Abstract
Yra1p is an essential RNA-binding protein that couples transcription to export. The YRA1 gene is one of only ∼5% of genes that undergo splicing in budding yeast, and its intron is unusual in several respects, including its large size and anomalous branchpoint sequence. We showed previously that the intron is required for autogenous regulation of Yra1p levels, which cause a dominant negative growth phenotype when elevated. The mechanism of this regulation, however, remains unknown. Here we demonstrate that growth is inversely correlated with splicing efficiency. Substitution of a canonical branchpoint moderately improves splicing but compromises autoregulation. Shortening the intron from 766 to ∼350 nt significantly improves splicing but abolishes autoregulation. Notably, proper regulation can be restored by insertion of unrelated sequences into the shortened intron. In that the current paradigm for regulated splicing involves the binding of protein factors to specific elements in the pre-mRNA, the regulation of YRA1 expression appears to occur by a novel mechanism. We propose that appropriate levels of Yra1p are maintained by inefficient cotranscriptional splicing.
Keywords: THO, cotranscriptional, XRN1
INTRODUCTION
One of the most fundamental questions in biology is how the cell maintains homeostasis. Autoregulatory feedback loops are an efficient mechanism for coupling the expression levels of a particular protein with its functional demand. A particularly elegant strategy is provided when the function of a given protein is to bind to a nucleic acid; when the primary target is titrated, binding to a secondary target within the (pre-) mRNA can shut off new synthesis by inhibiting gene expression at the transcriptional or post-transcriptional level. The prototype of such a mechanism is the case of ribosomal protein gene expression in bacteria. Ribosomal proteins, when present in excess of available rRNA, bind to their own mRNA, thereby inhibiting translation (Draper 1989). In eukaryotes, additional strategies are available. For example, excess amounts of the 28S rRNA-binding protein L30 bind to its intron-containing mRNA and inhibit the splicing of the L30 intron (Eng and Warner 1991). A similar mechanism has been suggested for the autogenous regulation of another ribosomal protein, S14. This protein directly interacts with both 18S rRNA and a stem–loop in its own pre-mRNA (Fewell and Woolford 1999). All of these examples have in common the binding of the autoregulatory protein to specific sequences in the mRNA or pre-mRNA.
We have previously reported that Yra1p is indeed autoregulated by a negative feedback mechanism dependent on its intron (Preker et al. 2002). This regulation is important for viability, as overexpression of Yra1p is toxic. Yra1p is a small RNA-binding protein first identified on the basis of its potent RNA annealing activity in vitro (Portman et al. 1997). It was subsequently shown that Yra1p is essential for mRNA export (Sträßer and Hurt 2000) and is recruited to the mRNA cotranscriptionally (Sträßer and Hurt 2000; Lei et al. 2001; Lei and Silver 2002; Abruzzi et al. 2004; Kim et al. 2004).
Cotranscriptional recruitment of Yra1p depends on an assembly of at least four proteins termed the THO complex (Sträßer et al. 2002; Zenklusen et al. 2002). The THO complex is associated with chromatin. Mutants in THO components were first described due to their phenotypes in transcription-dependent hyper-recombination (Aguilera 2005). Later, roles in transcription elongation (Jimeno et al. 2002; Huertas and Aguilera 2003) and transcription-coupled mRNA quality control (Libri et al. 2002) were proposed. Notably, deletions of any of the THO complex components also resulted in an mRNA export defect, consistent with the idea that cotranscriptional loading of Yra1p is required for mRNA export (Sträßer et al. 2002). A complex termed TREX (transcription and export) was identified that consists of THO, Yra1p, and at least two additional factors, Tex1p and Sub2p, a member of the DEAD-box ATPase family (Sträßer and Hurt 2001). Interestingly, Sub2p was initially described as a factor required for mRNA splicing (Kistler and Guthrie 2001; Libri et al. 2001; Zhang and Green 2001). Its essential role in mRNA export was identified subsequently (Jensen et al. 2001; Sträßer and Hurt 2001). Taken together, there is increasing evidence for a coupling between transcription, processing of the primary transcript, and export of the mature mRNA. Even though the precise role of Yra1p in any of these processes has not been established, this essential protein accompanies the transcript throughout all these steps in gene expression.
We have previously shown that the presence of an intron in the YRA1 gene is required for maintaining steady-state levels of the Yra1 protein (Preker et al. 2002). This regulation is important for optimal cell growth. In this study, we initially examined the biological significance of the highly unusual features of the YRA1 intron. These include (1) its large size—at 776 nt, it is the second largest intron in budding yeast; (2) its location within the transcript—most yeast introns are located close to the AUG initiation codon, while the YRA1 intron is almost 300 nt downstream; and (3) its nonconsensus branchpoint (BP) sequence—unlike the majority of yeast BPs, the YRA1 BP sequence is Gacuaac rather than uacuaac (Preker et al. 2002). Here we present phylogenetic evidence for the conservation of each of these elements throughout ascomycetous fungi. By mutational analysis, we demonstrate that the large intron size and the nonconsensus BP are critical for appropriate autoregulation. Finally, we use biochemical analyses to show that the degree of autoregulation is inversely correlated with Yra1p splicing efficiency and that cotranscriptional loading of Yra1p is required for autoregulation. Taken together, our results support a model in which appropriate levels of Yra1 protein are maintained by inefficient cotranscriptional splicing of its own pre-mRNA.
RESULTS
The YRA1 intron and its main features are conserved throughout fungi
We have previously shown that autoregulation of YRA1 depends on the presence of an intron, which is positioned roughly in the middle of the gene (Preker et al. 2002). To assess the biological significance of this regulation and to gain information about the mechanism of regulation, we first identified putative orthologs in other yeast species. We cloned the YRA1 intron from three additional budding yeasts, Saccharomyces unisporus, Kazachstania telluris, and Pichia canadensis (see Materials and Methods). In addition, we performed tBLASTn searches (Altschul et al. 1990) using the Saccharomyces cerevisiae Yra1 protein sequence as a query. Coding sequences for several proteins with significant similarity were identified from public databases. These sequences were used in iterative searches for additional homologs. In total, we identified putative full-length Yra1p homologs from 17 hemiascomycota (budding yeasts) and six pezizomycota (filamentous fungi). Most of these sequences have not previously been annotated as protein-coding genes, presumably because of their unconventional gene structure (see below). Sequences were aligned and a dendrogram was constructed (Fig. 1). The sequence relationship closely resembles the phylogenetic relationship of the different yeast species based on ribosomal DNA sequences (e.g., Souciet et al. 2000), suggesting that these proteins are true orthologs and share a common function. As noted previously for the REF family of proteins, which includes Yra1p, mammalian Aly/REF1, and other metazoan proteins (Stutz et al. 2000), sequence conservation is highest at the extreme termini and within the central RNA-binding domain. Those regions are separated by more variable regions that, however, do share a high content of basic amino acids. Notably, the pezizomycota Yra1 proteins identified all have a ∼30–amino acid extension of their C termini that is not found in hemiascomycota (Fig. 1). Because mutations in the C terminus of S. cerevisiae have the most profound effect on function (Preker et al. 2002), this extension might be functionally relevant.
FIGURE 1.
Dendogram constructed from putative yeast homologs of S. cerevisiae Yra1p. Homologs were identified as described in Materials and Methods. Only sequences that appeared to be full-length were used to create an unrooted, Neighbor-Joining tree (http://evolution.genetics.washington.edu/phylip.html). Sordariomycocetes and eurotiomycetes are different classes of pezizomycota (filamentous fungi). Schematic representations of the protein structure are shown to the right. The most highly conserved sequences are shaded in darker colors. Triangles point to introns that are found at conserved positions in several or all species. (Not shown are the hemiascomycota Candida tropicalis, C. dubliniensi, C. lusitaniae, C. parapsilosis, and Pichia guilliermondii.)
