Allelism is one of the most striking characteristics of the S locus, which controls self-incompatibility (SI) of flowering plants. The deceptively simple biology of SI requires some degree of allelism: styles reject those pollen grains that express an S allele that they themselves express. Even though a population expressing gametophytic SI can theoretically persist with only three S alleles, natural populations generally contain many more.
How do new S alleles evolve? Despite progress in the identification of genes involved in SI, answers to this apparently straightforward question remain elusive. Attempts to change the specificity of an S allele by mutation or meiotic recombination have been unsuccessful. The most likely explanation for this failure is that the S locus contains at least two genes: a style gene that encodes a factor to disable incompatible pollen and a pollen gene that encodes a factor to control recognition of the disabling style factor. Because mutations that alter allelic specificity while preserving allelic recognition are unlikely to arise simultaneously in both genes, S alleles have probably arisen by stepwise changes, first in one gene and then in the other, with self-incompatibility presumably not an intermediate state. The conceptual challenge has therefore been to describe a pathway in which a new specificity might evolve such that each step maintains allelic recognition and all intermediates are self-incompatible.
In a recent research article in THE PLANT CELL, Matton et al. (1999) describe an experimentally produced style factor that rejects pollen bearing either of two S alleles. The authors argue that such a dual-specificity style factor may play a pivotal role in the generation of new S alleles, and suggest a pathway in which all intermediates are self-incompatible. Here, we consider the evolutionary fate of new S alleles that arise by this pathway and argue that selection would eliminate them from the population. We propose alternative scenarios that would permit the maintenance of new S alleles.
In Solanum chacoense, the species studied by Matton et al. (1999), the style factor is an extracellular ribonuclease (the S RNase) and the pollen factor is an unknown molecule commonly called pollen S. In the following discussion, we refer to the genes that encode these factors as A and B, respectively, and designate particular alleles by integer subscripts. For example S allele S1 corresponds to haplotype A1B1, in which the pollen S encoded by B1 causes recognition of the S RNase encoded by A1. We assume that selection disfavors self-fertilization and removes from the population mutations that disrupt recognition between A and B of the same haplotype. It is important to note that allele and haplotype are not used here as synonymous terms: mutations that change a haplotype but preserve allelic recognition may segregate in the population as neutral variants. Positive selection to maintain such intermediates need not be invoked as Matton et al. (1999) appear to do.
Mutually distinct S alleles may arise through coordinated mutations in A and B. For example, haplotype A1B1 may give rise to A2B2 through mutation in A followed by mutation in B (pathway I: A1B1→ A1B2→ A2B2) or in the reverse order (pathway II: A1B1→ A2B1→ A2B2). The model of Matton et al. (1999) resembles pathway I, with the addition of an extra step in which a (dual-specificity) style factor recognizes two different pollen factors. In our nomenclature, we represent this dual-specificity factor as A1,2 and the proposed pathway as A1B1 → A1,2 B1→ A1,2 B2→ A2B2. By regarding A1,2 as a neutral variant of A1, we subsume this pathway under pathway I.
In pathway I, positive selection of gametophytic SI requires that A1 be recognized by both B1 and B2 (i.e., A1 is a dual-specificity style factor) and that B2 recognize both A1 and A2 (i.e., B2 is a dual-specificity pollen factor). Because A2 and B1 have never occurred in the same haplotype, selection has not constrained their interaction. Consequently, B1 pollen tubes may fail to recognize the A2 style factor, permitting compatibility between A1B1 pollen and styles carrying A2B2. In contrast, because B2 arose in an A1 haplotype, styles expressing A1 reject A2B2 pollen.
Alternatively, in pathway II, A2 is retained only if B1 recognizes A2 in addition to A1, and B2 is retained only if it recognizes A2. Because A1 and B2 have never occurred in the same haplotype, A2B2 may possibly fertilize a style carrying A1B1, whereas the converse may not occur.
Table 1 summarizes the compatibility relationships among the haplotypes in the two pathways. Both pathways show asymmetric compatibility between pairs of haplotypes: it is the original haplotype A1B1 that can pollinate styles expressing the derived form A2B2 in pathway I, whereas the converse holds in pathway II.
Table 1.
