Okubo et al. 1994; Salhany et al. 1995). Lys559 is located in EL2, the largest EL of NBC1 whereas in AE1 this region is located in TM5. It has been shown that AE1-Lys539 is not a part of the AE1 Cl− binding site (Falke & Chan, 1986a; Salhany et al. 1994; Salhany, 2001). Mutations of Lys539 and Lys891 in the DIDS binding site significantly reduced the DIDS affinity to AE1 but did not significantly alter the Km for chloride self-exchange (Wood et al. 1992). As the mutation of human kNBC1-Lys559 and Lys924 significantly inhibited the cotransporter function, it is clear that these residues play a principally different role than the homologous Lys539 and Lys891 residues in human AE1. The data suggest that Lys559 and Lys924 are involved in HCO3− recognition in the outward-facing conformation of kNBC1. These kNBC1 residues, located extracellularly are potential targets for DIDS inhibition. As stilbenes were shown to inhibit kNBC1 when applied both extra- and intracellularly (Heyer et al. 1999), intracellularly located Lys667, Lys668 and Lys681 adjacent to TM4 and TM5 together with Lys854 in TM8 could be targets of DIDS. Alternatively Lys559 (and possibly Lys924) could be accessible to DIDS in the inward-facing conformation. This assumption requires that Lys559 (and Lys924) migrates from the extracellular compartment to a location near the cotransporter ITS in the inward-facing conformation to stabilize this conformation by potentially interacting with negatively charged amino acids (e.g. Asp647 or/and Asp705). This interaction could also prevent access of ions to the ITS from the extracellular compartment. It should be noted that the Lys559 homologue in AE1 (Lys539) is located in TM5 and therefore could be more easily accessible from both sides than kNBC1-Lys559. However, inhibition of AE1 by DIDS has been reported only from the extracellular compartment (Kaplan et al. 1976; Cabantchik & Greger, 1992). Asp555, Ala556 and Lys558 probably assist Lys559 and Lys924 in HCO3− recognition. The region where these amino acids are located in kNBC1-EL2 is homologous to AE1 TM5. We have specifically shown that the kNBC1 amino acid residues from 539 to 564 do not possess any membrane insertion activity (Tatishchev et al. 2003). The second half of EL2 containing naturally glycosylated Asn597 and Asn617 (Choi et al. 2003) is located extracellularly. The movement of Lys924 should be more constrained than Lys559 because of the small size of EL5 where this residue is located. Nevertheless because Lys924 is located very close to the plasma membrane, its small movement into the plane of the membrane could be enough to maintain binding of bicarbonate to both lysines and to make Lys924 accessible to intracellular DIDS.
When Arg510 in the first IL1 was mutated to aspartate, plasma membrane targeting of kNBC1 was blocked. It should be mentioned that a patient has been described wherein a mutation of kNBC1-Arg510 to histidine caused renal proximal tubular acidosis with ocular abnormalities (Igarashi et al. 1999). The authors demonstrated that the function of the R510H mutant expressed in ECV304 cells was decreased by 43%. In the current study, mutation of Arg510 to acidic aspartic acid completely prevented plasma membrane expression of the cotransporter. Abnormal plasma membrane targeting of the R510H mutant may underlie the transport defect in these patients. Mutations of two additional charged amino acids in this loop, including conserved Glu492 (E492Q) and Glu508 (E508N), also blocked plasma membrane targeting. These data indicate that IL1 plays an important role in plasma membrane expression of kNBC1. EL1 is highly homologous in the SLC4 family and like IL1 is important for membrane targeting, as mutations of Asp449, Ala450 and Ser460 blocked kNBC1 plasma membrane expression.