We next inspected the coding sequences for the presence of introns. Remarkably, all homologs of S. cerevisiae YRA1 contain an intron at the exact same nucleotide position in the region encoding the central RNA-binding domain (Fig. 1; Table 1). We will refer to this intron as the YRA1 intron. We also found evidence for the existence of a YRA1 intron in at least one putative basidomycota homolog. In this case, however, the 5′ splice site could not be identified unambiguously. In contrast, the fission yeast (Saccharomyces pombe) homolog, mlo3, does not contain any introns, and in the putative metazoan YRA1 homologous introns that bear no resemblance to the YRA1 intron are found at different positions within the coding sequence (data not shown).
TABLE 1.
Conversation of the YRA1 intron across yeast species
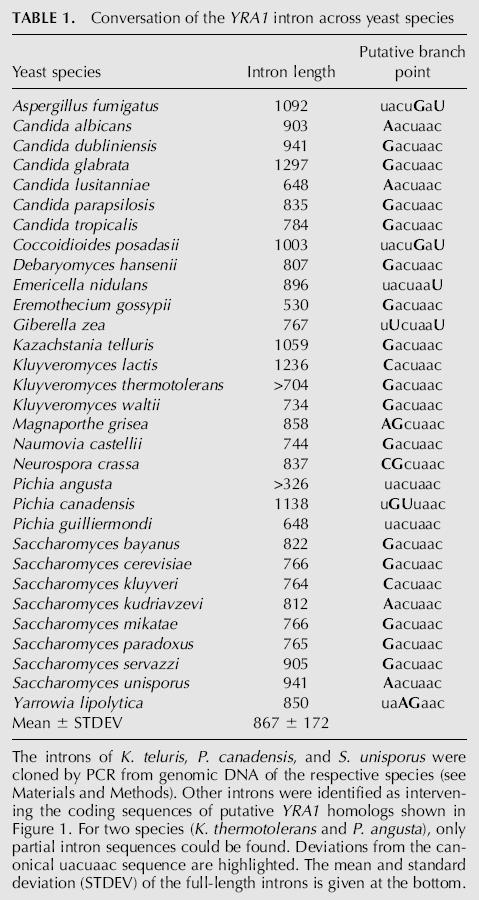
The YRA1 introns range in size from 418 (Candida lusitaniae) to 1297 nt (Candida glabrata). The average length of 867 nt (with a standard deviation of ±172 nt; n = 29) far exceeds the average intron size in any yeast species analyzed to date (e.g., S. cerevisiae, 264 ± 177 nt) (Bon et al. 2001). In addition to the YRA1 intron, filamantous fungi YRA1 genes also contain two or three short introns whose positions are conserved within their group (Fig. 1). An interesting exception is the budding yeast Yarrowia lipolytica, which shares an intron a short distance downstream of the YRA1 intron with the pezizomycota. As has been suggested elsewhere, this provides further evidence that Y. lipolytica is only distantly related to S. cerevisiae and other budding yeasts (Souciet et al. 2000). We also identified introns in two more putative YRA1 homologs (Pichia angusta and Kluyveromyces thermotolerans) for which only partial sequences were available. In total, we compiled YRA1 introns from 31 species representing a wide variety of ascomycota (Table 1).
Of the splicing signals within these 30 introns, the 5′ and 3′ splice sites mostly conform to the yeast consensus splice sites. In stark contrast, the putative branchpoint sequences diverge from the canonical uacuaac sequence in all but two species. Most often, the first residue is altered to a purine residue (Table 1). Interestingly, the two exceptions that confer to the canonical branchpoint sequence, Pichia angusta and Pichia guilliermondii, have an extremely short branchpoint to 3′ splice site distance. In all other species, this distance is highly variable (data not shown).
Intron size is a major determinant in YRA1 autoregulation
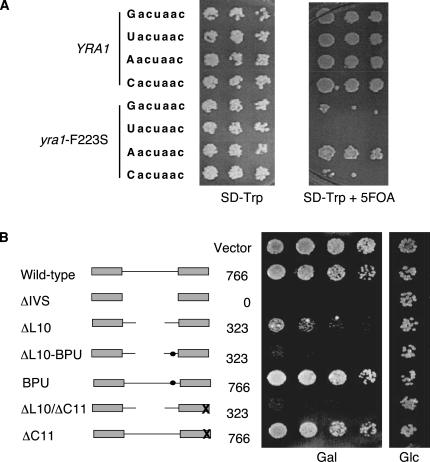
The striking conservation of the unusual features of the YRA1 intron led us to investigate their significance with respect to autoregulation of Yra1p expression. We previously showed that five introns from unrelated S. cerevisiae genes failed to restore autoregulation when replacing the YRA1 intron (Preker et al. 2002). We interpreted these results as evidence that the presence of an intron is not in itself sufficient to confer autoregulation, and that specific sequences of the YRA1 intron are required, possibly by serving as binding sites for Yra1p. With the initial goal of identifying such sites, we performed an extensive mutational analysis of the YRA1 intron by creating internal deletions, as well as by linker-scanning analysis. Surprisingly, however, no single short deletion or substitution affected cell viability or Yra1 protein levels (data not shown). Instead, we found that YRA1 introns deleted to a length of <400 nt exert a dominant negative effect on cell growth that gradually worsened upon further shortening. To confirm that this effect depends on intron length rather than the removal of regulatory sequence(s), we inserted unrelated sequences into a shortened YRA1 intron. Random DNA fragments of phage λ DNA were inserted into an intron deleted to 323 bp (ΔL10) under the control of the GAL1 promoter (see Materials and Methods). Any of three different sequences tested (denoted λ5, λ10, and λ21 in Fig. 2) rescues the dominant negative growth phenotype of the ΔL10 deletion on galactose-containing medium. Importantly, all constructs could fully complement a deletion of YRA1, indicating that they restore the regulation of YRA1 expression rather than prevent splicing nonspecifically.
FIGURE 2.
Size is a major determinant of YRA1 regulation. PJP168-32 (relevant genotype yra1rHIS3, <pRS316–YRA1>) cells were transformed with plasmids bearing the indicated YRA1 wild-type and mutant constructs under control of the inducible GAL1 promoter (see Table 2), as well as the empty vector. Cells were grown at 30°C in glucose-containing (noninducing) medium lacking tryptophan to select for the presence of the plasmids. When the cultures reached an OD600 of ∼1.0, serial dilutions were transferred to solid medium lacking tryptophan and containing either 2% galactose (“Gal,” left panel) or the same amount of glucose (“Glc,” middle panel) and allowed to grow at 30°C for 2–3 d. Subsequently, the galactose-containing plates were replica-plated on medium containing galactose and 5FOA (right panel) to counter-selects for cells dependent on the pRS316–YRA1 plasmid. Introns and their deletions are approximately drawn to scale. The bold gray lines represent heterologous sequences. Absolute intron sizes in number of nucleotides are given to the right of the schematic drawings.
The introns tested for their ability to substitute for the YRA1 intron in the previous study (Preker et al. 2002) all had consensus splice sites and ranged in size from 77 (APS3) to 308 nt (ACT1). Notably, the ACT1 intron partially alleviated the dominant negative growth phenotype of an intron-less allele of YRA1. To test whether sequences that are more closely related to the YRA1 intron could confer regulation, we inserted the 906-nt intron from its Candida albicans homolog. The C. albicans YRA1 gene can complement a deletion of its S. cerevisiae counterpart at all temperatures tested (data not shown), and its intron alleviates the dominant negative effect of a YRA1 cDNA completely (Fig. 2). The 413-nt intron of the S. cerevisiae RPS9B gene was also able to confer proper regulation. Similar intron swap experiments by Hurt and colleagues (Rodriguez-Navarro et al. 2002) have shown that the 414-nt RPL25 intron can also partially restore regulation of YRA1 expression. Taken together, it appears that intron size is an important determinant in the regulation of YRA1 expression.
