Abstract
Dividing cells, including human cancers, organize processes necessary for their duplication according to circadian time. Recent evidence has shown that disruption of central regulation of circadian rhythms can increase the rate at which a variety of cancers develop in rodents. In order to study circadian rhythms in liver tumors, we have chemically induced hepatocellular carcinoma in transgenic rats bearing a luciferase reporter gene attached to the promoter of a core circadian clock gene (Period 1). We explanted normal liver cells and hepatomas, placed them into short-term culture, and precisely measured their molecular clock function by recording light output. Results show that isolated hepatocellular carcinoma is capable of generating circadian rhythms in vitro. Temporally restricting food availability to either day or night altered the phase of the rhythms in both healthy and malignant tissue. However the hepatomas were much less sensitive to this signal resulting in dramatically different phase relationships between host and tumor tissue as a function of mealtime. These data support the conclusion that hepatoma is differentially sensitive to circadian timing signals, although it maintains the circadian organization of the non-malignant cells from which it arose. Because circadian clocks are known to modulate the sensitivity of many therapeutic cytotoxic targets, controlling mealtiming might be used to increase the efficacy of treatment. Specifically, meal and treatment schedules could be designed that take advantage of coincident times of greatest tumor sensitivity and lowest sensitivity of host tissue to damage.
Keywords: Cancer, circadian rhythm, peripheral oscillator
INTRODUCTION
Circadian rhythmicity provides a critically important temporal framework for many molecular, cellular and organismal functions. Cell division1-3, immune function4 and behavior5 are all regulated by endogenous circadian clocks that are synchronized to the 24h environment by external cues. The molecular mechanism driving the cell-autonomous circadian rhythm is comprised of a number of ‘clock’ genes that when disrupted, alter or abolish clock function6. Virtually all cells and tissues have the molecular machinery capable of generating circadian rhythms7 8.
Dividing cells organize all of their processes necessary for their duplication within circadian time9. Even hepatocytes, cells which ordinarily do not divide, use circadian timing to organize mitotic activity during recovery from an injury10, 11. This organized cell division is also observed in human cancers12, 13. Therefore it is not surprising that organizing treatment with cytotoxic agents according to circadian time in some cases improves drug tolerance14and outcome15.
Recent reports suggest that experimental manipulation of host circadian rhythmicity affects tumor growth and mortality in rodents with cancer. Arrhythmicity induced by suprachiasmatic nucleus (SCN) lesions16, 17 or chronic jet-lag18, 19 in mice, or chronic bright light in rats20 increases the rate of tumor growth and hastens death. Disruption of the clock gene Period 2 increases both spontaneous and radiation-induced tumor frequency21. Conversely, enhancement of host rhythms can inhibit tumor progression. Circadian rhythms in liver function and circadian clock gene expression synchronize to food-restriction schedules22, 23 and mice on daytime restricted feeding exhibit slower tumor development than mice fed at night or ad-libitum19, 24. Therefore it is important to understand the biological mechanism(s) that underlie the relationship between circadian rhythms in healthy and malignant hepatic tissue and the progression of cancer. In this study we examined expression patterns of a circadian clock gene, Period 1, in hepatocellular carcinoma (HCC) and healthy host tissues following photic resetting and timed food restriction in transgenic Per1-luciferase rats.
METHODS
Subjects and experimentally induced carcinogenesis
Male and female 6-week old transgenic rats bearing a luciferase reporter gene for mPer125 were subjects in the experiment. Except where noted, all rats were housed in a 12:12 light-dark cycle (lights on from 0500h-1700h). Animals were administered diethylnitrosamine (DENA) (N-Nitrosodiethylamine; Sigma N0258-1G) for six weeks in drinking water at a concentration of 80mg/l for an estimated daily dose of 10mg/kg20. Following the 6-week treatment rats were allowed 4 days to clear the DENA, then placed in a clean cage and transferred to our behavioral facility where they participated in the following two experiments.
Experiment I: Restricted feeding
Immediately following DENA treatment, 23 rats were placed on one of 3 feeding regimens: 12g of chow at Zeitgeber Time (ZT) 04:00 (ZT 00:00 = lights-on; ZT 12:00 = lightsoff) (RF Day); 12g of chow at ZT 16:00 (RF Night), or ad libitum (AL) chow. Rats were fed less food for the first 3 days of RF to insure they ate all the chow that was provided within a few hours. Food restriction was continued until rats were sacrificed. Body weights for all animals were assessed weekly and did not differ statistically among groups (Initial weights [Mean ± SEM]: AL 385 ± 21g, RF Day 391.9 ± 23.3g, RF Night 356.3 ± 12.8g; Single factor ANOVA p = 0.45; Final weight measured during the last week all animals were alive: AL 475.3 ± 19.8g, RF Day 437.4 ± 14.9g, RF Night 414.5 ± 4.4g; Single Factor ANOVA p=0.08). Rats were sacrificed for culture preparation at ZT 11:00 if they failed to eat their daily meal (indicating illness) (n=8; cultures were made from 7 of these rats) or after 9 weeks of restricted feeding, whichever came first. Fifteen of 23 animals survived until this point of the experiment; of these 15, 4 rats had no macroscopic liver lesions. The remaining rats, including those that were sacrificed before 9 weeks of RF had elapsed, all had multiple discrete macroscopic liver foci that were white or gray in color. Histological analysis (Antech diagnostics, Lake Success, NY) verified that the lesions were hepatocellular carcinoma. Final group sizes were: Ad lib: n=8; RF Night: n=4; RF Day: n=6 for a total number of 18 rats. Since all rats had multiple foci, at least 2 hepatoma and healthy liver explant cultures were prepared from each rat. All healthy tissue explants were taken from the same DENA-treated rats, but were isolated from liver tissue that appeared macroscopically normal.