In AE1, it has been hypothesized that charged lysine, histidine and glutamic acid residues lining the ITS have the potential to form a chain of hydrogen bonds traversing the thickness of the plasma membrane (Müller-Berger et al. 1995a, b). In addition to its different topological structure and different pattern of DIDS inhibition, kNBC1 has significantly fewer charged amino acids in its TMs than AE1 (Fig. 1) suggesting that some differences exist between these two members of the SLC4 family in their transport mechanisms. There are five acidic/basic amino acids in kNBC1 TMs (Tatishchev et al. 2003). Human AE1 homologues of these amino acids are also located in TMs. One acidic residue (Asp685) and two basic residues (Lys854 and His907) were shown in this study to be involved it kNBC1 function. Asp685 (TM5) and His907 (TM9) are conserved in members of the SLC4 family and therefore are likely to participate in translocation of bicarbonate. A human AE1 homologue of kNBC1-His907 (His834) has been hypothesized to participate in AE1-mediated ion transport (Müller-Berger et al. 1995b). Lys854 (TM8) that is not conserved in AE transporters but is conserved in Na+-dependent members of the SLC4 family, could therefore play a role in the Na+ binding of the cotransporter. Phe656 in TM4 and Ser684 in TM5 specific for Na+-dependent SLC4 transporters also could participate in Na+ translocation whereas Tyr433 that is conserved in the SLC4 family could participate in HCO3− translocation.
Our results suggest that TMs 1, 4, 5, 8 and 9 participate in the ITS of kNBC1. The importance of TM8 and TM9 is supported by the data of Inatomi et al. (2004) who described a new kNBC1 mutation (deletion of adenine 2311) leading to permanent isolated renal tubular acidosis because of a frame shift at codon 721 eliminating the last five TMs. Xenopus oocytes transfected with this mutant did not demonstrate any electrogenic Na+–HCO3− cotransport. The importance of TM1 is supported by the data of Dinour et al. (2004) who described a kNBC1-S427L mutation with significantly decreased kNBC1 function. We cannot exclude involvement of additional TMs in the ITS, for example TM6 and TM7, as EL3, EL4 and IL3 flanking these TMs have residues that are very important for kNBC1 function.
Based on our results, we have developed a preliminary structural and mechanistic model for kNBC1-mediated transport that provides a basis for future study. Our model is based on the assumptions that: (1) the kNBC1 monomer can mediate Na+–HCO3− cotransport; (2) the cotransporter exists in inward-facing or outward-facing conformations (Wright, 2001; Forster et al. 2002; DeFelice, 2004); and (3) simultaneous binding of both Na+ and HCO3− ions is necessary for the ion transport in both directions (Jentsch et al. 1986; Gross & Hopfer, 1998).
In our model the inward-facing conformation is stabilized by basic Lys559 in EL2 and possibly Lys924 in EL5 that form salt bridges with acidic Asp647 and Asp705 flanking TM4 and TM5 and prevent extracellular ion binding to the ITS. HCO3− and Na+ transferred through the ISF comprised of amino acids located in IL2–4 subsequently are translocated to the ITS (Ser427, Tyr433, Phe656, Ser684, Asp685, Lys854, His907 and potentially Asp699). The order of ion binding is currently unknown. When both Na+ and HCO3− are bound to the ITS, the cotransporter switches to an outward-facing conformation and the ions are released extracellularly. This change in conformation destroys the salt bridges between Lys559, Lys924, Asp647 and Asp705. Electrostatic interactions between acidic Asp754 and Asp778 with basic lysine and arginine residues flanking IL4 probably stabilize this conformation. In the outward-facing confirmation Na+ and HCO3− ions are transferred to the translocation site and are subsequently released intracellularly. The cotransporter then returns to its inward-facing conformation and is ready for another transport cycle. The kNBC1 amino acid residues important for Na+ and HCO3− transport are shown in Fig. 7.
For simplicity, our model does not include interaction of kNBC1 with carbonic anhydrase II, which has been shown to bind to the C-terminus of the cotransporter and increase kNBC1-mediated transport (Gross & Kurtz, 2002; Gross et al. 2002, 2003; Pushkin et al. 2004). It has been shown that kNBC1 is expressed on the plasma membrane of HEK-293T cells as a mixture of monomers, dimers and tetramers (Pushkin et al. 2001). Oligomers of anion exchangers AE1 (Clarke, 1975; Nakashima & Makino, 1983; Schubert et al. 1983; Casey & Reithmeier, 1991; Dodler et al. 1993; Wang et al. 1993) and AE3 (Pushkin et al. 2000) have been previously described, and it is currently accepted that the active form of AE1 is a dimer (Alper, 2002). Oligomerization could be a characteristic feature of SLC4 transporters (DeFelice, 2004). Additional studies are required to resolve whether oligomerization of kNBC1 is necessary and/or modulates its properties.