The noncanonical branchpoint sequence is required for optimal regulation of YRA1 expression
Interestingly, both the C. albicans YRA1 intron and the S. cerevisiae RPS9B intron have a noncanonical branchpoint sequence (Aacuaac and Gacuaac, respectively), as does S. cerevisiae YRA1 and virtually all of its homologs (see Table 1). To assess a possible contribution of the noncanonical branchpoint to the regulation of Yra1p expression, we converted the wild-type, noncanonical G at the first position into a T to create a consensus uacuaac branchpoint sequence. Substitutions into either C or A were also created. All of these alleles supported growth when present as the only copy in the cell (Fig. 3A, left panel) and none had a dominant negative growth phenotype, as would have been expected if the mutation interfered greatly with the regulation of YRA1 expression.
FIGURE 3.
A contribution of the branchpoint sequence to autoregulation. (A) The noncanonical branchpoint is required for viability in the context of a temperature-sensitive YRA1 mutation. Single point mutations were introduced into the first position of the branchpoint sequence of either wild-type YRA1 (rows 2–4) or the yra1-F223S allele (rows 6–8) on plasmids. After transformation into PJP168-32C <pRS316–YRA1>, cells were grown and serial dilutions were applied to either synthetic medium lacking tryptophan or onto the same medium supplied with 5FOA, to select for the absence of the URA3-marked wild-type plasmid. Incubation was at 30°C for 2–3 d. (B) A consensus branchpoint exacerbates the dominant negative effect of a shortened intron. PJP168-32 cells were transformed with plasmids bearing the indicated YRA1 wild-type and mutant constructs under control of the GAL1 promoter (see Table 2), as well as the empty vector, and their phenotypes were assayed as described in the legend to Figure 2. The consensus uacuaac branchpoint in the schematic drawings of rows 5 and 6 is marked by a dot. The crosses mark deletion of the C terminus of yra1-ΔC11. The sizes of the introns are also given.
While not essential for regulation, the noncanonical branchpoint might still contribute to regulation. To evaluate this possibility, we tested the individual mutations in the context of a temperature-sensitive allele of YRA1, yra1-F223S. We had previously shown that the yra1-F223S allele affects regulation, resulting in elevated levels of mutant protein (Preker et al. 2002). Interestingly, deletion of the entire intron in the context of the yra1-F223S allele does not support growth when present as the only copy in the cell, suggesting that the mutant is hypersensitive to a loss of regulation. As shown in Figure 3A, the combination of the consensus branchpoint uacuaac and the yra1-F223S mutation also rendered cells inviable. In contrast, all other variations support growth to some extent. Similar results were obtained in combination with a deletion of the 11 most C-terminal amino acids of Yra1p, which includes residue F223 (yra1-ΔC11; data not shown). Thus, the identity of the nucleotide at the first position of the branchpoint contributes to regulation. Specifically, it is the absence of the consensus U that is important.
In support of this conclusion, the mutant consensus branchpoint also enhanced the dominant negative effect of a shortened YRA1 intron (Fig. 3B; cf. ΔL10 and ΔL10-BPU). The yra1-ΔC11 mutation had a similar effect when combined with the ΔL10 allele. Taken together, at least three elements seem to contribute to regulation of Yra1p expression: a long intron, a noncanonical branchpoint, and an intact Yra1p C terminus. Any combination of defects in two of these elements results in a synergistic effect on regulation of Yra1p expression.
Evidence for a conserved stem–loop that acts as a splicing enhancer
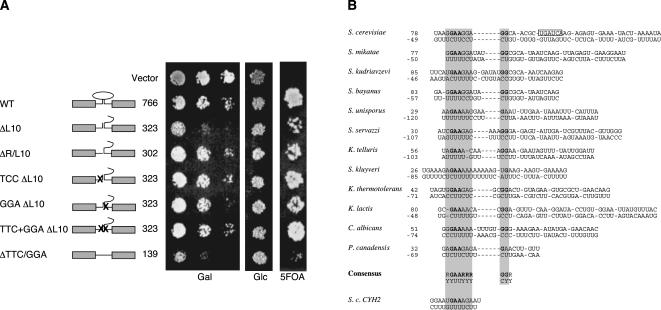
During our deletion analysis of the YRA1 intron, we were surprised to find that a slightly longer deletion of the ΔL10 intron that extended a further 21 nt to the 5′ end (“ΔR/L10” in Fig. 4A) almost completely reverted the dominant negative growth defect of that mutant. Closer inspection of this region revealed that it has the potential to base-pair with a region close to the 3′ splice site to form a stem–loop. The same stem–loop was also found by computational RNA-structure prediction using the mfold algorithm (Zuker 2003). This algorithm also predicted similar structures in almost all other budding yeast YRA1 introns, but the proposed structure was less apparent in filamantous fungi. Representative structures are shown in Figure 4B. The 5′ portions of the predicted stems were always rich in purines and contained an invariant GAA sequence close to the base. Because similar structures have been shown to enhance splicing previously (Newman1987; Libri et al. 1995), we disrupted the predicted stem–loop by site-directed mutagenesis. The expectation was that this would reduce splicing and thus possibly rescue the phenotype of an otherwise dominant negative variant of YRA1. Indeed, a mutation of this sequence to TCC (“TCC ΔL10” in Fig. 3A), which should disrupt the stem–loop, rescued the dominant negative effect of the ΔL10 intron, as did a mutation in its putative pairing region (“GGA ΔL10”), located ∼600 nt downstream. We next constructed a double mutant (“TCC + GGA ΔL10”) that would restore complementarity. Surprisingly, this double mutant also rescued the ΔL10 phenotype. It thus appears that complementarity is insufficient to enhance splicing under these conditions but that the actual sequence plays a role (see Discussion).
FIGURE 4.
Evidence for a conserved stem–loop in the YRA1 intron that acts as a positive regulator for YRA1 expression. (A) Disruption of a proposed secondary structure elevates the dominant negative effect of a shortened intron. Constructs under GAL1 promoter control were introduced and assayed as described in the legend to Figure 2. The putative secondary structure of wild-type YRA1 and its derivatives is also shown. Crosses to the left and the right of the structure symbolize substitutions of the “GAA” and “TTC” motifs, respectively. (B) Alignment of putative secondary structures involving both the “GAA” and “TTC” motifs. Full-length introns were subjected to analyses with the mfold algorithm for secondary structure prediction of RNA. Manual alignment of the highest scoring structures revealed striking similarities between virtually all introns (not shown: Saccharomyces paradoxus, Naumovia castellii, Eremothecium gossypii, Kluyveromyces waltii, C. tropicalis, C. dubliniensis, and P. guilliermondii). The “GAA/TTC” motifs are highlighted. The BclI restriction site used to create the ΔBS series of deletions is boxed. The numbers indicate the position of the nucleotide preceding and following the putative stem–loop. For comparison, a structure that has been experimentally verified to be important for the splicing of an unrelated S. cerevisiae intron (CYH2; Newman 1987) is shown below the consensus sequence.
Finally, deletion of the entire sequence between the two elements to generate an intron of only 139 nt (“ΔTCC/GGA”) exhibit a strong dominant negative effect, presumably by making splicing entirely independent of the GAA and TCC motifs (Fig. 4A). We conclude that these sequence elements promote splicing of the YRA1–pre-mRNA, and that mutations in either element suppress the effect of a shortened intron.
