Abstract
We have developed a series of fluorescent splicing reporter minigenes for the establishment of cell-based screens to identify splicing regulatory proteins. A key technical advance in the application of these reporters was the use of two different fluorescent proteins: EGFP and monomeric Red Fluorescent Protein (mRFP). Through establishment of stable cell lines expressing such dual color fluorescent reporters, these minigenes can be used to perform enhanced screens for splicing regulatory proteins. As an example of such applications we generated fluorescent minigenes that can be used to determine the splicing of mutually exclusive FGFR2 exons IIIb and IIIc by flow cytometry. One minigene contained a coding sequence for EGFP whose translation was dependent on splicing of exon IIIb, whereas a second minigene required exon IIIc splicing for translation of an mRFP coding sequence. Stable incorporation of both minigenes into cells that express endogenous FGFR2-IIIb or FGFR2-IIIc resulted in EGFP or mRFP fluorescence, respectively. Cells stably transfected with both minigenes were used to screen a panel of cDNAs encoding known splicing regulatory proteins, and several were identified that induced a switch in splicing that could be detected specifically by an increase in green, but not red, fluorescence. We further demonstrated additional minigenes that can be used in dual color fluorescent screens for identification of splicing regulatory proteins that function through specific intronic splicing enhancer elements (ISEs). The methods and minigene designs described here should be adaptable for broader applications in identification of factors and mechanisms involved in alternative splicing of numerous other gene transcripts.
Keywords: alternative splicing, fluorescent minigenes, exons, introns, RNA-binding proteins
INTRODUCTION
Alternative splicing is a highly utilized process in mammalian cells whereby multiple mRNAs can be produced from the same gene transcript, and in many cases the products of these different mRNAs have divergent, or even opposing, functions (Black 2003). In recent years, several computational and experimental studies have suggested progressively higher estimates of the percentage of mammalian gene transcripts that undergo alternative splicing; at present, it appears likely that at least 60%–70% of human genes are alternatively spliced (Modrek and Lee 2002; Johnson et al. 2003). While the precise functional relevance of different spliced forms of specific transcripts has thus far been delineated in a limited number of cases, there is no question that alternative splicing yields variants with differential functions that are critical for development as well as for maintenance of normal cellular homeostasis and intercellular communication pathways (Matlin et al. 2005). As such, it has also become evident that alterations in the regulation of splicing can have drastic consequences resulting in human diseases, including cancer (Xu and Lee 2003; Brinkman 2004; Hui et al. 2004; Venables 2004; Relogio et al. 2005).
Our current understanding of alternative splicing mechanisms in mammals has largely been derived from focused studies of specific alternatively spliced genes using transfected minigene models and in vitro splicing assays (Black 2003). These studies have led to the identification of auxiliary RNA cis-elements that, together with consensus sequences at 5′ and 3′ splice sites, collectively influence splicing of alternative exons. These cis-elements either enhance or suppress splicing and are generally located within or in close proximity to regulated exons and are thereby classified as exonic splicing enhancers (ESEs), exonic splicing silencers (ESSs), intronic splicing enhancers (ISEs), or intronic splicing silencers (ISSs). It is believed that the ability of the auxiliary cis-elements to regulate splicing is generally mediated by RNA-binding proteins that interact with them and either promote or hinder splicing at adjacent splice sites. Such splicing regulatory proteins include ubiquitously expressed factors as well as those with a more tissue or cell-type limited pattern of expression (Bourgeois et al. 2004; Ladd et al. 2004; Ule et al. 2005; Underwood et al. 2005). The best characterized elements have been the ESEs and ESSs, which have been shown to play a role in splicing of both constitutive as well as alternatively spliced exons (Zheng 2004). In addition to modulating the splicing of alternative exons, it appears that ESEs and ESSs also play a prominent role in maintaining the general fidelity of splicing of mammalian transcript by directing splicing only of “authentic” splice sites as opposed to “decoy” splice sites (Pozzoli and Sironi 2005; Zhang et al. 2005a,b).
Intronic cis-elements have been less extensively catalogued and mechanistically characterized than exonic elements. While intronic regulatory elements may play a role in constitutive splicing, they have largely been characterized in the regulation of alternatively spliced exons (Ladd and Cooper 2002). Intronic sequences flanking exons that are alternatively spliced in both human and mouse tissues often display extensive sequence identity, especially in the 100 nt adjacent to the 3′ or 5′ splice sites (Sorek and Ast 2003; Yeo et al. 2005). This same degree of sequence identity is not seen in introns flanking constitutive exons, suggesting that these conserved sequences contain functional cis-elements that bind splicing regulatory factors that regulate alternative exons. A challenge to our understanding of the mechanisms that regulate mammalian alternative splicing has been that multiple exonic and intronic elements influence splicing of regulated exons. As such, the splicing outcome often appears to result from the combinatorial effects of regulatory factors that bind these elements (Black 2003; Matlin et al. 2005).