Experiment II: Experimental Jet-lag
Twenty-six transgenic rats were treated with DENA, and then remained under a 12:12 L:D cycle with ad-libitum food and water for 10 weeks. Of the 26 initial animals, 6 were removed from the protocol early due to health concerns. The 20 healthy rats that remained were then transferred to our behavioral facility and placed on a shifted light-dark cycle to simulate jet-lag conditions. Nine rats were housed in a 6h-delayed light-dark cycle (lights on between 1100h and 2300h), accomplished by lengthening the light-period by 6 hours on the day of tranfer. Separately, eight rats were housed in an advanced light-cycle (lights on between 2300 and 1100h) accomplished by decreasing the dark-period by 6 hours on the day of transfer. Only one phase-shift was performed for each rat in these groups. The remaining three rats remained in the pre-shift light regime. Subgroups of rats were then sacrificed at ZT 1100 on the first, third or sixth day in the new (or unshifted) light cycle in order to track the resetting of the circadian rhythm in both tumors and healthy liver. One rat of these 20 (Phase-delayed group) had no macroscopic lesions. The remaining 19 all had multiple foci. At least 2 tumors and 2 adjacent pieces of healthy liver were explanted into culture from each rat.
Assessment of PER1 gene expression
Rats were sacrificed by decapitation under CO2 anesthesia. Liver was exposed and samples of both healthy liver and hepatocellular carcinoma were removed and placed in chilled Hank's basic salt solution containing HEPES buffer and antibiotics. Small (1-2 mm2) slices of these tissues were made with scalpels and placed on a culture membrane (Millicell-CM PICMORG50) in a 35mm dish containing culture medium (DMEM [Sigma D2902] supplemented with B27 [2%; Gibco 15630-088], 10 mM HEPES buffer [Gibco 15630-088] and antibiotics) containing 0.1 mM luciferin [Promega E1602]. While the explants were not analyzed histologically, they were macroscopically homogeneous such that hepatoma cultures contained only tissue that was of a consistent color and texture. The dishes were sealed with cover glass and vacuum grease and placed into a Lumicycle bioluminescence recording apparatus (Actimetrics, Evanston Il) housed inside an environmental chamber held at 35.5°C. Measurements of bioluminescence (1 min. duration) were made every 10min for a minimum of 4 days.
To characterize the circadian rhythms in healthy liver and HCC explants, a detailed analysis of the waveforms was performed for tissues harvested from animals that were not subjected to experimental manipulation (ad-libitum-fed rats in Experiment I and unshifted rats in Experiment II, pooled). Briefly, a detrending algorithm (DTRNDANL) was applied to remove the considerable mean (baseline drifting) and variance (damping oscillatory amplitude) nonstationarities consistently encountered in the observed experimental data. Then FFT-NLLS analysis26, 27 was applied. This analysis permits assignment of values to parameters characterizing the period and amplitude of circadian oscillation exhibited by the data sets. Next, we applied a comparative analysis by FFT-NLLS of the oscillatory amplitude among sequentially isolated 24-hour subseries of the detrended data, followed by analysis of the rate of amplitude decay. This measure of amplitude decay, or damping rate, indicates the number of elapsed hours in vitro during which the rhythm reached or exceeded an amplitude of 30% of that measured on the first cycle 28.
These analyses resulted in means and 95% confidence limits for period, initial amplitude and rate of amplitude decay for each tissue culture. Variance-weighted averaging29, followed by Welch's t-test was then used to determine if there were differences between tumors and healthy liver tissue with respect to these rhythmic parameters.
The phase of the circadian rhythm in luciferase-reported mPer1 gene expression was determined by first detrending (subtraction of the 24h running mean from the raw data) then smoothing (2h running mean) the time-series data23, 25. The first peak in the bioluminescence data after the initial 24h in culture was used as the phase marker (Fig. 1A). In our experience data from the first 24h in culture are inconsistent due to masking by the exponential decay of initial bioluminescence. All cultures were included in the phase analysis.
Fig. 1.