Inverse correlation between pre-mRNA accumulation and the severity of dominant negative growth defects
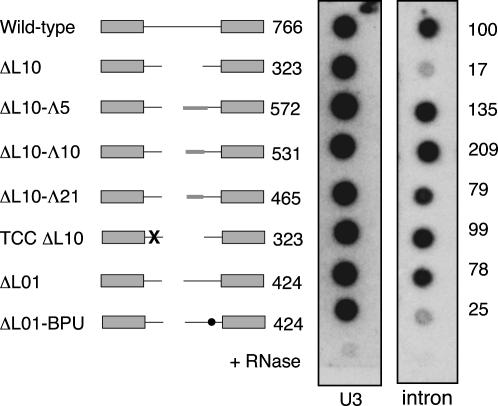
Our previous analysis showed a significant accumulation of YRA1 pre-mRNA when YRA1 was overexpressed either from a high-copy number vector (Preker et al. 2002) or when the gene was placed under the highly active GAL1 promoter, as expected for negative-feedback regulation (data not shown). We predicted that the growth defects caused by perturbations of the YRA1 intron are the direct consequences of increasing splicing efficiency. To test this directly, we assayed pre-mRNA levels in representative YRA1 intron mutants. We introduced them into yeast cells as the sole copy of the gene. Following isolation of total RNA, YRA1 pre-mRNA levels were assayed by dot-blotting onto nylon membranes and hybridization with an oligonucleotide complementary to the intron (see Materials and Methods). When normalized to an unrelated transcript (U3 snRNA), pre-mRNA levels were greatly reduced in the ΔL10 mutant, suggesting that this shorter intron is spliced more efficiently than the wild-type pre-mRNA. The introduction of sequences derived from bacteriophage λ DNA restored pre-mRNA accumulation to wild-type levels or higher, as did a mutation of the GAA motif. We were not able to test the effect of a consensus uacuaac branchpoint in the context of the ΔL10 deletion, because this strain is barely viable (Fig. 3B). Therefore, we tested the branchpoint mutation in the background of a shorter deletion, ΔL01, that on its own doesn't exhibit a phenotype, and found it to reduce pre-mRNA levels at least threefold (Fig. 5). No significant change in the amount of total YRA1 RNA was detected with a probe complementary to the second exon (data not shown), presumably because the fraction of pre-mRNA is much lower than the total RNA. Thus, a long intron appears to be refractory to splicing while a consensus branchpoint favors splicing.
FIGURE 5.
Dominant negative mutations in the YRA1 intron correlate with reduced accumulation of pre-mRNA. PJP168-32C was transformed with the indicated constructs under GAL1 promoter control. After loss of the residual copy of YRA1, cells were grown in liquid medium containing 2% galactose to early log phase. Total RNA was isolated, applied to membrane and probed for the amount of a control transcript (“U3,” left panel) or YRA1 pre-mRNA (right panel). See Materials and Methods for more details. The percentage of pre-mRNA relative to wild type (= 100%, top row) is given to the right. To control for cross-hybridization with any residual DNA in the preparation, an equal amount of the wild type sample was treated with RNaseA before blotting (“+ RNase,” bottom row).
The RNA-polymerase associated THO complex is required for regulation
To assess whether proteins other than Yra1p itself are involved in the regulation of YRA1 expression, we screened a set of selected factors for their ability to regulate expression properly. For this, yeast strains carrying gene deletions that perturb various aspects of gene expression were transformed with plasmids carrying wild-type YRA1 under the control of the inducible GAL1 promoter or the empty vector. While wild-type cells are tolerant to overexpression of YRA1, all three of the four members of the THO complex tested (MFT1, HPR1, and THP2) were hypersensitive to YRA1 overexpression (Fig. 6A). The THO complex has been implicated in transcription elongation, recruitment of Yra1p to nascent transcripts, and mRNA export (see Introduction). We, therefore, tested deletions of factors involved in either of these processes for their sensitivity to YRA1 overexpression. The products of the PPR2 (yTFIIS), SPT4, and RFT1 genes are all required for transcription elongation. None, however, showed an increased sensitivity to YRA1 overexpression from the GAL1 promoter (Fig. 6A; data not shown). Likewise, deletions of the nuclear pore complex component NUP100 and NUP84, which are both required for mRNA export, were unaffected (data not shown). Thus, neither the impairment of transcription by RNA polymerase II nor a defect in general mRNA export per se appears to affect regulation. Also unaffected for growth were deletions of factors involved in degradation of (pre-) mRNA and mRNA surveillance, namely upf1, pan2, rrp6, mlp1, mlp2, ccr4, xrn1, edc2, and ski2. Thus, the dominant negative growth defect can best be explained by a failure to recruit Yra1p to its own transcript.
FIGURE 6.
Factors affecting YRA1 regulation in trans. (A) The THO complex is hypersensitive to overexpression of YRA1. Selected deletions of nonessential factors were transformed with a construct (pIA558) bearing wild-type, intron-containing YRA1 under the control of the GAL1 promoter or the empty vector. After growth of the transformants under noninducing conditions, equal amounts of cells were transferred onto synthetic medium containing galactose and lacking leucine to select for the presence of the plasmid. Cells were incubated for 3–4 d at the temperature indicated below the two panels. (B) YRA1 pre-mRNA levels are decreased in deletions of THO-complex subunits. The indicated strains were transformed with the same constructs as in A and grown in galactose-containing medium to early log-phase. Total RNA was applied to membrane and probed for the amount of total YRA1 (left panel), YRA1 pre-mRNA (middle panel), or a normalization control (U3 snRNA, right panel). Two serial dilutions are shown. (C) Yra1 protein levels are greatly elevated in deletions of THO-complex components. Protein extracts from wild-type and the indicated deletion strains were separated by electrophoresis on a 12% polyacrylamide/SDS gel and blotted to nitrocellulose, and the relative amounts of endogenous Yra1p were determined with a polyclonal αYra1p antibody. The positions of protein size markers (in kDa) are indicated on the right. Polyclonal antiserum against Brr3p/Gle1p was used to verify equal loading of the gel (lower panel). (D) Deletion of the cytoplasmic exonuclease Xrn1p overaccumulates YRA1 pre-mRNA approximately fivefold compared with wild type. Strains were analyzed for their endogenous pre-mRNA levels as in B. See text for more details.
In agreement with the observed growth phenotypes, Yra1 protein levels were several times higher in THO-complex mutants (Fig. 6C), while pre-mRNA levels were reduced about twofold (Fig. 6B). This was in contrast to all other mutants tested, with one notable exception: In the Δxrn1 mutant, YRA1 pre-mRNA levels were four- to fivefold higher than in an isogenic wild-type strain (Fig. 6D). Xrn1p is a 5′ exonuclease involved in the cytoplasmic turnover of RNAs (Hilleren and Parker 2003 and references therein). Yra1 protein levels were unchanged in the xrn1 deletion strain, because the YRA1 pre-mRNA contains a translational stop codon early in its intron that would prevent the production of a full-length protein (data not shown). Interestingly, mutations in the nonsense-mediated decay pathway did not affect YRA1 pre-mRNA levels (Δupf1 in Fig. 6D; data not shown).
We conclude that mutants of the THO complex favor YRA1 splicing, resulting in overexpression of the protein and a dominant negative growth defect. In addition, YRA1 pre-mRNA is greatly stabilized in a deletion of the cytoplasmic exonuclease Xrn1p, suggesting that a sizable fraction of pre-mRNA escapes the nucleus (see Discussion).
DISCUSSION
A novel mechanism of splicing regulation
Maintenance of Yra1 protein concentrations at appropriate levels is critical for viability. Indeed, the YRA1 gene was initially identified as an anonymous cDNA in a screen for transcripts that, when overexpressed from an inducible promoter, would arrest cell growth (Espinet et al. 1995). Upon the discovery of the requirement for Yra1p in mRNA export, it was shown that transient overexpression of this essential protein can block mRNA export (Preker et al. 2002; Rodriguez-Navarro et al. 2002). We demonstrated that functional Yra1 protein is necessary to maintain wild-type levels of expression by a mechanism that requires the presence of the intron. This is consistent with a negative feedback loop in which excess levels of Yra1p inhibit the splicing of its own pre-mRNA (Preker et al. 2002). Here we present evidence that it is the unusual features of the YRA1 intron, including its large size and nonconsensus branchpoint sequences that are critical for this autoregulatory mechanism.