A more comprehensive characterization of alternative splicing pathways and mechanisms that control them will require new tools to study functionally relevant examples of splicing regulation. Recently, microarray platforms that can distinguish different splice isoforms have shown the capability to reveal global patterns of splicing regulation (Yeakley et al. 2002; Johnson et al. 2003; Lee and Roy 2004; Pan et al. 2004; Srinivasan et al. 2005; Ule et al. 2005). In addition, more elegant genetic strategies using mouse models to delineate targets of mammalian splicing regulatory factors have been used to dissect their functions (Ule et al. 2003; Ding et al. 2004; Ladd et al. 2005; Lin et al. 2005; Xu and Fu 2005; Xu et al. 2005). Another tool that has recently been applied to studies of splicing regulation has been use of fluorescent reporter constructs that recapitulate the splicing pattern of the endogenous gene for alternatively spliced exons, facilitating identification of splicing differences via fluorescence analysis (Sheives and Lynch 2002; Toba et al. 2002; Ellis et al. 2004; Wagner et al. 2004). Such constructs have been used to identify splicing patterns in animal models but also can be used in cultured cell lines to establish genetic screens for splicing regulatory factors and cis-elements (Sheives and Lynch 2002; Wang et al. 2004). While different designs used in such constructs have been shown to allow recognition of splicing patterns through fluorescence analysis, some limitations in previously described fluorescent reporters may hinder their use in genetic screening applications. For example, some reporter constructs have been described in which an alternatively spliced exon and flanking intron sequences are inserted into a heterologous intron that disrupts an EGFP coding sequence. Inclusion of the alternative exon is designed to interrupt the EGFP reading frame, whereas skipping restores the EGFP open reading frame (ORF). This design can be used to study silencing of the internal exon by monitoring increases in fluorescence that correlate with a switch from its inclusion to skipping. However, background levels of skipping of the exon can complicate use of fluorescence as a direct indicator of exon skipping (Ellis et al. 2004; Wagner et al. 2004; R.P. Carstens, unpubl.). Furthermore, differences or changes in fluorescence may not always reflect differences in splicing. For example, differences in transcription of the fluorescent reporter as well as in the stability or translation of the transcribed RNA could also affect the level of fluorescence. In addition, we have found that when using traditional eukaryotic expression vectors with selectable markers to isolate cell lines carrying such fluorescent reporters, there is often a gradual decline in expression of the reporter even from cloned cell lines maintained under selective conditions. Here we describe a series of fluorescent splicing reporter constructs designed to circumvent some of these technical limitations in order to establish cell-based screens for splicing regulatory proteins.
RESULTS
Generation of FGFR2 splicing reporter minigenes for analysis of alternative splicing using fluorescence
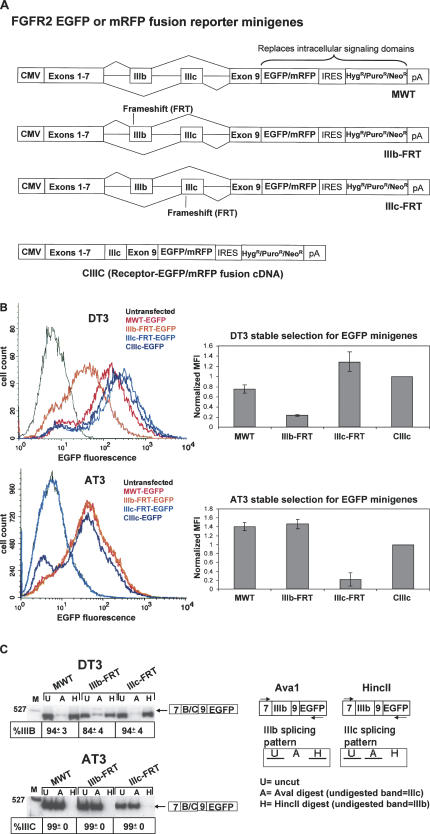
These studies were initiated toward a goal of identification of trans-acting splicing regulatory proteins that are required for the highly cell-type-specific splicing of mutually exclusive exons IIIb and IIIc of FGFR2 (Fig. 1). These exons encode different peptides in the extracellular domain of FGFR2 and yield receptors with dramatic differences in ligand-binding specificity that have important functional consequences during development (Xu et al. 1998; De Moerlooze et al. 2000). Our studies of FGFR2 splicing have primarily used the rat prostate cancer cell lines, DT3 and AT3, which exclusively splice exon IIIb or IIIc, respectively, to yield FGFR2-IIIb or FGFR2-IIIc. In addition, human cell lines C4-2 (Wu et al. 1994) and 293 have been used, which yield exclusively FGFR2-IIIb and FGFR2-IIIc, respectively. Several intronic and exonic cis-elements within and flanking exons IIIb and IIIc have been described, including several elements in intron 8 that enhance splicing of exon IIIb, as shown schematically in Figure 1 (Hovhannisyan and Carstens 2005 and references therein). Although several ubiquitously expressed proteins that bind these elements have been described, the mechanisms that give rise to distinct cell-type-specific expression of these isoforms remain poorly understood. To complement traditional approaches toward identification of splicing regulators, we also sought to establish functional screens for factors that regulate FGFR2 splicing in vivo. One rationale was that a functionally relevant protein might regulate splicing without directly interacting with the RNA transcript itself. For example, changes in phosphorylation of splicing factors by kinases and phosphatases have been shown to modulate binding, function, or localization of several splicing regulators and would be difficult to identify using traditional biochemical approaches (Kanopka et al. 1998; Matter et al. 2002; Patel et al. 2005). To establish these screens, we adapted previously described FGFR2 minigenes in which FGFR2 cDNA sequences encoding the exons upstream of exon IIIb, including the natural translational start codon, were fused with a cassette containing intron 7, exon IIIb, intron 8, exon IIIc, intron 9, and exon 9 (Jones et al. 2001). This minigene was joined