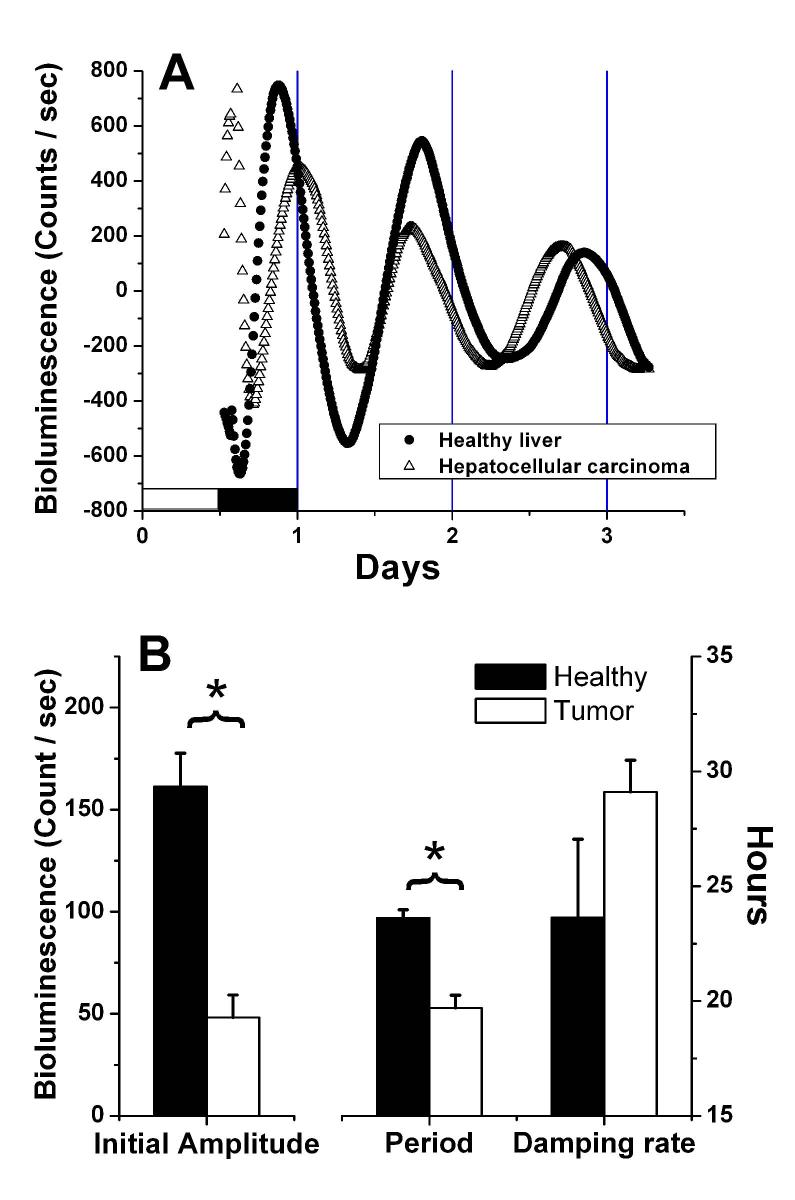
Bioluminescence measured in vitro from healthy liver and hepatocellular carcinoma tissue explants isolated from a PER1-LUC transgenic rat. A. Detrended bioluminescence counts smoothed with a 2h running average. The beginning of the record is the time of lights-on on the day of sacrifice, and the lighting cycle for that day is indicated by the white and shaded bars. The rat was killed about one hour before lights-off (ZT 11:00). B. Variance-weighted mean values for initial amplitude, circadian period and damping rate of healthy liver and hepatocellular carcinoma explants measured in vitro. Amplitude, in bioluminescence counts, is plotted on the left axis, while period and damping rate, in hours, are plotted on the right axis. * p<0.0001; Two-tailed Welch's t-test. Healthy n=14; Tumor n=21.
RESULTS
Damped circadian rhythms of PER1-promoter-driven luciferase (Per1-luc) activity were detected in both healthy liver and hepatocellular carcinoma (HCC) explants (Figure 1A). Analysis of the rhythms measured in tissues from ad-libitum fed (control) rats revealed that the initial rhythmic amplitude was lower in the tumor explants than in healthy liver (Fig. 1B). Furthermore, the period of the oscillation was shorter in tumors (23.6 ± 0.36h in healthy tissue vs. 19.7 ± 0.55h in HCC explants; Welch's t-test p< 0.0001; Fig. 1B) However the rate at which the amplitude of the rhythm decayed, or damped, in vitro was not significantly different between the tissue types. To increase the sample size at least 2 cultures were made from hepatomas and normal liver from each rat. Therefore the results reflect data from a variety of tumor sizes and from tissues originating in all 5 lobes of the liver. Results did not appear to differ as a function of tumor size or tissue source, however the data were insufficient for statistical analysis using these independent variables.
The phase of peak Per1-luc expression during ad-libitum feeding occurred during the middle of the night in both HCC and healthy liver explants (Fig. 1A; Group phase data summarized in Fig. 2B). If food was restricted to a single daily meal at ZT 1600 (4h after lights-off), Per1-luc expression in tumor explants peaked about 3h earlier than did healthy liver (Fig. 2A) In contrast Per1-luc expression in tumor explants from rats fed at ZT 0400 (4h after lights-on) peaked late in the light phase, about 3h later than did healthy liver tissue from the same animals.