In the two best-studied cases of regulated splicing in budding yeast, protein factors bind to specific sequences near the 5′ splice site and influence splicing either positively or negatively. The MER2 pre-mRNA has a suboptimal 5′ splice site, which makes it a poor substrate for the splicing machinery during vegetative growth (Engebrecht et al. 1991). The MER1 gene is transcribed only during meiosis; the Mer1 protein, together with the U1 snRNP protein Nam8p, promotes the use of this nonconsensus splice site (Nandabalan and Roeder 1995; Spingola and Ares 2000). In the case of the RPL30 gene, the ribosomal protein product autoregulates its own splicing by binding to a stem–loop structure that is formed between the 5′ splice side and the first exon (Eng and Warner 1991). This inhibits splicing when the levels of protein reach a critical level. Thus we initiated a mutational analysis of the YRA1 intron with the initial goal of identifying a binding site for Yra1p. This simple model predicted that deletion of such a binding site would result in the up-regulation of Yra1 protein levels, conferring a dominant negative growth phenotype. Surprisingly, however, deletion and linker-scanning analyses failed to identify any such sequences. Instead, the length of the intron, regardless of its sequence, appears to be a primary determinant. Introns shorter than ∼400 nt exert a strong dominant negative effect (Fig. 2). Intriguingly, the shortest YRA1 intron identified among 30 different yeast species is 418 nt in length (C. lusitaniae), suggesting that this might be close to the minimal length required for regulation. In S. cerevisiae, the dominant negative effect increases upon further shortening, but the phenotype can be suppressed by the insertion of random sequences. Importantly, quantitation revealed that the shorter introns were spliced more efficiently than the wild-type intron. Because splicing efficiency is inversely correlated with the strength of the dominant negative growth phenotype, we conclude that appropriate autoregulation requires inefficient splicing of the long YRA1 intron. In summary, to our knowledge, this largely sequence-independent mechanism constitutes a novel mode of splicing regulation. Interestingly, preliminary experiments using splicing microarrays (J. Pleiss, P.J. Preker, and C. Guthrie, unpubl.) indicate that when Yra1p is overexpressed, other genes with large introns are unaffected.
A nonconsensus branchpoint contributes to autoregulation
All yeast species analyzed to date show a strong bias for the sequence uacuaac at their intron branchpoints (Spingola et al. 1999; Bon et al. 2003). In addition, the complementary branchpoint-recognition sequence in U2 snRNA is invariable (data not shown). It is thus highly significant that our phylogenetic comparisons of YRA1 homologs in 20 budding yeasts and six filamentous fungi reveal that in all but one case the YRA1 intron has a noncanonical branchpoint sequence. Most often the first position is altered to a purine. This pattern is especially apparent in the budding yeast species (Table 1) and prompted us to investigate the effect of variations in this sequence. Substitution of the consensus sequence did not impact growth in an otherwise wild-type strain background. Notably, however, when autoregulation is compromised by C-terminal mutations of Yra1p, this substitution causes inviability. The consensus substitution also strongly enhanced the dominant negative effect caused by shortening the YRA1 intron. Moreover, RNA analyses demonstrated that the introduction of the consensus branchpoint to a shortened intron improved splicing approximately threefold. Thus the presence of a noncanonical branchpoint is important for optimal autoregulation.
The branchpoint is sequentially recognized during the earliest steps of spliceosome assembly, first by the heterodimer of branchpoint-binding protein (BBP) and Mud2 (yU2AF65) and subsequently by base-pairing with U2 snRNA (for review, see Brow 2002). In principle, the reduced splicing efficiency of the YRA1 intron with its nonconsensus sequence could be due to the lowered affinity of the branchpoint for one or more of these interaction partners. Indeed, chromatin-immunoprecipitation experiments indicate that BBP has a reduced affinity for the YRA1 gene as compared with other intron-containing genes (G. Wilmes and C. Guthrie, unpubl.).
A short sequence element as a splicing enhancer
We were initially surprised to find that two 3-nt elements in the YRA1 intron that are separated by >600 nt positively affect splicing (Fig. 4A). That is, mutation of either of these elements alleviates the dominant negative phenotype due to a shortened intron. Interestingly, these elements are part of sequences that have the potential to form extensive base-pairing interactions, and, notably, this putative stem–loop is conserved in virtually all budding yeasts. Base-pairing between these elements could bring the 5′ and 3′ splice sites closer together, resulting in an “effective” intron length of 130 nt ± 17 (average for all budding yeast homologs), which is close to the average length of nonribosomal protein genes (Bon et al. 2003). Similar structures have been shown experimentally to be required for efficient splicing of larger introns, including those of the CYH2 and RPB51 genes (Newman 1987; Libri et al. 1995) and have been predicted for many more (Spingola et al. 1999). Complementary mutations predicted to restore base-pairing did not revert the phenotype of the individual mutations, as would be expected if the precise sequences, as well as base-pairing potential, were important. In support of this idea, the conserved GAA trinucleotide in the 5′ part of the predicted stem is also found at a corresponding position in the CYH2 intron (Fig. 4B). Alternatively, it is possible that the two elements affect splicing independently of secondary structure. Structure probing of the YRA1 pre-mRNA would be required to rule out this possibility.
Our results suggest that the YRA1 intron is a composite of negative and positive features. The apparent negative effects due to the suboptimal branchpoint and, in particular, the unusual length of the intron could be offset by long-range folding nucleated by the GAA “enhancer.” In this view, autoregulation might be accomplished by a partitioning of the intron structure in response to Yra1 protein concentration, as we discuss below. It would be interesting to see whether overexpression of Yra1p would affect splicing of either the ΔL10 intron or a deletion of the GAA “enhancer,” as we would predict.
A coupling-dependent mechanism
In searching for molecular mechanisms that might underlie length-dependence, we tested mutations in selected trans-acting factors for dominant negative phenotypes. Interestingly, we found that deletions in each of three components of the tetrameric THO complex are strongly hypersensitive to overexpression of the YRA1 gene. The strong dominant negative growth defects of these mutants are accompanied by a decrease in YRA1 pre-mRNA and accumulation of Yra1 protein. Importantly, chromatin-immunoprecipitation experiments have established that the THO complex is involved in the recruitment of Yra1p to nascent transcripts and that this cotranscriptional recruitment might explain the requirement of the THO complex for efficient mRNA export (Sträßer et al. 2002; Zenklusen et al. 2002). Sub2p, an essential ATPase that interacts with both Yra1p and the THO complex, has also been implicated in Yra1p loading onto the DBP2 transcript (Lei and Silver 2002). Intriguingly, this gene also contains a large intron. In fact, certain alleles of Sub2p are also hypersensitive to YRA1 overexpression (data not shown). The interpretation of these results, however, is complicated by the fact that Sub2p is itself also involved in splicing (Kistler and Guthrie 2001; Libri et al. 2001; Zhang and Green 2001). Further experiments are needed to differentiate the effect of different mutations in Sub2p on loading of Yra1p versus splicing of the YRA1 pre-mRNA.