with a downstream coding sequence for either EGFP or mRFP in which the usual start codon at the 5′-end of the ORF was changed from ATG (Met) to ATC (Ile) to preclude translational initiation at this position (Fig. 2A). This design thus replaces the intracellular FGFR2 tyrosine kinase domains with sequences encoding either fluorescent protein. Because the number of nucleotides in exon IIIb and exon IIIc (148 and 145 nt, respectively) are not multiples of three, maintenance of an ORF that will translate the fluorescent component of the fusion protein requires that one or the other exon be included in the spliced mRNA. Skipping of both exons, or inclusion of both exon IIIb and IIIc, results in a frame that terminates at stop codons upstream of either fluorescent coding sequence. In addition, the presence of numerous stop codons in the introns precludes the possibility of generating a fluorescent product from unspliced or partially spliced pre-mRNAs. We also generated minigenes in which a frameshifting nucleotide was inserted into either exon IIIb or exon IIIc. These minigenes, IIIb-FRT and IIIc-FRT, generate a frame that ends in a stop codon immediately preceding the fluorescent protein-coding sequence if the respective exon is included in the spliced transcript. As a result, production of a fluorescent fusion protein only occurs if the exon without the frameshift mutation is spliced. As a control, we also generated a “prespliced” cDNA control containing exon IIIc (CIIIc) that would generate an FGFR2-fluorescent protein fusion independent of splicing. In all minigenes and controls we also deleted an extracellular peptide required for receptor dimerization to prevent direct association of the minigene products with each other or with the products of the endogenous FGFR2 gene (Wang et al. 1997). These minigenes were inserted in bicistronic expression vectors upstream of an Internal Ribosome Entry Site (IRES) directing expression of antibiotic selectable markers (e.g., neomycin-, hygromycin-, or puromycin-resistance genes). This expression system facilitated establishment of pools of stably transfected cell lines whereby nearly all cells that survived in selective media expressed the minigenes since the selectable gene was expressed under the control of the same promoter. This system also ensured that the expression of the fluorescent minigenes was maintained during prolonged culture. In previous experiments using the same minigenes in vectors containing selectable markers expressed under the control of an independent promoter, we noted a gradual loss of minigene expression over time despite maintenance of selective conditions. Furthermore, in these cases, only one-quarter to one-third of the cells that survived the initial selection also expressed the minigene based on flow cytometric analysis.
FIGURE 1.
Schematic demonstrating alternative splicing of exons IIIb and IIIc. At top is a schematic of the protein showing the extracellular region in which exons IIIb and IIIc encode different peptides. Below is a map demonstrating the pre-mRNA region containing these alternative exons together with auxiliary cis-elements in intron 8 previously shown to promote exon IIIb inclusion (hatched boxes). (TM) Transmembrane domain, (TK) tyrosine kinase domains, (Ig) immunoglobulin-like domain. Shaded boxes represent exons, and solid lines represent introns.
FIGURE 2.
Fluorescent FGFR2 minigenes can be used to detect the alternative splicing pattern of exons IIIb and IIIc in live cultured cells. (A) Schematic of the fluorescent minigenes. The MWT minigene has no alterations in either exon IIIb or exon IIIc. Introduction of a single additional nucleotide (frameshift) in either exon eliminates fluorescent protein expression when the affected exon is spliced. A control, the CIIIc minigene, expresses the fluorescent fusion protein containing exon IIIc without splicing and, similar to the MWT minigenes, fluorescence cells that splice exon IIIb or IIIc. (B) Determination of cell-type-specific splicing by flow cytometry in pooled stably transfected cells. Introduction of a frameshift (FRT) mutation in exon IIIb or IIIc abrogates green fluorescence in DT3 or AT3 cells, respectively. Raw data from a representative experiment are shown at left. Mean fluorescence intensities (MFI) from three independent experiments were determined as the computed mean of each experimental cell population (without a gate). Data from each experiment were normalized by dividing the MFI obtained with each minigene by that of CIIIc. These data are shown graphically at right with error bars to represent the standard deviation from three independent experiments. (C) RT-PCR to analyze the splicing pattern from transfected minigenes. Average percentage of IIIc inclusion for AT3 cells and IIIb inclusion for DT3 cells is shown with standard deviations from three independent experiments. Exon IIIb inclusion was determined as the percentage of the RT-PCR product band remaining after HincII digestion divided by the sum of this band and the band remaining after AvaI digestion. Exon IIIc inclusion was determined as the percentage of the RT-PCR product band remaining after AvaI digestion divided by the sum of this band and the band remaining after HincII digestion. Shown at right is a schematic to illustrate results expected following digestion of RT-PCR products. (U) Uncut, (A) AvaI digestion, (H) HincII digestion, (M) molecular weight marker lane. Arrows indicate the locations of primers used for RT-PCR.