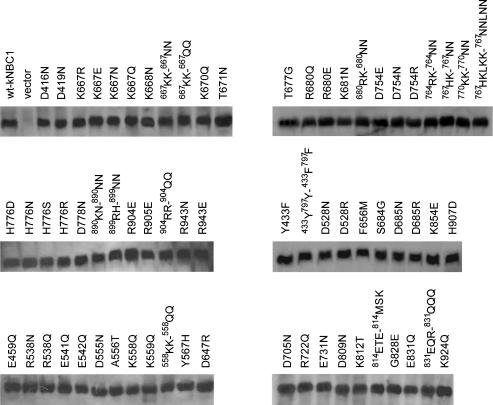
Figure 3. Determination of wt-kNBC1 and mutant-kNBC1 construct plasma membrane expression in HEK-293T cells.
Cell surface biotinylation and immunoblotting with kNBC1-specific antibody was used. Shown are the constructs that were studied functionally.
Acknowledgments
This work was supported by National Institutes of Health grants DK63125, DK58563 and DK07789, the Max Factor Family Foundation, the Richard and Hinda Rosenthal Foundation, the Fredricka Taubitz Fund, the National Kidney Foundation of Southern California J891002 and AHA (Western State Affiliate) grant 0365022Y.
References
- Abuladze N, Lee I, Newman D, Hwang J, Boorer K, Pushkin A, Kurtz I. Molecular cloning, chromosomal localization, tissue distribution, and functional expression of the human pancreatic sodium bicarbonate cotransporter. J Biol Chem. 1998;273:17689–17695. doi: 10.1074/jbc.273.28.17689. [DOI] [PubMed] [Google Scholar]
- Abuladze N, Song M, Pushkin A, Newman D, Lee I, Nicholas S, Kurtz I. Structural organization of the human NBC1 gene: kNBC1 is transcribed from an alternative promoter in intron 3. Gene. 2000;251:109–122. doi: 10.1016/s0378-1119(00)00204-3. [DOI] [PubMed] [Google Scholar]
- Alper SL. Genetic deseases of acid-base transporters. Annu Rev Physiol. 2002;64:899–923. doi: 10.1146/annurev.physiol.64.092801.141759. [DOI] [PubMed] [Google Scholar]
- Bok D, Schibler MJ, Pushkin A, Sassani P, Abuladze N, Naser Z, Kurtz I. Immunolocalization of electrogenic sodium-bicarbonate cotransporters pNBC1 and kNBC1 in the rat eye. Am J Physiol Renal Physiol. 2001;281:F920–F935. doi: 10.1152/ajprenal.2001.281.5.F920. [DOI] [PubMed] [Google Scholar]
- Burnham CE, Amlal H, Wang Z, Shull GE, Soleimani M. Cloning and functional expression of a human kidney Na+:HCO3− cotransporter. J Biol Chem. 1997;272:19111–19114. doi: 10.1074/jbc.272.31.19111. [DOI] [PubMed] [Google Scholar]
- Cabantchik ZI, Greger R. Chemical probes for anion transporters of mammalian cell membranes. Am J Physiol. 1992;262:C803–C827. doi: 10.1152/ajpcell.1992.262.4.C803. [DOI] [PubMed] [Google Scholar]
- Casey JR, Reithmeier RAF. Analysis of the oligomeric state of band 3, the anion transport protein of the human erythrocyte membrane, by size exclusion high performance liquid chromatography. J Biol Chem. 1991;266:15726–15737. [PubMed] [Google Scholar]
- Chernova MN, Jiang L, Crest M, Hand M, Vandorpe DH, Strange K, Alper SL. Electrogenic sulfate/chloride exchange in Xenopus oocytes mediated by murine AE1 E699Q. J Gen Physiol. 1995;109:345–360. doi: 10.1085/jgp.109.3.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi I, Hu L, Rojas JD, Schmidtt BM, Boron WF. Role of glycosylation in the renal electrogenic Na+–HCO3− cotransporter (NBCe1) Am J Physiol Renal Physiol. 2003;284:F1199–F1206. doi: 10.1152/ajprenal.00131.2002. [DOI] [PubMed] [Google Scholar]
- Clarke S. The size and detergent binding of membrane proteins. J Biol Chem. 1975;250:5459–5469. [PubMed] [Google Scholar]
- DeFelice LJ. Transporter structure and mechanism. Trends Neurosci. 2004;27:352–359. doi: 10.1016/j.tins.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Dinour D, Chang M-H, Satoh J-I, Smith BL, Angle N, Knecht A, Serban I, Holtzman EJ, Romero MF. A novel missense mutation in the sodium bicarbonate cotransporter (NBCe1/SLC4A4) causes proximal tubular acidosis and glaucoma through ion transport defects. J Biol Chem. 2004;279:52238–52246. doi: 10.1074/jbc.M406591200. [DOI] [PubMed] [Google Scholar]