Fig. 2.
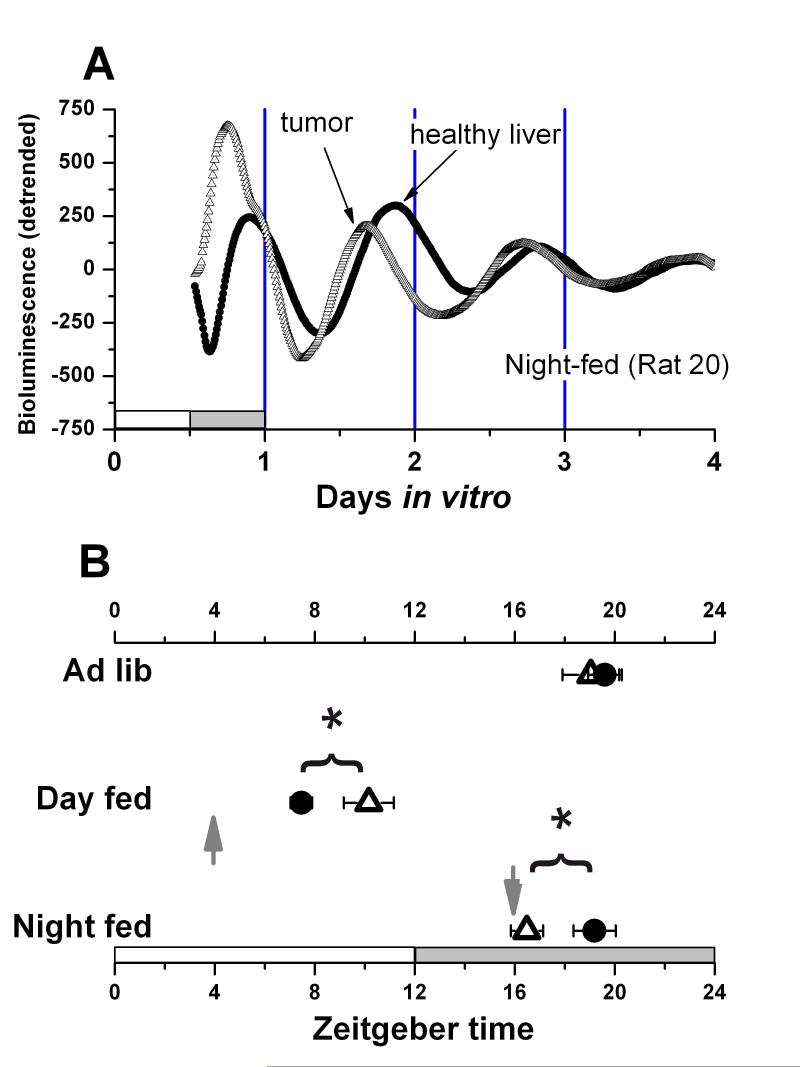
A. Examples of detrended bioluminescence records made from an HCC and healthy liver explant both taken from a rat fed at ZT 1600 (night-fed). The arrows indicate the peak that was used for phase analysis. B. Mean (± SEM) peak phase in vitro for HCC (Open triangle) and healthy liver (closed circle) explants from rats fed ad-libitum, at ZT 0400 (RF Day), or at ZT 1600 (RF Night). The arrows indicate the mealtime for the restricted fed groups. White and dark bars on the bottom of the chart indicate the alternating light-dark cycle. Each datapoint indicates results from a minimum of 9 (RF Night) and a maximum of 21 (Ad-libitum) cultures. * p< 0.025.
Figure 3 shows peak phases Per1-luc expression for tissues explanted from rats that have been exposed to 1, 3 or 6 days of an advanced (Fig. 3A) or delayed (Fig. 3B) light cycle. In contrast to the phase differences we observed in day or night food-restricted rats, no significant differences were detected in the rate at which HCC and healthy host liver reset to the shifted light schedule.
Fig. 3.