Fluorescence of cells stably transfected with FGFR2 minigenes can be used to determine the splicing pattern of the endogenous FGFR2 gene transcript
We stably transfected DT3 and AT3 cells with the set of minigenes shown in Figure 2A, and pooled cells were subjected to flow cytometric analysis, which is presented in Figure 2B. As expected, the minigenes with the wild-type sequences (MWT) and the cDNA control (CIIIc) both exhibited green fluorescence in both DT3 and AT3 cells that was easily distinguishable from that in cells transfected with an empty vector control. In DT3 cells, an equivalent level of fluorescence was seen with the IIIc-FRT-EGFP minigene, but a markedly lower level of fluorescence was observed with IIIb-FRT-EGFP. Conversely, in AT3 cells, reduced fluorescence was observed in cells stably transfected with the IIIc-FRT-EGFP minigene, whereas IIIb-FRT-EGFP yielded results equivalent to those with the MWT minigene or cDNA (CIIIc) control (Fig. 2B). These results were consistent with our predicted outcomes and suggested that the frame-dependent fluorescence indeed required cell-type-specific inclusion of exon IIIb or IIIc. To confirm that cell-type-specific splicing of exons IIIb and IIIc in these minigenes recapitulated that obtained from the endogenous gene, we performed an RT-PCR assay to distinguish exon IIIb from IIIc inclusion. RT-PCR confirmed that splicing of the exons from the wild-type minigene as well as minigenes containing frameshift mutations yielded predominantly exon IIIb inclusion in DT3 cells and exon IIIc inclusion in AT3 cells (Fig. 2C). However, in contrast to similar analysis of the endogenous transcript, a small but detectable amount of products containing exon IIIc was detected in DT3 cells. We also stably transfected these rat FGFR2 minigenes in the human C4-2 and 293 cell lines (that splice exon IIIb or IIIc, respectively) and confirmed that they also recapitulated the splicing pattern of endogenous FGFR2 in a manner that can be determined by fluorescence (data not shown). Taken together, the RNA analysis indicates that the relative fluorescence observed with the minigenes containing the frameshift mutations in exons IIIb or IIIc can be used to determine the inclusion level of the unmodified exon. Because this level of fluorescence can be determined in live cultured cells, these minigenes can be used to directly assess the level of exon IIIb or IIIc inclusion without the need to lyse cells and harvest RNA. Transfection of the same set of minigenes using mRFP in place of EGFP yielded similar results, indicating that flow cytometric analysis for this fluorescent protein can similarly be used as an indicator of exon IIIb or exon IIIc splicing (data not shown).
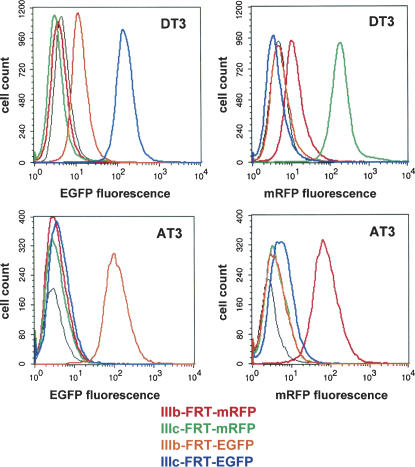
Establishment of flow cytometry parameters that enable specific detection of fluorescence from minigenes containing EGFP or mRFP
Because of the distinctly different excitation and emission wavelengths of mRFP and EGFP, microscopic or flow cytometric analysis can be carried out in which expression of either fluorescent protein can be distinguished, even in the same cell. As a result, we expected that it would be possible to independently assess splicing of two different reporters present in the same cell, provided that one of the minigenes contained the EGFP coding sequence and the other included the mRFP coding sequence. Prior to generating cell lines containing these reporters, we evaluated our ability to distinguish fluorescence generated by these minigenes using fluorescence analysis of live stably transfected DT3 and AT3 cells by flow cytometric analysis. Pools of DT3 and AT3 were stably transfected with the IIIb-FRT or IIIc-FRT minigenes containing either the EGFP or mRFP coding sequence (four different minigenes). By using advanced flow cytometry systems, such as the LSR II (BD Biosciences), in which precise wavelengths of excitation and detection can be specified, conditions have been established that permit sensitive detection of both EGFP and mRFP without any bleeding through of one fluorescent color to the other. This permits independent flow cytometric analysis for expression of either fluorescent protein in the same cell.
We subjected DT3 and AT3 cells transfected with the minigenes described above to fluorescent analysis using conditions set up for detection of each fluorescent protein, and the results are shown in Figure 3. In DT3 cells, using settings for EGFP (FL1) we noted the highest level of fluorescence with IIIc-FRT-EGFP and a significantly lower level of fluorescence with IIIb-FRT-EGFP, consistent with the relative differences in exon IIIb and IIIc inclusion described previously. No fluorescence was detected in cells transfected with IIIb-FRT-mRFP or IIIc-FRT-mRFP. However, when settings optimal for mRFP (532-nm green laser, 585/42-nm detection settings) were used, IIIc-FRT-mRFP yielded the highest fluorescence, with lower levels from IIIb-FRT-mRFP and none from either EGFP containing minigene. As described previously, DT3 cells exhibit a low level of IIIc inclusion, and thus the low level of fluorescence observed with the IIIb-FRT minigenes is attributable to its inclusion. Similar results were also observed in AT3 cells, with the significant difference being that in both cases fluorescence was higher with the IIIb-FRT than IIIc-FRT minigenes. Thus, both fluorescent proteins can be detected separately from one another.