Taken together, these observations suggest that proper autoregulation requires cotranscriptional loading of the Yra1 protein onto its own pre-mRNA. Indeed, Yra1p coimmunoprecipitates with its own pre-mRNA, as well as mRNA (K. Kim-Guisbert and C. Guthrie, unpubl.). A long transcript comprising a large 5′ exon and intron might provide more time for the THO complex to load Yra1p during the transcription process, just as increasing the levels of Yra1 protein should favor its cotranscriptional loading. An important implication of these results is that cotranscriptional loading of Yra1p is also required for the inhibition of splicing. That is, since high concentrations of Yra1 protein per se are unable to confer appropriate regulation, the splicing effect is presumably itself cotranscriptional. Although splicing can be shown to occur post-transcriptionally under certain experimental conditions in vivo (Lopez and Seraphin 2000), most reports are generally consistent with the cotranscriptional splicing of the several introns assayed (Howe et al. 2003). It is important to note that Yra1p is neither required for splicing nor is it a general splicing repressor (Wang and Rymond 2003; J. Pleiss, G. Whitworth, M. Bergkessel, and C. Guthrie, in prep.).
Taken together, our observations are consistent with a model in which appropriate autoregulation requires a careful balance between positive and negative factors. The natural YRA1 intron is intrinsically inefficient because it is long and has a suboptimal branchpoint. These negative features are partially compensated by a long-range folding structure that is promoted and/or stabilized at physiological concentrations of Yra1p. At high levels of Yra1 protein, RNA binding may become highly cooperative, inhibiting splicing in the same way that mammalian hnRNP proteins are thought to coat the introns that they negatively regulate (Black 2003). Interestingly, studies of the mammalian ortholog, Aly/REF, suggest that it can readily oligomerize (Virbasius et al. 1999).
A link to export
What is the fate of unspliced YRA1 transcript? A large proportion of the RPL30 pre-mRNA, which is also controlled by autoregulation, is found in the cytoplasm (Vilardell et al. 2000). In addition, Parker and coworkers (Hilleren and Parker 2003) have recently shown that incompletely spliced pre-mRNAs can be exported to the cytoplasm where they will eventually be degraded by the 5′ → 3′ exonuclease Xrn1p. Indeed, our data show that YRA1 pre-mRNA strongly accumulates in strains deleted for this nuclease, suggesting that the unspliced transcript is exported from the nucleus (Fig. 6D). Furthermore, microarray experiments have shown that the yra1-1 mutant that is deficient for regulation (Preker et al. 2002) accumulates its own pre-mRNA (J. Pleiss, G. Whitworth, M. Bergkessel and C. Guthrie, in prep.), suggesting that regulation may involve Yra1p promoting the export of its unspliced pre-mRNA. This would allow regulation to be achieved at least in part by competition between splicing and premature export of the pre-mRNA (Fig. 7). In this model, elevated levels of Yra1p would favor export of the unspliced YRA1 RNA. In the absence of functional Yra1p, the pre-mRNA might remain in the nucleus, and a fraction of it might eventually get spliced. This would be consistent with the kinetic competition model between splicing and nuclear export proposed by Hilleren and Parker (2003).
FIGURE 7.
Model for YRA1 autoregulation. In this model cotranscriptional recruitment of Yra1p by the THO complex as well as an inefficiently spliced intron are essential for regulation. Inefficient splicing results from an unusually long intron, which could alter formation of a long-range secondary structure required for productive splicing. A short intron might allow for more rapid secondary structure formation, while a consensus branchpoint sequence might bypass its need altogether. In this model, transcripts that do not undergo rapid cotranscriptional splicing can be exported to the cytoplasm, possibly through the action of Yra1p itself.
The branchpoint-binding protein (BBP/Msl5p) has been implicated in the retention of unspliced RNAs (Rutz and Seraphin 2000). Recent results from our laboratory (G. Wilmes and C. Guthrie, unpubl.) indicate that BBP interacts poorly with the YRA1 gene as compared with other intron-containing genes. Thus, in addition to its role in slowing down splicing, the noncanonical branchpoint might also facilitate “escape” of the YRA1 pre-mRNA into the cytoplasm by lowering the affinity of the pre-mRNA for BBP.
Other protein factors are also likely involved in regulation of YRA1 expression. Possibly, this system might serve to fine-tune Yra1p levels under different growth conditions with important consequences for the export rates of subsets of mRNAs. Yra1p regulation and its function in mRNA metabolism are a further illustration of the extensive coupling of transcription, processing and export. It will thus be very informative to elucidate the molecular mechanism of this process in detail.
MATERIALS AND METHODS
Cloning and identification of S. cerevisiae YRA1 homologs
To clone putative YRA1 homologs of Pichia canadiensis (strain IH207), S. unisporus (ARS Culture Collection no. NRRL Y-1556), and K. telluris (also known as Saccharomyces telluris; NRRL YB-4302), we recovered DNA from these strains using a standard protocol for the isolation of nucleic acids from S. cerevisiae (Guthrie and Fink 2002). Next, “guessmers” were designed based on regions of highest conservation between the YRA1 homologs known at that time (Souciet et al. 2000) and used to amplify partial YRA1 sequences by PCR. All PCR reactions used a degenerate reverse primer (SFO136, TCIAAITAGTCAGCCATYTC) complimentary to the extreme C terminus. The forward primer SFO135 (AAATGTCTGCWAAYTTIGATAAITCWTTWGA) is complementary to the extreme N terminus and together with SFO136 yielded sequences for S. unisporus and P. canadiensis. A primer annealing to the central RNA-binding domain and overlapping the 5′ splice site (underlined: SFO146, GACATTAAICAAGATGCYGTTAIGTATGT) was successful in amplifying the putative YRA1 homolog of K. teluris. The intron sequences have been submitted to the GenBank database under accession numbers DQ302761-DQ302763.
Putative homologs of S. cerevisiae Yra1p were identified by tBLASTn (Altschul et al. 1990) searches of public databases (including http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi?organism=fungi; http://www.broad.mit.edu/cgi-bin/annotation/fgi/blast_page.cgi; http://cbi.labri.fr/Genolevures/blast.php; http://agd.unibas.ch/; http://tigrblast.tigr.org/ufmg/; http://www.sanger.ac.uk/DataSearch/blast.shtml; and http://www.genome.wustl.edu/blast/yeast_client.cgi). Default search parameters were used, except that the filter for low complexity sequences was disabled. In some cases, only partial sequences were retrieved that could later be assembled into contigs by iterative BLAST searches. A dendogram was constructed using the ClustalW (http://clustalw.genome.jp/) and phylip (Version 3.6; http://evolution.genetics.washington.edu/phylip.html) algorithms.
Strains and plasmids constructions
Yeast media and manipulations were done following standard procedures (Guthrie and Fink 2002). G418 and 5-fluoro orotic acid (5FOA) were included in solid growth media at 0.2 and 1 mg/mL, respectively, as indicated.
The YRA1 shuffle strain PJP168-32C (MATa, ade2, his3, leu2, trp1, ura3, yra1rHIS3 <pRS316–YRA1>) has been described previously (Preker et al. 2002). Strains harboring deletions of specific genes were purchased from Research Genetics (Invitrogen) and are based on BY4741 (MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0).
DNA recombinant work was done according to standard protocols. Enzymes were purchased from New England Biolabs and Roche. The pRS series of shuttle vectors was used for expression of YRA1 (Christianson et al. 1992). Plasmids pRS424 and pRS425 are 2-μ-based (high-copy-number) vectors marked by TRP1 and LEU2, respectively. Plasmids pIA300 (pRS424–YRA1), pIA304 (pRS424–YRA1-ΔIVS), and pIA309 (pRS424–yra1-F223S) have been described previously (Preker et al. 2002). A 2047-bp XhoI/SacI fragment containing the entire YRAI gene was cloned from pIA300 into pRS425 yielding plasmid pIA557. Other plasmids used in this study are listed in Table 2. The sequences of oligonucleotides, where not given in this publication, are available upon request.
TABLE 2.
Plasmids used in this study not named in Materials and Methods
Mutations of the branchpoint sequence were introduced by site-directed mutagenesis and confirmed by sequencing. Plasmids pIA351, pIA377, and pIA378 are derivatives of pIA300 that encode nucleotides U, A, and C, respectively, at the first position of the branchpoint. Likewise, pIA427–429 are the equivalent mutations in the context of mutant YRA1 (pIA309).