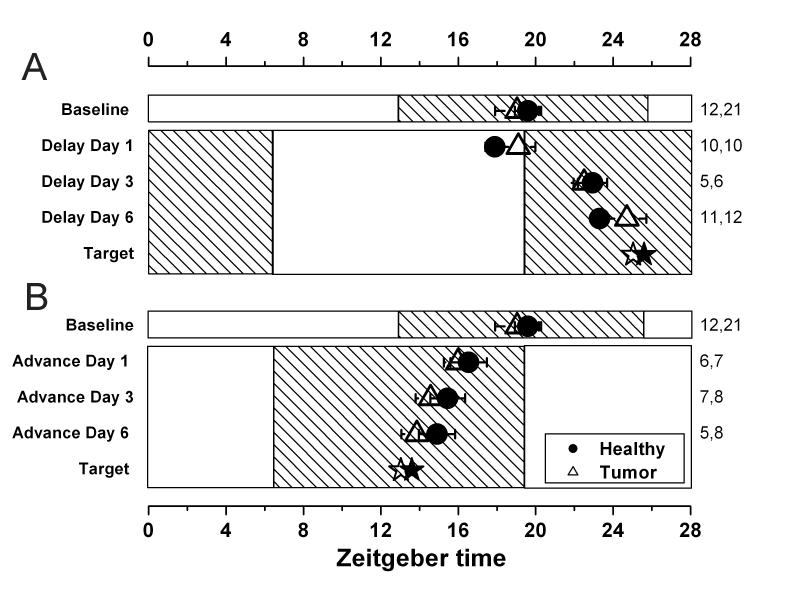
Photic resetting of healthy liver (closed circles) and HCC (open triangles). Mean ± SEM peak phase is shown for rats that have undergone a 6h phase delay (A), or a 6h phase advance (B) and then been killed for assessment of the circadian phase on either the first, third or sixth day in the new lighting conditions. The number of cultures that contribute to each datapoint are shown in the right margin (healthy, tumor). The hatching indicates the dark phase of the lighting cycle. The target, shown by a star, represents the theoretical phase of the tissue after the shift has been completed. It is calculated by adding or subtracting 6h from the baseline phase.
DISCUSSION
In this study we demonstrate that induced hepatocellular carcinoma maintains the capacity to generate circadian rhythms of Period 1 gene expression. While diurnal (24h) rhythms in cell division in tumors are well-documented (for review see30 and 12), most of those rhythms were measured in tumors ex vivo (i.e. in tissue from animals killed at different times around the clock). While it is critically important to understand how rhythms in tumors interact with the host, it is also important to study the molecular clock in the tumor in isolation from other circadian oscillators in the organism. Such an approach may reveal important alterations in circadian control of cell division11 or other functions. To our knowledge our study represents the first demonstration of endogenous circadian rhythmicity in a primary tumor. Furthermore, spontaneous tumors that have occasionally arisen in our colony of Per1-luc transgenic rats (mammary adenosarcoma, mammary fibroadenoma, and lymphosarcoma) also express circadian rhythms in vitro (data not shown).
The analysis of rhythms generated by HCC, as measured via a luciferase reporter in vitro, revealed two differences between HCC and healthy liver. First, the mean period of the rhythm was shorter for tumors than for healthy liver by nearly 4h. This finding suggests that the transformation of healthy tissue to malignancy may have effects on the molecular mechanisms generating circadian rhythmicity. In support of this possibility, Nagoshi and colleagues 31 suggested that cell division may influence circadian period by transiently up- or down-regulating the Per and Cry circadian clock proteins during mitosis. It is possible that, even in the absence of serum in our media, the HCC explants are dividing in vitro. If this cell division occurred consistently during the phase of the cycle in which Per and Cry degradation occurs, a shorter circadian period might result.
The second change we observed was that initial rhythmic amplitude was reduced in tumors compared with healthy liver. We interpret this result with caution, however, since amplitude in vitro can be affected by several variables, including the exact size and shape of the explant. However if amplitude differences we observe in vitro reflect systematic biological alteration, it could arise as a result of reduced coupling among tumor cells, or as a result of a decreased oscillatory amplitude in individual liver cells. The fact that damping rate is similar between the tissue types suggests that while differences exist in the rhythmic parameters, hepatoma still possesses a robust circadian clock.
Cell division is regulated by the circadian clock. First reported by Scheving32, 33 in the 1970s, mitotic activity in the rat and mouse GI tract and cornea occurs with a circadian rhythm. Human oral34 and rectal35, 36 mucosa also divide rhythmically. Furthermore key stages in the cell cycle correlate with rhythms in clock gene expression. For example in humans, G1 correlates with peak expression of hper1, and M phase correlates with peak hbmal1 expression2, 3. One potential mechanism for such regulation (elucidated in regenerating liver) is that Cyclin B1-Cdc2 kinase, a regulator of mitosis, is linked to the clock mechanism through rhythmic expression of wee111. Specifically, it is suggested that the CLOCK/BMAL1 heterodimeric transcription factor, a rhythmically active and critical component of the core clock mechanism, binds to one or several of 3 E-box elements that are located upstream but near wee1. The increased transcriptional activity that results causes the up-regulation of WEE1 kinase, which then phosphorylates the Cyclin B1-Cdc2 complex kinase, inhibiting its activity during the phase of the cycle in which CLOCK/BMAL1 is active.
Interestingly circadian regulation of cell division is found widely in organisms as diverse as cyanobacteria37, 38 and zebrafish39. Abnormal or absent gating of cell division is one mechanism by which alterations in circadian regulation may affect tumor development. It is tempting to speculate that some cancers have abnormal rhythms, or are poorly regulated by the organisms' central circadian system. In our case, it appears that the molecular machinery necessary for the generation of rhythms is largely intact in rat hepatoma. However the sensitivity of the tumor to external signals does appear to be altered.