- Dodler M, Walz T, Hefti A, Engel A. Human erythrocyte band 3. Solubilization and reconstitution into two-dimensional crystals. J Mol Biol. 1993;231:119–132. doi: 10.1006/jmbi.1993.1261. [DOI] [PubMed] [Google Scholar]
- Falke JJ, Chan SI. Molecular mechanism of band 3 inhibitors. 1. Transport site inhibitors. Biochemistry. 1986a;25:7888–7894. doi: 10.1021/bi00372a015. [DOI] [PubMed] [Google Scholar]
- Falke JJ, Chan SI. Molecular mechanism of band 3 inhibitors. 2. Channel blockers. Biochemistry. 1986b;25:7895–7898. doi: 10.1021/bi00372a016. [DOI] [PubMed] [Google Scholar]
- Forster IC, Köhler K, Biber J, Murer H. Forcing the link between structure and function of electrogenic cotransporters: the renal type IIa Na+/Pi cotransporter as a case study. Prog Biophys Mol Biol. 2002;80:69–108. doi: 10.1016/s0079-6107(02)00015-9. [DOI] [PubMed] [Google Scholar]
- Frohlich O, Gunn RB. Erythrocyte anion transport: the kinetics of a single-site obligatory exchange system. Biochim Biophys Acta. 1986;864:169–194. doi: 10.1016/0304-4157(86)90010-9. [DOI] [PubMed] [Google Scholar]
- Fujinaga J, Tang XB, Casey JR. Topology of the membrane domain of human erythrocyte anion exchange protein, AE1. J Biol Chem. 1999;274:6626–6633. doi: 10.1074/jbc.274.10.6626. [DOI] [PubMed] [Google Scholar]
- Grinstein S, Ship S, Rothstein A. Anion transport in relation to proteolytic dissection of band 3 protein. Biochim Biophys Acta. 1978;507:294–304. doi: 10.1016/0005-2736(78)90424-8. [DOI] [PubMed] [Google Scholar]
- Gross E, Fedotoff O, Pushkin A, Abuladze N, Newman D, Kurtz I. Phosphorylation-induced modulation of pNBC1 function: distinct roles for the amino- and carboxy-termini. J Physiol. 2003;549:673–682. doi: 10.1113/jphysiol.2003.042226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross E, Hawkins K, Pushkin A, Abuladze N, Hopfer U, Sassani P, Dukkipati R, Kurtz I. Phosphorylation of Ser982 in kNBC1 shifts the HCO3−: Na+ stoichiometry from 3:1 to 2:1 in proximal tubule cells. J Physiol. 2001;537:659–665. doi: 10.1111/j.1469-7793.2001.00659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross E, Hopfer U. Voltage and cosubstrate dependence of the Na-HCO3 cotransporter kinetics in renal proximal tubule cells. Biophys J. 1998;75:810–824. doi: 10.1016/S0006-3495(98)77570-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross E, Kurtz I. Structural determinants and significance of regulation of electrogenic Na+-HCO3− cotransporter stoichiometry. Am J Physiol Renal Physiol. 2002;283:F876–F887. doi: 10.1152/ajprenal.00148.2002. [DOI] [PubMed] [Google Scholar]
- Gross E, Pushkin A, Abuladze N, Fedotoff O, Kurtz I. Regulation of the sodium bicarbonate cotransporter kNBC1 function: role of Asp986, Asp988 and kNBC1-carbonic anhydrase II binding. J Physiol. 2002;544:679–685. doi: 10.1113/jphysiol.2002.029777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves JD, Tanner MJ. Co-expressed complementary fragments of the human red cell anion exchanger (band 3, AE1) generate stilbene disulfonate-sensitive anion transport. J Biol Chem. 1995;270:9097–9105. doi: 10.1074/jbc.270.16.9097. [DOI] [PubMed] [Google Scholar]
- Groves JD, Tanner MJ. Structural model for the organization of the transmembrane spans of the human red-cell anion exchanger (band 3; AE1) Biochem J. 1999;344:699–711. [PMC free article] [PubMed] [Google Scholar]