For the creation of the ΔL01 and ΔL10 deletions, sequences within the intron were first replaced by site-directed mutagenesis. In the intermediate L01 (“linker 01”; pIA438) nt 443–454 of the intron were replaced by the sequence CCTCCCGGGTCC containing a unique SmaI site (underlined). In L10 (“linker 10”; pIA481), nt 545–557 were replaced by the sequence ACTAGTCCCGGG CGCC, containing a SmaI and an overlapping NarI site (italicized). Subsequently, deletions were created by digestion with BclI (targeting a unique restriction site located in the YRA1 intron 103 nt distal to the 5′ splice site), blunt-ending with T4 DNA polymerase, digestion with SmaI, and religation. ΔH/L10 is a shorter deletion of the region between a unique HpaI site 286 nt downstream from the 5′ splice site and the SmaI site.
Bacteriophage λ DNA was partially digested with TaqI, which creates ends that are compatible with NarI. Next, DNA fragments ranging in size from 150 to 300 bp were gel-purified, followed by ligation into NarI-restricted ΔL10. DNA sequencing of three isolates confirmed that they are derived from different regions of the λ genome. All constructs conferred viability when present as the only source of Yra1p in the cell (Fig. 2).
The YRA1 intron was replaced by heterologous introns, namely those from the S. cerevisiae RPS9B and SEC14 genes, as well as from the homologous C. albicans YRA1 gene, as detailed in Preker et al. (2002). The following oligonucleotide combinations were used: SFO176/284 (RPS9B), SFO196/197 (SEC14), and SFO178/179 (C. albicans YRA1). Recombinant was selected for growth on synthetic growth medium lacking tryptophan and confirmed by whole-cell PCR.
YRA1 alleles (yra1-ΔC11) were created by insertion of an in-frame stop codon that results in truncation of the last 11 amino acids from Yra1p as described in Preker et al. (2002).
The GAL1 promoter was introduced in front of pRS424-derived YRA1 construct (see Table 2 ) by homologous in vivo recombination of plasmids that had been linearized with XhoI and KpnI in the polylinker sequence upstream of the YRA1 gene and a PCR fragment containing the GAL1 promoter preceded by the kanamycin resistance gene. The PCR fragment was generated using pFa6-kanMX6-PGAL1 (Longtine et al. 1998) as a template and oligonucleotides SFO132 and SFO133. Recombinants were identified by its ability to grow on medium containing kanamycin but lacking tryptophan, recovered into E. coli, and retransformed into yeast. Plasmid pIA558 was constructed accordingly, except that the parental plasmid, pIA557 (pRS425–YRA1), was only linearized with KpnI.
Mutations of the conserved “GAA” motif (nt position 82–84) and the “TTC” motif (nt 712–714) of the predicted stem–loop into “TCC” and “GGA,” respectively, were created by site-directed mutagenesis. Both mutations contained BspEI sites. A construct containing the double mutant was also created, and the BspEI fragment was subsequently deleted from it (pIA535 in Table 2).
Isolation of total yeast RNA and dot-blot analysis
Total RNA from exponentially growing cells was isolated by extraction with hot acidic phenol as described (Preker et al. 2002). Next, 2.5 μL of a 1 μg/μL solution of RNA in water as well as serial dilutions were spotted on nylon membranes (Hybond N+, Amersham) in triplicate and UV-cross-linked at 80 mJ/cm2 in a GS Gene Linker (Biorad). Following prehybridization in “Rapid-hyb” buffer (Amersham) at 42°C for 90 min, the blots were separately probed with DNA oligonucleotides that had been labeled at their 5′ ends with ATP [γ32P] and T4 polynucleotide kinase. Oligonucleotides complementary to the intron and second exon of YRA1 were CCTTACAAAGAATATTTCTCGTATCC (SFO159) and CGGTACCAGTAGATTGGCCCCTTTC (SFO131), respectively. As a control, the third blot was probed with an oligonucleotide (CCAAGTTGGATTCAGTGGCTC, SFO082) complementary to U3 snoRNA. After incubation at 42°C overnight, the blots were washed once in 5× SSC/0.1% SDS and twice in 1× SSC/0.1% SDS for 30 min each at 25°C. The blots were exposed to a PhosphorImager. After subtraction of background, the signal from the YRA1 pre-mRNA and total RNA were normalized to U3 snoRNA levels.
Antibodies and Western blot analysis
To prepare protein extracts, pellets from logarithmically growing cells were resuspended in buffer (20 mM Tris/HCl at pH 7.4, 50 mM ammonium acetate, 2 mM EDTA). After addition of glass beads and trichloroacetic acid to a final concentration of 10%, cells were lyzed by vortexing for 5 min in the cold. Proteins were precipitated by centrifugation, washed with 80% acetone. and resuspended in SDS-PAGE loading buffer. Equivalent amount of extract (normalized to OD600 of the starting cultures) was separated by electrophoresis on SDS-polyacrylamide gels, blotted to nitrocellulose membrane, and probed with antibodies against Gle1p and Yra1p (Kashyap et al. 2005) at dilutions of 1:25′000 and 1:2′000, respectively. Secondary antibodies (goat anti-rabbit, Biorad) conjugated to horseradish peroxidase were used at a dilution of 1:2′000 and detected by enhanced chemoluminescence (Amersham) and exposure to film.
ACKNOWLEDGMENTS
We thank W. Gilbert and H. Madhani, as well as many members of the Guthrie laboratory for critical reading of the manuscript and helpful suggestions. We are indebted to D. Kellogg (University of California, Santa Cruz) for his generous gift of Yra1p antibodies. The Pichia canadiensis strain IH207 was obtained from I. Hershkowitz (University of California, San Francisco). M. Dinglasan provided excellent technical assistance. This work was supported by a grant from the NIH (GM21119) to C.G., who is also an American Cancer Society Research Professor of Molecular Genetics. P.J.P. was the recipient of a Research Special Fellowship Award from the Leukemia and Lymphoma Society during parts of this study.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.6706.