FIGURE 3.
Green and red fluorescence can be independently detected by flow cytometry. (Upper left) DT3 cells display significantly greater green fluorescence with the IIIb-FRT-EGFP than the IIIc-FRT-EGFP minigenes, but none with mRFP containing minigenes at these settings. (Upper right) DT3 cells display greater red fluorescence with the IIIb-FRT-mRFP than the IIIc-FRT-mRFP minigenes, but none with EGFP containing minigenes using these settings. (Lower left) Fluorescence is only detected in AT3 cells stably expressing minigene IIIb-FRT-EGFP. (Lower right) Under conditions established for RFP detection, fluorescence is only detected in AT3 cells expressing IIIb-FRT-mRFP. The black line represents results from control untransfected cells.
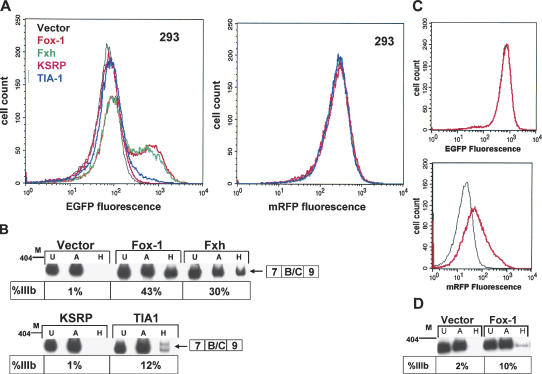
Stable cell lines containing dual color minigene reporters facilitate detection of changes in FGFR2 splicing regulation induced by trans-acting protein factors
The results described thus far confirmed that splicing outcomes from minigenes containing either EGFP or mRFP can independently be analyzed by flow cytometry. Therefore, cells can be identified and sorted using fluorescence activated cell sorting (FACS) on the basis of the relative fluorescence of either (or both) of these fluorescent markers. Since fluorescence of cells expressing these minigenes can be used to monitor splicing of FGFR2 exon IIIb (using the IIIc-FRT minigenes) or exon IIIc (using the IIIb-FRT minigenes), cells expressing these minigenes represent an ideal substrate for the establishment of genetic screens for the identification of functional splicing regulators. A simple screen using cells stably expressing a single fluorescent reporter could be used for these purposes. However, changes in fluorescence of a single reporter could be achieved through gain or loss of factors that would not necessarily be due to changes in splicing. For example, changes in transcription, transport, mRNA stability, or translational efficiency of minigene-derived transcripts could likewise affect fluorescence. To obviate this possibility and establish more elegant genetic strategies, we established cell lines stably expressing two minigenes in which exon IIIb or exon IIIc inclusion is reflected by relative EGFP or mRFP expression from the respective minigenes. This was achieved through use of a IIIc FRT-EGFP minigene containing a puromycin-resistance cassette and a IIIb-FRT-mRFP minigene linked to hygromycin resistance. Cells stably expressing both minigenes were obtained by sequential transfection and selection with the appropriate antibiotic. After stable insertion of both minigenes, DT3 cells displayed easily detectable green fluorescence, but only low levels of red fluorescence (data not shown). Predictably, AT3 cells instead displayed intense red fluorescence but green fluorescence that was near background levels (data not shown). 293 Cells, which also express FGFR2-IIIc, also displayed intense red fluorescence, and significantly lower green fluorescence. Thus, cells expressing this combination of minigenes enabled use of either EGFP or mRFP expression to independently determine the level of exon IIIb or IIIc splicing.
The resulting cell lines expressing both EGFP and mRFP containing splicing reporters can be used in screens in which an induced increase in the level of only one fluorescent protein is highly predictive of a change in splicing. Because both minigenes contain the same CMV promoter and 5′- and 3′-untranslated regions (UTRs), any induced changes in the level of transcription, stability, or translational efficiency would be expected to yield parallel changes in EGFP and mRFP. Therefore, through use of FACS-based sorting to select cells that only cause a shift in fluorescence of one marker, we can more robustly and elegantly screen for cellular mutants that switch splicing or for the ability of exogenously expressed regulatory factors to induce a splicing switch. To validate the feasibility of using these cell lines for this purpose, we tested the ability of a panel of previously described splicing regulatory proteins to induce changes in FGFR2 splicing in a 293 cell clone stably expressing both fluorescent minigenes. Although splicing regulation of FGFR2 is complex and involves combinatorial functions of several regulatory factors, several observations are consistent with the function of at least one protein required for exon IIIb inclusion, but not for exon IIIc inclusion (Carstens et al. 1998). Therefore, overexpression of such factors in 293 cells might result in a shift from predominant inclusion of exon IIIc to exon IIIb that can be detected as an increase in EGFP, but not mRFP. Although expression of a single factor may not be sufficient to cause a complete switch in exon IIIc or exon IIIb splicing, an added benefit of use of fluorescence to establish these screens is that even a partial switch in splicing can be detected. As shown in Figure 4A, transient transfection of plasmids expressing cDNAs for several regulatory proteins resulted in detectable increases in EGFP without parallel changes in mRFP. Analysis of endogenous FGFR2 splicing by RT-PCR confirmed that the relative level of EGFP obtained after transfection of each protein corresponded to partial switches from inclusion of exon IIIc to exon IIIb (Fig. 4B). The most robust switch was observed in response to overexpression of the related Fox-1 and Fxh RNA-binding proteins (Lieberman et al. 2001; Jin et al. 2003). While we anticipated that a decrease in mRFP expression might result, we suspect this reduction was not detected, because it would not only require a reduction in production of exon IIIc containing spliced