- Heyer M, Müller-Berger Romero MF, Boron WF, Frömter E. Stoichiometry of the rat kidney Na+-HCO3− cotransporter expressed in Xenopus laevis oocytes. Eur J Physiol. 1999;438:322–329. doi: 10.1007/s004240050916. [DOI] [PubMed] [Google Scholar]
- Igarashi T, Inatomi J, Sekine T, Cha SH, Kanai Y, Kunimi M, et al. Mutations in SLC4A4 cause permanent isolated proximal tubular acidosis with ocular abnormalities. Nat Genet. 1999;23:264–266. doi: 10.1038/15440. [DOI] [PubMed] [Google Scholar]
- Inatomi J, Horita S, Braverman N, Sekine T, Yamada H, Suzuki Y, et al. Mutational and functional analysis of SLC4A4 in a patient with proximal renal tubular acidosis. Eur J Physiol. 2004;448:438–444. doi: 10.1007/s00424-004-1278-1. [DOI] [PubMed] [Google Scholar]
- Jennings ML, Whitlock J, Shinde A. Pre-steady state transport by erythrocyte band 3 protein: uphill countertransport induced by the impermeant inhibitor H2DIDS. Biochem Cell Biol. 1998;76:807–813. doi: 10.1139/bcb-76-5-807. [DOI] [PubMed] [Google Scholar]
- Jentsch TJ, Schwartz P, Schill B, Langner B, Lepple AP, Keller SK, Wiederholt M. Kinetic properties of the sodium bicarbonate (carbonate) symport in monkey kidney epithelial cells (BSC–1). Interaction between Na+, HCO3− and pH. J Biol Chem. 1986;23:10673–10679. [PubMed] [Google Scholar]
- Kang D, Okubo K, Hamasaki N, Kuroda N, Shiraki H. A structural study of the membrane domain of band 3 by tryptic digestion. Conformational change of band 3 in situ induced by alkali treatment. J Biol Chem. 1992;267:19211–19217. [PubMed] [Google Scholar]
- Kaplan JH, Scorah K, Fasold H, Passow H. Sidedness of the inhibition action of disulfonic acids on chloride equilibrium exchange and net transport across the human erythrocyte membrane. FEBS Lett. 1976;62:182–185. doi: 10.1016/0014-5793(76)80048-8. [DOI] [PubMed] [Google Scholar]
- Knauf PA, Spinelly LJ, Mann NA. Flufenamic acid senses conformation and asymmetry of human erythrocyte band 3 anion transport protein. Am J Physiol. 1989;257:C277–C289. doi: 10.1152/ajpcell.1989.257.2.C277. [DOI] [PubMed] [Google Scholar]
- Kopito RR, Lee BS, Simmons DM, Lindsay AE, Morgans CW, Schneider K. Regulation of intracellular pH by a neuronal homolog of the erythrocyte anion exchanger. Cell. 1989;59:927–937. doi: 10.1016/0092-8674(89)90615-6. [DOI] [PubMed] [Google Scholar]
- Kuma H, Shinde AA, Howren TR, Jennings ML. Topology of the anion exchange protein AE1: the controversial sidedness of lysine 743. Biochemistry. 2002;41:3380–3388. doi: 10.1021/bi015879p. [DOI] [PubMed] [Google Scholar]
- Kurtz I, Nagami G, Yanagawa N, Li L, Emmons C, Lee I. Mechanism of apical and basolateral Na+-independent Cl−/base exchange in the rabbit superficial proximal straight tubule. J Clin Invest. 1994;94:173–183. doi: 10.1172/JCI117304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz I, Petrasek D, Tatishchev S. Molecular mechanisms of electrogenic sodium bicarbonate cotransport: structural and equilibrium thermodynamic considerations. J Membr Biol. 2004;197:77–90. doi: 10.1007/s00232-003-0643-x. [DOI] [PubMed] [Google Scholar]