Our study shows that the phase of peak Per1-luc expression in HCC is not different from that in healthy host liver under normal feeding conditions. Daytime restricted feeding caused phase-shifts of this peak in both healthy and malignant tissue. Most importantly, while the peak phase in healthy liver differs by 11.5h in day versus night-fed groups of animals, the phase of HCC tissue explants differed by only 6.3 hours among the same groups of animals. Therefore tumor tissue is far less sensitive to the phase-resetting stimulus of restricted feeding than is healthy liver. This important result is consistent with the results of a study by Lakatua and colleagues40 showing that restricted feeding reset the rhythm in DNA synthesis in healthy colon, but not in a transplanted Harding-Passey melanoma. While the sensitivity of the HCC to restricted feeding was reduced relative to normal liver, the response to light was not changed. The trajectory of phase-resetting after an advanced or delayed light-dark cycle revealed that HCC and healthy liver are equally responsive to light induced circadian signals. Therefore it appears that the differential response to RF is specific to that modality and that the molecular circadian clock mechanism in this malignant tissue is largely intact and otherwise functioning normally.
Wu, Filipski and colleagues recently demonstrated that daytime food restriction can inhibit tumor development in mice inoculated with Glasgow osteosarcoma19, 24. Nighttime food restriction also inhibited tumor progression, but to a much lesser extent. This finding helped to differentiate between two competing explanations for the long-reported effects of caloric restriction on cancer: reduced caloric intake per se (as reviewed in41) versus time of feeding. The authors of the study suggested that both caloric restriction and the temporal restriction of food intake to the light-phase act to inhibit tumor growth. The mechanisms responsible for these effects are unknown. However our results suggest one possible mechanism for alterations of tumor development by daytime food restriction. In day-fed rats HCC peaked 3h later than healthy tissue while in night-fed rats the tumors peaked 3h earlier. Some specific phase relationships between tumors and host may promote tumor proliferation, and other phase relationships may support host-mediated defense against the cancerous tissue. Although our study did not address the rate of tumor development, future experiments will address this hypothesis by combining feeding schedules with assessment of molecular rhythms in tumor and host tissue while tracking the rate of tumor development.
The goal of chronomodulated chemotherapy is to adjust the temporal pattern of drug delivery to improve the toxic-therapeutic ratio. Adjustment of drug dosage by time-of-day may serve two independent purposes: 1) To reduce doses at circadian times when healthy tissue is most susceptible to drug-induced toxicity and 2) To increase doses during circadian times at which the tumor is most susceptible to the effects of the drug. Clinical control over phase and amplitude of circadian rhythms in peripheral organs and tumors has the potential to dramatically improve outcomes in patients receiving chronotherapy. In this study we have shown that timed meals exert phase control over both healthy host liver and cancer, and that meal timing alters the phase-relationship between tumor and host. It should be possible to design feeding regimes that differentially affect host and tumor rhythms and thereby allow drug administration at circadian times that minimize toxic side-effects to healthy tissue while maximizing therapeutic effects on the tumor.
Acknowledgements:
This work was supported by NIA grant F32 AG22741-01 to AJD, NSBRI grant NCC9-58-167 and NIMH grant RO1 MH56647 to MM, and NIMH grant RO1 MH062517 to GDB. The authors appreciate technical assistance and advice provided by Deanna Arble, Amy O'Coin, Jeffry Wimsatt, and Sanford Feldman. The authors also appreciate the helpful comments on this manuscript provided by Mike Sellix, Oscar Castañón-Cervantes and Shin Yamazaki.