mRNAs from the mRFP minigene but also turnover of pre-existing FGFR2-mRFP fusion proteins. We also noted a small but reproducible increase in exon IIIb splicing in response to TIA-1. A TIA-1 effect is consistent with previous reports demonstrating that it binds to uridine-rich sequences downstream of exon IIIb and enhances its inclusion (Del Gatto-Konczak et al. 2000). In addition to the factors shown in Figure 4, several additional RNA-binding proteins (FBP, Etr-3, CELF4, CELF6, and TLS/FUS) were also tested that did not result in changes in splicing of either reporter or the endogenous FGFR2 transcript (data not shown). While these results demonstrated the ability of several proteins to increase exon IIIb splicing, the supra-physiologic level of expression often achieved in transient transfections might suggest that the observed effects of these proteins do not reflect a normal biological role in FGFR2 splicing. Therefore, we also determined the feasibility of using 293 cells containing both reporters to detect changes in splicing following stable integration of a cDNA expressing such factors. To further demonstrate that these experiments can also be performed with the respective fluorescent proteins swapped, we used a 293 cell clone stably expressing IIIc-FRT-mRFP and IIIb-FRT-EGFP minigenes. The Fox-1 cDNA in a bicistronic plasmid encoding blasticidin resistance was transfected in these cells, and selection was carried out using blasticidin. This bicistronic design is expected to yield a population of cells in which nearly all stably express Fox-1. As shown in Figure 4C, stable expression of Fox-1 was also capable of inducing a partial switch from exon IIIc to exon IIIb splicing, although in this case the magnitude was lower. In this case, an increase in red fluorescence was observed without a change in green fluorescence (Fig. 4C). Again, the increase in fluorescence correlated with changes in splicing of the endogenous transcript (Fig. 4D). Taken together, the results in 293 cells indicate that use of two color splicing assays should facilitate larger scale screens for FGFR2 splicing regulators using either loss- or gain-of-function approaches.
FIGURE 4.
Changes in splicing of endogenous FGFR2 in response to overexpression of splicing regulatory factors can be detected specifically using cell lines stably expressing dual color fluorescent reporter minigenes. (A) A 293 clonal cell line expressing both IIIc-FRT-EGFP and IIIb-FRT-mRFP was transiently transfected with constructs expressing the indicated RNA binding proteins (RBPs) or the same expression construct without an inserted cDNA (Vector). (B) Confirmation that the induced increases in green fluorescence represents changes in splicing of endogenous FGFR2. RNAs from the same cells analyzed by flow cytometry were analyzed by RT-PCR. Percentage IIIb inclusion was determined as described in the legend to Figure 2C. (C) An increase in red, but not green, fluorescence was observed in response to stable transfection with a Fox-1 cDNA in a 293 clonal cell line containing the opposite combination of minigene/fluorescent protein. (D) RT-PCR analysis demonstrating increased exon IIIb inclusion in response to stable expression of Fox-1.
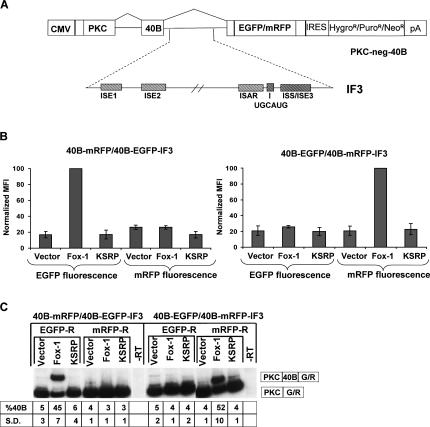
Dual color fluorescent reporters can be used to detect induced splicing changes via defined ISEs
The results shown in Figure 4 show the applicability of cell lines containing these reporters for identification of trans-acting factors involved in regulation of FGFR2 exons IIIb and IIIc. While identification of such regulators should be highly informative, these minigenes contain numerous regulatory cis-elements in all three introns as well as both exons. As a result, several regulators may be identified without elucidating mechanisms or cis-elements that mediate their effects on splicing. Therefore, we also established dual color fluorescence assays that allow us to discover factors that enhance exon IIIb splicing through the ISEs located in intron 8. For this purpose, we adapted minigenes from a reporter construct that was successfully used to detect cell-type-specific functions of ISE/ISS-3 using fluorescence (Hovhannisyan et al. 2006). As shown in Figure 5A, these minigenes contain an ORF consisting of a 159-nt ORF derived from rat protein kinase C (PKC)-γ in a 5′-terminal exon. In the parental PKC-neg-40B minigenes, a single 40-nt artificial exon (exon 40B), together with flanking introns, separates this ORF from a 3′-terminal exon containing a coding sequence for either EGFP or mRFP. Splicing of the 40B exon maintains an ORF that yields green or red fluorescence, whereas skipping of the exon generates a frame terminating in a stop codon upstream of the fluorescent protein reading frame. Due to to its small size and weak splice sites, this artificial exon is included in only 3%–5% of spliced transcripts in transfected 293 cells; the predominant splicing pathway leads to skipping of the exon and thus low levels of green or red fluorescence. However, insertion of FGFR2 intron 8 ISE elements downstream of this exon results in substantial activation of exon 40B splicing in DT3 cells but not in AT3 or 293 cells (data not shown). This activation is most robust when an intronic fragment (intron fragment 3, IF3) containing all five of the ISE elements shown in Figure 5A is inserted downstream of the test exon. In contrast, insertion of an intron 8 fragment that has been truncated at the 3′-end to exclude both the UGCAUG element and ISE/ISS-3 does not result in increased exon inclusion in any cell lines tested (data not shown). Although factors that bind ISE/ISS-3 and regulate splicing have not been