- Li J, Quilty J, Popov M, Reithmeier RA. Processing of N-linked oligosaccharide depends on its location in the anion exchanger, AE1, membrane glycoprotein. Biochem J. 2000;349:51–57. doi: 10.1042/0264-6021:3490051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohi H, Lamprecht G, Markovich D, Heil A, Kujala M, Seidler U, Kere J. Isoforms of SLC26A6 mediate anion transport and have functional PDZ interaction domains. Am J Physiol Cell Physiol. 2003;284:C769–C779. doi: 10.1152/ajpcell.00270.2002. [DOI] [PubMed] [Google Scholar]
- Müller-Berger S, Karbach D, Kang D, Aranibar N, Wood PG, Ruterjans H, Passow H. Roles of histidine 752 and glutamate 699 in the pH dependence of mouse band 3 protein-mediated anion transport. Biochemistry. 1995a;34:9325–9332. doi: 10.1021/bi00029a007. [DOI] [PubMed] [Google Scholar]
- Müller-Berger S, Karbach D, Konig J, Lepke S, Wood PG, Appelhans H, Passow H. Inhibition of mouse erythroid band 3-mediated chloride transport by site-directed mutagenesis of histidine residues and its reversal by second site mutation of Lys 558, the locus of covalent H2DIDS binding. Biochemistry. 1995b;34:9315–9324. doi: 10.1021/bi00029a006. [DOI] [PubMed] [Google Scholar]
- Nakashima H, Makino S. Purification and characterization of band 3, the major intrinsic membrane protein of the bovine erythrocyte membrane. J Biochem. 1983;87:899–910. doi: 10.1093/oxfordjournals.jbchem.a132820. [DOI] [PubMed] [Google Scholar]
- Okubo K, Kang D, Hamasaki N, Jennings ML. Red blood cell band 3. Lysine 539 and lysine 851 react with the same H2DIDS (4,4′-diisothiocyanodihydrostilbene-2,2′-disulfonic acid) molecule. J Biol Chem. 1994;269:1918–1926. [PubMed] [Google Scholar]
- Parker MD, Boron WF, Tanner MJ. Characterization of human ‘AE4’ as an electroneutral sodium bicarbonate cotransporter. FASEB J. 2002;16:A796. [Google Scholar]
- Pushkin A, Abuladze N, Gross E, Newman D, Tatishchev S, Lee I, Fedotoff O, Bondar G, Azimov R, Ngyuen M, Kurtz I. kNBC1 and CAII form a transport metabolon in renal proximal tubule cells. J Physiol. 2004;559:55–65. doi: 10.1113/jphysiol.2004.065110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushkin A, Abuladze N, Lee I, Newman D, Hwang J, Kurtz I. Cloning, tissue distribution, genomic organization, and functional characterization of NBC3, a new member of the sodium bicarbonate cotransporter family. J Biol Chem. 1999;274:16569–16575. doi: 10.1074/jbc.274.23.16569. [DOI] [PubMed] [Google Scholar]
- Pushkin A, Sassani P, Abuladze N, Newman D, Tatishchev S, Kurtz I. Oligomeric structure of electrogenic sodium bicarbonate cotransporters. J Am Soc Nephrol. 2001;12:8A. [Google Scholar]
- Pushkin AV, Tsuprun VL, Abuladze NK, Newman D, Kurtz I. Oligomeric structure of bAE3 protein. IUBMB Life. 2000;50:397–401. doi: 10.1080/713803739. [DOI] [PubMed] [Google Scholar]
- Romero MF, Fulton CM, Boron WF. The SLC4 family of HCO3− transporters. Eur J Physiol. 2004;447:495–509. doi: 10.1007/s00424-003-1180-2. [DOI] [PubMed] [Google Scholar]
- Romero MF, Hediger MA, Boulpaep EL, Boron WF. Expression cloning and characterization of a renal electrogenic Na+/HCO3− cotransporter. Nature. 1997;387:409–413. doi: 10.1038/387409a0. [DOI] [PubMed] [Google Scholar]
- Salhany JM. Mechanistic basis for site–site interactions in inhibitor and substrate binding to band 3 (AE1): evidence distinguishing allosteric from electrostatic effects. Blood Cells Mol Dis. 2001;27:901–912. doi: 10.1006/bcmd.2001.0464. [DOI] [PubMed] [Google Scholar]