REFERENCES
- Abruzzi K.C., Lacadie S., Rosbash M. Biochemical analysis of TREX complex recruitment to intronless and intron-containing yeast genes. EMBO J. 2004;23:2620–2631. doi: 10.1038/sj.emboj.7600261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera A. mRNA processing and genomic instability. Nat. Struct. Mol. Biol. 2005;12:737–738. doi: 10.1038/nsmb0905-737. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Black D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Bon E., Casaregola S., Blandin G., Llorente B., Neuveglise C., Munsterkotter M., Guldener U., Mewes H.W., Van Helden J., Dujon B., et al. Molecular evolution of eukaryotic genomes: Hemiascomycetous yeast spliceosomal introns. Nucleic Acids Res. 2003;31:1121–1135. doi: 10.1093/nar/gkg213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brow D.A. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 2002;36:333–360. doi: 10.1146/annurev.genet.36.043002.091635. [DOI] [PubMed] [Google Scholar]
- Christianson T.W., Sikorski R.S., Dante M., Shero J.H., Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Draper D.E. How do proteins recognize specific RNA sites? New clues from autogenously regulated ribosomal proteins. Trends Biochem. Sci. 1989;14:335–338. doi: 10.1016/0968-0004(89)90167-9. [DOI] [PubMed] [Google Scholar]
- Eng F.J., Warner J.R. Structural basis for the regulation of splicing of a yeast messenger RNA. Cell. 1991;65:797–804. doi: 10.1016/0092-8674(91)90387-e. [DOI] [PubMed] [Google Scholar]
- Engebrecht J.A., Voelkel-Meiman K., Roeder G.S. Meiosis-specific RNA splicing in yeast. Cell. 1991;66:1257–1268. doi: 10.1016/0092-8674(91)90047-3. [DOI] [PubMed] [Google Scholar]
- Espinet C., de la Torre M.A., Aldea M., Herrero E. An efficient method to isolate yeast genes causing overexpression-mediated growth arrest. Yeast. 1995;11:25–32. doi: 10.1002/yea.320110104. [DOI] [PubMed] [Google Scholar]
- Fewell S.W., Woolford J.L., Jr. Ribosomal protein S14 of Saccharomyces cerevisiae regulates its expression by binding to RPS14B pre-mRNA and to 18S rRNA. Mol. Cell. Biol. 1999;19:826–834. doi: 10.1128/mcb.19.1.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C., Fink G.R. Guide to yeast genetics and molecular and cell biology. Academic Press; San Diego, CA: 2002. [Google Scholar]
- Hilleren P.J., Parker R. Cytoplasmic degradation of splice-defective pre-mRNAs and intermediates. Mol. Cell. 2003;12:1453–1465. doi: 10.1016/s1097-2765(03)00488-x. [DOI] [PubMed] [Google Scholar]
- Howe K.J., Kane C.M., Ares M., Jr. Perturbation of transcription elongation influences the fidelity of internal exon inclusion in Saccharomyces cerevisiae . RNA. 2003;9:993–1006. doi: 10.1261/rna.5390803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P., Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell. 2003;12:711–721. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- Jensen T.H., Boulay J., Rosbash M., Libri D. The DECD box putative ATPase Sub2p is an early mRNA export factor. Curr. Biol. 2001;11:1711–1715. doi: 10.1016/s0960-9822(01)00529-2. [DOI] [PubMed] [Google Scholar]
- Jimeno S., Rondon A.G., Luna R., Aguilera A. The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J. 2002;21:3526–3535. doi: 10.1093/emboj/cdf335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap A.K., Schieltz D., Yates J., III, Kellogg D.R. Biochemical and genetic characterization of Yra1p in budding yeast. Yeast. 2005;22:43–56. doi: 10.1002/yea.1185. [DOI] [PubMed] [Google Scholar]
- Kim M., Ahn S.H., Krogan N.J., Greenblatt J.F., Buratowski S. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J. 2004;23:354–364. doi: 10.1038/sj.emboj.7600053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler A.L., Guthrie C. Deletion of MUD2, the yeast homolog of U2AF65, can bypass the requirement for sub2, an essential spliceosomal ATPase. Genes & Dev. 2001;15:42–49. doi: 10.1101/gad.851301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei E.P., Silver P.A. Intron status and 3′-end formation control cotranscriptional export of mRNA. Genes & Dev. 2002;16:2761–2766. doi: 10.1101/gad.1032902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei E.P., Krebber H., Silver P.A. Messenger RNAs are recruited for nuclear export during transcription. Genes & Dev. 2001;15:1771–1782. doi: 10.1101/gad.892401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri D., Stutz F., McCarthy T., Rosbash M. RNA structural patterns and splicing: Molecular basis for an RNA-based enhancer. RNA. 1995;1:425–436. [PMC free article] [PubMed] [Google Scholar]
- Libri D., Graziani N., Saguez C., Boulay J. Multiple roles for the yeast SUB2/yUAP56 gene in splicing. Genes & Dev. 2001;15:36–41. doi: 10.1101/gad.852101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri D., Dower K., Boulay J., Thomsen R., Rosbash M., Jensen T.H. Interactions between mRNA export commitment, 3′-end quality control, and nuclear degradation. Mol. Cell. Biol. 2002;22:8254–8266. doi: 10.1128/MCB.22.23.8254-8266.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae . Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lopez P.J., Seraphin B. Uncoupling yeast intron recognition from transcription with recursive splicing. EMBO Rep. 2000;1:334–339. doi: 10.1093/embo-reports/kvd065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandabalan K., Roeder G.S. Binding of a cell-type-specific RNA splicing factor to its target regulatory sequence. Mol. Cell. Biol. 1995;15:1953–1960. doi: 10.1128/mcb.15.4.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman A. Specific accessory sequences in Saccharomyces cerevisiae introns control assembly of pre-mRNAs into spliceosomes. EMBO J. 1987;6:3833–3839. doi: 10.1002/j.1460-2075.1987.tb02720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portman D.S., O'Connor J.P., Dreyfuss G. YRA1, an essential Saccharomyces cerevisiae gene, encodes a novel nuclear protein with RNA annealing activity. RNA. 1997;3:527–537. [PMC free article] [PubMed] [Google Scholar]
- Preker P.J., Kim K.S., Guthrie C. Expression of the essential mRNA export factor Yra1p is autoregulated by a splicing-dependent mechanism. RNA. 2002;8:969–980. doi: 10.1017/s1355838202020046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Navarro S., Strasser K., Hurt E. An intron in the YRA1 gene is required to control Yra1 protein expression and mRNA export in yeast. EMBO Rep. 2002;3:438–442. doi: 10.1093/embo-reports/kvf091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutz B., Seraphin B. A dual role for BBP/ScSF1 in nuclear pre-mRNA retention and splicing. EMBO J. 2000;19:1873–1886. doi: 10.1093/emboj/19.8.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souciet J., Aigle M., Artiguenave F., Blandin G., Bolotin-Fukuhara M., Bon E., Brottier P., Casaregola S., de Montigny J., Dujon B., et al. Genomic exploration of the hemiascomycetous yeasts: 1. A set of yeast species for molecular evolution studies. FEBS Lett. 2000;487:3–12. doi: 10.1016/s0014-5793(00)02272-9. [DOI] [PubMed] [Google Scholar]
- Spingola M., Ares M., Jr. A yeast intronic splicing enhancer and Nam8p are required for Mer1p-activated splicing. Mol. Cell. 2000;6:329–338. doi: 10.1016/s1097-2765(00)00033-2. [DOI] [PubMed] [Google Scholar]
- Spingola M., Grate L., Haussler D., Ares M., Jr. Genome-wide bioinformatic and molecular analysis of introns in Saccharomyces cerevisiae . RNA. 1999;5:221–234. doi: 10.1017/s1355838299981682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sträßer K., Hurt E. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 2000;19:410–420. doi: 10.1093/emboj/19.3.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sträßer K., Hurt E. Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature. 2001;413:648–652. doi: 10.1038/35098113. [DOI] [PubMed] [Google Scholar]
- Sträßer K., Masuda S., Mason P., Pfannstiel J., Oppizzi M., Rodriguez-Navarro S., Rondon A.G., Aguilera A., Struhl K., Reed R., et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- Stutz F., Bachi A., Doerks T., Braun I.C., Seraphin B., Wilm M., Bork P., Izaurralde E. REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA. 2000;6:638–650. doi: 10.1017/s1355838200000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilardell J., Chartrand P., Singer R.H., Warner J.R. The odyssey of a regulated transcript. RNA. 2000;6:1773–1780. doi: 10.1017/s135583820000145x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virbasius C.M., Wagner S., Green M.R. A human nuclear-localized chaperone that regulates dimerization, DNA binding, and transcriptional activity of bZIP proteins. Mol. Cell. 1999;4:219–228. doi: 10.1016/s1097-2765(00)80369-x. [DOI] [PubMed] [Google Scholar]
- Wang Q., Rymond B.C. Rds3p is required for stable U2 snRNP recruitment to the splicing apparatus. Mol. Cell. Biol. 2003;23:7339–7349. doi: 10.1128/MCB.23.20.7339-7349.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D., Vinciguerra P., Wyss J.C., Stutz F. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol. Cell. Biol. 2002;22:8241–8253. doi: 10.1128/MCB.22.23.8241-8253.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Green M.R. Identification and characterization of yUAP/Sub2p, a yeast homolog of the essential human pre-mRNA splicing factor hUAP56. Genes & Dev. 2001;15:30–35. doi: 10.1101/gad.851701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]