described, Fox-1 and Fxh (also referred to as Fox-2) have been shown to bind UGCAUG sequence elements and mediate splicing enhancement (Jin et al. 2003; Underwood et al. 2005). Therefore, the most straightforward mechanism by which these factors can promote exon IIIb splicing in 293 cells would include binding to this sequence in intron 8 (Baraniak et al. 2003). Because the most robust activation of splicing requires the contribution of all five of the indicated FGFR2 cis-elements, we cotransfected 293 cells with combinations of these minigenes in which one fluorescent reporter contained IF3 (with all indicated ISEs including UGCAUG), whereas the second reporter with the other fluorescent marker contained no sequences from FGFR2. These reporters were also cotransfected with either an empty expression vector, or the same vector containing cDNAs for Fox-1 and KSRP. As shown in Figure 5B, when the mRFP reporter contained IF3 (40B-mRFP-IF3) and the EGFP reporter did not (40B-EGFP), Fox-1 induced an increase in red, but not green, fluorescence as predicted. KSRP, which was previously shown not to increase exon IIIb splicing, showed no change in fluorescence of either marker. When the opposite combination of minigene/fluorescent protein (40B-EGFP-IF5 and 40B-mRFP) was used, we noted an increase in green, but not red, fluorescence with Fox-1 overexpression. To confirm that the specific changes in either fluorescent reporter corresponded to changes in inclusion of the 40B exon, we also performed RT-PCR assays with RNAs from the same cells that were analyzed by flow cytometry. As shown in Figure 5C, Fox-1 induced an increase in exon 40B inclusion to >40% in minigenes containing the FGFR2 IF3 sequences. In contrast, neither the empty vector control nor KSRP elicited any change in splicing whether IF3 was present or not. Therefore, these findings validated the utility of this dual color fluorescent approach to identify factors that mediate splicing enhancement by a defined ISE or set of ISEs.
FIGURE 5.
Dual color fluorescent assays can be used to detect splicing changes mediated by specific auxiliary cis-elements. (A) Schematic of the heterologous parent PKC-neg-40B minigene (top) and FGFR2 intron 8 fragments (IF3) that were inserted downstream of exon 40B and tested for splicing activation. (B) 293 Cells were transiently cotransfected with PKC-40B minigenes with and without FGFR2 ISEs (containing opposing fluorescent markers) together with empty expression plasmid (Vector), Fox-1, or KSRP. Increased fluorescence is only observed when the fluorescent marker is present in minigenes containing FGFR2 intron fragment 3. Flow cytometry results from three experiments are shown as mean fluorescence intensities (MFI) normalized to Fox-1. Normalization was performed by dividing each MFI by that of Fox-1 and multiplying by 100. Error bars represent standard deviations. (C) RT-PCR confirmation that induced increases in fluorescence correspond to activation of exon 40B splicing only when FGFR2 auxiliary cis-elements are present. The percentage of spliced products containing exon 40B (vs. exon skipping) is indicated beneath each lane as well as the standard deviation (S.D.) compiled from three independent experiments.
DISCUSSION
We have developed several robust fluorescent splicing reporter minigenes that facilitate highly sensitive determination of alternative splicing outcomes in live cells. We show that these minigene reporters can be used to determine cell-type-specific patterns of splicing using flow cytometric analysis. In addition, these minigenes can be used to detect changes in alternative splicing that are induced by trans-acting splicing regulatory proteins. Furthermore, we describe cell-based systems that can be generated using these minigenes that should be highly effective for the establishment of either gain- or loss-of-function genetic screens for the identification of regulatory proteins. Furthermore, minigenes have been developed that enable fine-tuned screens for factors that mediate regulation via specific intronic cis-elements. Several design features of these constructs are described that significantly enhance their ability to establish cell-based screens. First, we found that use of an IRES-based bicistronic expression system substantially facilitated establishment and maintenance of stably transfected cell lines expressing the fluorescent minigenes. Second, the positions of the introns as well as the sequences located between the alternatively spliced regions and the fluorescent coding regions were carefully designed to eliminate the likelihood that cryptic splicing events could also lead to fluorescence. Third, the design in which the default splicing pathway (exon skipping) does not elicit fluorescence reduces background and enhances the power for detecting changes in splicing as a positive selection for increased fluorescence. Fourth, by placing the fluorescent coding sequences well downstream of the 5′-end of the transcript and eliminating the ATG start codon of the fluorescent protein coding sequence, the possibility of alternative translation start sites leading to fluorescence is effectively abrogated. Finally, the use of two different fluorescent proteins with expression coupled to different selective markers enables the establishment of genetic screening strategies that are highly sensitive and specific for induced changes in splicing pathways. As previously discussed, the derivation of cell lines expressing two different fluorescent minigene variants can essentially eliminate the possibility that factors affecting transcription or other post-transcriptional steps could complicate screens for splicing regulators.