REFERENCES CITED
- 1.Scheving LE, Tsai TH, Scheving LA. Chronobiology of the intestinal tract of the mouse. The American Journal of Anatomy. 1983;168(4):433–65. doi: 10.1002/aja.1001680405. [DOI] [PubMed] [Google Scholar]
- 2.Bjarnason GA, Jordan RC, Sothern RB. Circadian variation in the expression of cell-cycle proteins in human oral epithelium. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10027418. Am J Pathol. 1999;154(2):613–22. doi: 10.1016/S0002-9440(10)65306-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bjarnason GA, Jordan RC, Wood PA, Li Q, Lincoln DW, Sothern RB, Hrushesky WJ, Ben-David Y. Circadian expression of clock genes in human oral mucosa and skin: association with specific cell-cycle phases. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11337377. Am J Pathol. 2001;158(5):1793–801. doi: 10.1016/S0002-9440(10)64135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haus E, Smolensky MH. Biologic rhythms in the immune system. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10513884. Chronobiol Int. 1999;16(5):581–622. doi: 10.3109/07420529908998730. [DOI] [PubMed] [Google Scholar]
- 5.Stephan FK, Zucker I. Circadian rhythms in drinking behavior and locomotor activity of rats are eliminated by hypothalamic lesions. Proc. Natl. Acad. Sci. U. S. A. 1972;69(6):1583–6. doi: 10.1073/pnas.69.6.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12198538. Nature. 2002;418(6901):935–41. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 7.Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15485355. Annu Rev Genomics Hum Genet. 2004;5:407–41. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davidson AJ, Yamazaki S, Menaker M. SCN: ringmaster of the circadian circus or conductor of the circadian orchestra? http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=14712917. Novartis Found Symp. 2003;253:110–21. [PubMed] [Google Scholar]
- 9.Hrushesky WJ, Bjarnason GA. Circadian cancer therapy. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=8315438. J Clin Oncol. 1993;11(7):1403–17. doi: 10.1200/JCO.1993.11.7.1403. [DOI] [PubMed] [Google Scholar]
- 10.Barbason H, Bouzahzah B, Herens C, Marchandise J, Sulon J, Van Cantfort J. Circadian synchronization of liver regeneration in adult rats: the role played by adrenal hormones. Cell Tissue Kinet. 1989;22(6):451–60. doi: 10.1111/j.1365-2184.1989.tb00228.x. [DOI] [PubMed] [Google Scholar]
- 11.Matsuo T, Yamaguchi S, Mitsui S, Emi A, Shimoda F, Okamura H. Control mechanism of the circadian clock for timing of cell division in vivo. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12934012. Science. 2003;302(5643):255–9. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- 12.Hrushesky WJ, Lannin D, Haus E. Evidence for an ontogenetic basis for circadian coordination of cancer cell proliferation. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9776414. J Natl Cancer Inst. 1998;90(19):1480–4. doi: 10.1093/jnci/90.19.1480. [DOI] [PubMed] [Google Scholar]
- 13.Klevecz RR, Shymko RM, Blumenfeld D, Braly PS. Circadian gating of S phase in human ovarian cancer. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=3677075. Cancer Res. 1987;47(23):6267–71. [PubMed] [Google Scholar]
- 14.Hrushesky WJ. Circadian timing of cancer chemotherapy. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=3883493. Science. 1985;228(4695):73–5. doi: 10.1126/science.3883493. [DOI] [PubMed] [Google Scholar]
- 15.Levi F, Zidani R, Misset JL. Randomised multicentre trial of chronotherapy with oxaliplatin, fluorouracil, and folinic acid in metastatic colorectal cancer. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9291901. International Organization for Cancer Chronotherapy. Lancet. 1997;350(9079):681–6. doi: 10.1016/s0140-6736(97)03358-8. [DOI] [PubMed] [Google Scholar]
- 16.Filipski E, King VM, Li X, Granda TG, Mormont MC, Claustrat B, Hastings MH, Levi F. Disruption of circadian coordination accelerates malignant growth in mice. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12852994. Pathol Biol (Paris) 2003;51(4):216–9. doi: 10.1016/s0369-8114(03)00034-8. [DOI] [PubMed] [Google Scholar]
- 17.Filipski E, King VM, Li X, Granda TG, Mormont MC, Liu X, Claustrat B, Hastings MH, Levi F. Host circadian clock as a control point in tumor progression. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11983758. J Natl Cancer Inst. 2002;94(9):690–7. doi: 10.1093/jnci/94.9.690. [DOI] [PubMed] [Google Scholar]
- 18.Filipski E, Delaunay F, King VM, Wu MW, Claustrat B, Grechez-Cassiau A, Guettier C, Hastings MH, Francis L. Effects of Chronic Jet Lag on Tumor Progression in Mice. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15520194. Cancer Res. 2004;64(21):7879–85. doi: 10.1158/0008-5472.CAN-04-0674. [DOI] [PubMed] [Google Scholar]
- 19.Filipski E, Innominato PF, Wu M, Li XM, Iacobelli S, Xian LJ, Levi F. Effects of light and food schedules on liver and tumor molecular clocks in mice. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15812076. J Natl Cancer Inst. 2005;97(7):507–17. doi: 10.1093/jnci/dji083. [DOI] [PubMed] [Google Scholar]
- 20.van den Heiligenberg S, Depres-Brummer P, Barbason H, Claustrat B, Reynes M, Levi F. The tumor promoting effect of constant light exposure on diethylnitrosamine-induced hepatocarcinogenesis in rats. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10403512. Life Sci. 1999;64(26):2523–34. doi: 10.1016/s0024-3205(99)00210-6. [DOI] [PubMed] [Google Scholar]