Cross-Compatibility between Haplotypes Expressed in Style and Pollen
| Pathway I | ||||
|---|---|---|---|---|
| Pollen
|
||||
| Style
|
A1B1
|
A1B2
|
A2B2
|
|
| A1B1 | −a | − | ||
| A1B2 | − | − | ||
| A2B2 | +b | − | ||
| Pathway II | ||||
| Pollen
|
||||
| Style
|
A1B1
|
A2B1
|
A2B2
|
|
| A1B1 | − | + | ||
| A2B1 | − | − | ||
| A2B2 | − | − | ||
(−) denotes incompatibility.
(+) denotes compatibility.
A simple argument shows that, in the absence of any selective forces other than the expression of gametophytic SI, haplotypes that escape rejection by haplotypes that they themselves reject drive the latter to extinction. First, consider that half of the gene pool in any generation is derived from parental egg cells and half from parental pollen cells. Each gene can be expected, assuming Mendelian segregation of mating type alleles, to transmit one copy of itself to the offspring generation through an egg cell, whereas transmission through pollen depends on access to compatible mates.
Let pi denote the frequency of the S locus haplotype i within any given generation; pi′, the frequency of i in the subsequent generation, will then be
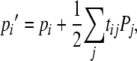 |
(1) |
where tij denotes the rate of production of pollen bearing Si by pollen incompatibility class j (i.e., 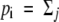 tij). Pj represents the pollination success of class j. Because pollen incompatibility class is determined under gametophytic SI by the S allele carried by the pollen itself, tii corresponds to pi, the frequency of Si in pollen, with tij equal to zero for all i different from j. Equation 1 reduces under gametophytic SI to
tij). Pj represents the pollination success of class j. Because pollen incompatibility class is determined under gametophytic SI by the S allele carried by the pollen itself, tii corresponds to pi, the frequency of Si in pollen, with tij equal to zero for all i different from j. Equation 1 reduces under gametophytic SI to
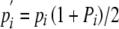 |
(2) |
We use pi to denote the frequency of the ith haplotype among the k haplotypes derived from and including the original S allele A1B1. For example, in pathway I these haplotypes include A1B1, A1B2, and A2B2, so that k equals three and i ranges between one and three. Suppose that pollen carrying a certain haplotype (arbitrarily designated α) can fertilize styles carrying at least one haplotype in this group, but that the reciprocal cross is incompatible. Some number of other S alleles, fully functionally distinct from this group of haplotypes and from each other, also segregate in the population, each with frequency q.
Equation 2 determines evolutionary changes in the frequencies of all haplotypes:
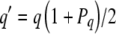 |
(3) |
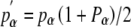 |
(4) |
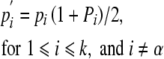 |
(5) |
If haplotype α can nonreciprocally fertilize a group of styles that includes at least one other haplotype derived from A1B1, its pollination success exceeds that of other haplotypes in the group (Pα > Pi). Consequently, as long as this advantage in transmission through pollen accrues to haplotype α, it increases relative to other members of the group (pα′/pi′ > pα /pi for 1⩽ i ⩽ k, and i ≠ α.).
Evolution favors style component mutations that expand the set of pollen factor alleles rejected by the style factor and favors pollen component mutations that restrict the style factors recognized. In pathway I, haplotype A1B1 is expected to cause the extinction of the new haplotypes, whereas in pathway II, the derived haplotype A2B2 is expected to replace A1B1. This analysis suggests that, in the pathway proposed by Matton et al. (1999), the new haplotype A2B2 can enter the population only if the original haplotype A1B1 were no longer present. In the absence of A1B1, however, the new haplotype A2B2 would simply segregate as a neutral variant of the intermediate A1B2 rather than constitute a functionally distinct S allele.
During the course of evolution, mutations in both the pollen and style components may arise, undergoing extinction or substitution as a consequence of genetic drift and selection. Preliminary studies of our model indicate that the rate of fission of S allele lineages, corresponding to the coexistence of functionally distinct S haplotypes derived from a common ancestral haplotype, depends strongly on population structure. In particular, subdivision into a number of partially isolated demes in which alternative descendant haplotypes may undergo substitution and subsequent evolution enhances the rate of S allele diversification.
References
- Matton, D.P., Luu, D.T., Qin, X., Laublin, G., O'Brien, M., Maes, O., Morse, D., and Cappadocia, M. (1999). Production of an S RNase with dual specificity suggests a novel hypothesis for the generation of new S alleles. Plant Cell 11 2087–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]