- Salhany JM, Schopfer LM, Kay MMB, Gamble DN, Lawrence C. Differential sensitivity of stilbenedisulfonates in their reaction with band 3 HT (Pro-868 → Leu) Proc Natl Acad Sci U S A. 1995;92:11844–11848. doi: 10.1073/pnas.92.25.11844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salhany JM, Sloan RL, Cordes KA, Schopfer LM. Kinetic evidence for ternary complex formation and allosteric interactions in chloride and stilbenedisulfonate binding to band 3. Biochemistry. 1994;33:11909–11916. doi: 10.1021/bi00205a029. [DOI] [PubMed] [Google Scholar]
- Sassani P, Pushkin A, Gross E, Gomer A, Abuladze N, Dukkipati R, Carpenito G, Kurtz I. Functional characterization of NBC4: a new electrogenic sodium bicarbonate cotransporter. Am J Physiol Cell Physiol. 2002;282:C408–C416. doi: 10.1152/ajpcell.00409.2001. [DOI] [PubMed] [Google Scholar]
- Schmitt BM, Biemesderfer D, Romero MF, Boulpaep EL, Boron WF. Immunolocalization of the electrogenic Na+-HCO3− cotransporter in mammalian and amphibian kidney. Am J Physiol. 1999;276:F27–F38. doi: 10.1152/ajprenal.1999.276.1.F27. [DOI] [PubMed] [Google Scholar]
- Schubert D, Boss K, Dorst H-J, Flossdorf J, Pappert G. The nature of the stable noncovalent dimers of band 3 protein from erythrocyte membranes in solutions of Triton X-100. FEBS Lett. 1983;163:81–84. doi: 10.1016/0014-5793(83)81168-5. [DOI] [PubMed] [Google Scholar]
- Tatishchev S, Abuladze N, Pushkin A, Newman D, Liu W, Weeks D, Sachs G, Kurtz I. Identification of membrane topography of the electrogenic sodium bicarbonate cotransporter pNBC1 by in vitro transcription/translation. Biochemistry. 2003;42:755–765. doi: 10.1021/bi026826q. [DOI] [PubMed] [Google Scholar]
- Vincourt JB, Jullien D, Amalric F, Girard JP. Molecular and functional characterization of SLC26A11, a sodium-independent sulfate transporter from high endothelial venules. FASEB J. 2003;17:890–892. doi: 10.1096/fj.02-0787fje. [DOI] [PubMed] [Google Scholar]
- Wang DN, Kühlbrandt W, Sarabia VE, Reithmeier RAF. Two-dimensional structure of the membrane domain of human band 3, the anion transport protein of the erythrocyte membrane. EMBO J. 1993;12:2233–2239. doi: 10.1002/j.1460-2075.1993.tb05876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Groves JD, Mawby WJ, Tanner MJA. Complementation studies with co-expressed fragments of the human red cell anion transporter (band 3; AE1). The role of some exofacial loops in anion transport. J Biol Chem. 1997;272:10631–10638. doi: 10.1074/jbc.272.16.10631. [DOI] [PubMed] [Google Scholar]
- Wood PG, Muller H, Sovak M, Passow H. Role of Lys 558 and Lys 869 in substrate and inhibitor binding to the murine band 3 protein: a study of the effects of site-directed mutagenesis of the band 3 protein expressed in the oocytes of Xenopus laevis. J Membr Biol. 1992;127:139–148. doi: 10.1007/BF00233286. [DOI] [PubMed] [Google Scholar]
- Wright EM. Renal Na+-glucose transporters. Am J Physiol Renal Physiol. 2001;280:F10–F18. doi: 10.1152/ajprenal.2001.280.1.F10. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Casey JR. The substrate anion selectivity filter in the human erythrocyte Cl−/HC03− exchange protein, AE1. J Biol Chem. 2004;279:23565–23573. doi: 10.1074/jbc.M401380200. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Lee DW, Casey JR. Novel topology in C-terminal region of the human plasma membrane anion exchanger, AE1. J Biol Chem. 2003;278:3112–3120. doi: 10.1074/jbc.M207797200. [DOI] [PubMed] [Google Scholar]