- 21.Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12372299. Cell. 2002;111(1):41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- 22.Davidson AJ, Cantenon-cervantes O, Stephan FK. Daily oscillations in liver function: diurnal versus circadian rhythmicity. Liver International. 2004;24:179–86. doi: 10.1111/j.1478-3231.2004.00917.x. [DOI] [PubMed] [Google Scholar]
- 23.Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the Circadian Clock in the Liver by Feeding. http://www.ncbi.nlm.nih.gov/cgibin/Entrez/referer?http://www.sciencemag.org/cgi/content/abstract/291/5503/490. Science. 2001;291(5503):490–93. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- 24.Wu MW, Li XM, Xian LJ, Levi F. Effects of meal timing on tumor progression in mice. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15219806. Life Sci. 2004;75(10):1181–93. doi: 10.1016/j.lfs.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 25.Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, Block GD, Sakaki Y, Menaker M, Tei H. Resetting central and peripheral circadian oscillators in transgenic rats. http://www.ncbi.nlm.nih.gov/cgibin/Entrez/referer?http://www.ncbi.nlm.nih.gov/htbinpost/Omim/getmim%3ffield=medline_uid&search=10784453. Science. 2000;288(5466):682–5. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- 26.Millar AJ, Straume M, Chory J, Chua NH, Kay SA. The regulation of circadian period by phototransduction pathways in Arabidopsis. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=7855596. Science. 1995;267(5201):1163–6. doi: 10.1126/science.7855596. [DOI] [PubMed] [Google Scholar]
- 27.Plautz JD, Straume M, Stanewsky R, Jamison CF, Brandes C, Dowse HB, Hall JC, Kay SA. Quantitative analysis of Drosophila period gene transcription in living animals. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9181432. J Biol Rhythms. 1997;12(3):204–17. doi: 10.1177/074873049701200302. [DOI] [PubMed] [Google Scholar]
- 28.Abe M, Herzog ED, Yamazaki S, Straume M, Tei H, Sakaki Y, Menaker M, Block GD. Circadian rhythms in isolated brain regions. http://www.ncbi.nlm.nih.gov/cgibin/Entrez/referer?http://www.jneurosci.org/cgi/content/abstract/22/1/350. J Neurosci. 2002;22(1):350–6. doi: 10.1523/JNEUROSCI.22-01-00350.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bevington P. Data reduction and error analysis for the physical sciences. New York; McGraw-Hill: 1969. [Google Scholar]
- 30.Canaple L, Kakizawa T, Laudet V. The days and nights of cancer cells. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=14633665. Cancer Res. 2003;63(22):7545–52. [PubMed] [Google Scholar]
- 31.Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, Schibler U. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15550250. Cell. 2004;119(5):693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 32.Scheving LE, Burns ER, Pauly JE. Circadian rhythms in mitotic activity and 3 H-thymidine uptake in the duodenum: effect of isoproterenol on the mitotic rhythm. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=4673136. Am J Anat. 1972;135(3):311–7. doi: 10.1002/aja.1001350302. [DOI] [PubMed] [Google Scholar]
- 33.Scheving LE, Burns ER, Pauly JE, Tsai TH. Circadian variation in cell division of the mouse alimentary tract, bone marrow and corneal epithelium. Anatomical Record. 1978;191(4):479–86. doi: 10.1002/ar.1091910407. [DOI] [PubMed] [Google Scholar]
- 34.Warnakulasuriya KA, MacDonald DG. Diurnal variation in labelling index in human buccal epithelium. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=8141673. Arch Oral Biol. 1993;38(12):1107–11. doi: 10.1016/0003-9969(93)90173-j. [DOI] [PubMed] [Google Scholar]
- 35.Buchi KN, Moore JG, Hrushesky WJ, Sothern RB, Rubin NH. Circadian rhythm of cellular proliferation in the human rectal mucosa. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=2065918. Gastroenterology. 1991;101(2):410–5. doi: 10.1016/0016-5085(91)90019-h. [DOI] [PubMed] [Google Scholar]
- 36.Brandi G, Calabrese C, Pantaleo MA, Morselli Labate A, Di Febo G, Hakim R, De Vivo A, Di Marco MC, Biasco G. Circadian variations of rectal cell proliferation in patients affected by advanced colorectal cancer. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15142678. Cancer Lett. 2004;208(2):193–6. doi: 10.1016/j.canlet.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 37.Mori T, Johnson CH. Independence of circadian timing from cell division in cyanobacteria. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11274102. J Bacteriol. 2001;183(8):2439–44. doi: 10.1128/JB.183.8.2439-2444.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mori T, Johnson CH. Circadian control of cell division in unicellular organisms. Prog Cell Cycle Res. 2000;4:185–92. doi: 10.1007/978-1-4615-4253-7_16. [DOI] [PubMed] [Google Scholar]
- 39.Dekens MP, Santoriello C, Vallone D, Grassi G, Whitmore D, Foulkes NS. Light regulates the cell cycle in zebrafish. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=14653994. Curr Biol. 2003;13(23):2051–7. doi: 10.1016/j.cub.2003.10.022. [DOI] [PubMed] [Google Scholar]
- 40.Lakatua DJ, White M, Sackett-Lundeen LL, Haus E. Change in phase relations of circadian rhythms in cell proliferation induced by time-limited feeding in BALB/c X DBA/2F1 mice bearing a transplantable Harding-Passey tumor. Cancer Res. 1983;43(9):4068–72. [PubMed] [Google Scholar]
- 41.Kritchevsky D. Caloric restriction and cancer. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11349885. J Nutr Sci Vitaminol (Tokyo) 2001;47(1):13–9. doi: 10.3177/jnsv.47.13. [DOI] [PubMed] [Google Scholar]