Using 293 cell lines expressing these reporters, we have demonstrated the most direct manner in which a screen can be carried out to identify splicing regulators through a gain-of-function approach. A potential limitation to carrying out cDNA-library-based screens in this manner is the possibility that exogenous expression of a single factor may be insufficient to lead to changes in splicing. Nevertheless, the use of a fluorescent readout permits the identification of even small or partial changes in splicing pathway. Our results demonstrating that several factors could independently induce a partial switch in FGFR2 splicing indicates that these reporters can be used in larger screens to identify additional FGFR2 regulatory factors, including possible cell-type-specific factors that may promote expression of FGFR2-IIIb. However, the application of this method for large-scale screening should be undertaken with caution. Although fairly substantial changes in FGFR2 splicing were observed in response to Fox family proteins in transiently transfected 293 cells, it is unlikely that the same degree of change would be observed using standard screening methods. The high transfection efficiency and high transcriptional activity of the CMV promoter in 293 cells suggests that the expression levels of these proteins are likely supra-physiologic, and thus the degree of splicing switch observed is likely higher than these factors could achieve on their own at physiologic expression levels. In fact, the level of switch in response to stably incorporated Fox-1 was substantially lower. It is also noteworthy that we saw no increase in exon IIIb splicing from either the reporter minigenes or endogenous FGFR2 in AT3 cells in response to overexpression of Fox-1. Thus, even in cells that express predominantly FGFR2-IIIc, there may be differences in how “hardwired” this splicing choice is that may influence the feasibility of this type of screen.
An alternative approach would be a loss-of-function screen whereby mutagenesis is carried out and cells are selected using fluorescence to indicate an induced switch in splicing. One manner of performing such a screen would be through use of chemical mutagens in DT3 cells expressing the IIIb-FRT-EGFP and IIIc-FRT-mRFP minigenes and isolation of mutants displaying increased green fluorescence and decreased red fluorescence (Stark and Gudkov 1999). A similar approach using fluorescent reporters, in fact, generated mutants defective for signal-induced splicing of CD45 (Sheives and Lynch 2002). Isolation of recessive mutant DT3 cells could then be followed by complementation, or rescue of the mutant gene with a DT3-cell-derived cDNA library. Technically, such an approach may require extensive mutagenesis, as isolation of recessive mutants is likely to necessitate mutation of both alleles of functional FGFR2 splicing regulators. Nonetheless, an analogous strategy using chemical mutagenesis and rescue with a cDNA library was used successfully to identify a factor involved in IL-3 mRNA turnover by establishing a GFP reporter construct that reflected the stability of the encoded mRNA (Stoecklin et al. 2000, 2002). Interestingly, one difficulty encountered in these studies using a single fluorescent reporter was that most cellular mutants initially identified in the screen were due to changes in transcription of the reporter, complicating isolation of mutants with changes in mRNA stability (Stoecklin et al. 2000). Use of dual fluorescent reporters should preclude this difficulty. However, a more direct loss-of-function screen could also be performed using recently described RNA interference libraries. Although the efficacy of such libraries to effectively silence expression of most cellular genes remains to be established, such tools have already been used successfully in cell-based genome-wide screens (Kolfschoten et al. 2005; Westbrook et al. 2005). Further development of such libraries to provide full genome-wide silencing capability should make such an approach feasible and effective for identification of splicing regulators using fluorescent systems such as described here.
An additional application of fluorescent splicing reporters is to generate transgenic mouse models to identify splicing patterns in whole animals (Ellis et al. 2004). Such applications are of interest in studies to identify splicing patterns of certain genes during development as well as changes in splicing that occur in disease models, including cancer. Changes in FGFR2 splicing have been described in several models of cancer progression, including implication in epithelial–mesenchymal transitions (Yan et al. 1993; Thiery and Chopin 1999; Yasumoto et al. 2004). The FGFR2 splicing reporters described here would thus be applicable for studies to define stages of carcinogenesis at which splicing patterns are altered. The modular nature of the constructs described here should facilitate further development of additional reporter constructs for study of numerous alternative splicing events in cell culture as well as to study the role of alternative splicing in animal models of development and disease.
MATERIALS AND METHODS
Plasmid construction
The plasmid DNA constructs used in this study were made using standard cloning techniques. FGFR2 reporter minigenes and the cDNA control were derived from the previously described MWt and CIIIc minigenes (Jones et al. 2001). PCR was used to generate EGFP and mRFP cDNAs that substituted ATC in place of the normal start codon, and the resulting products were inserted downstream of FGFR2 exon 9 in the original minigenes, replacing a luciferase coding sequence present in the original plasmids. The EGFP template used was pEGFP-N1 (Clontech), and mRFP was derived from a construct kindly provided by Dr. Roger Tsien (Campbell et al. 2002). In all minigenes, sequences encoding the peptide ERSPHRPI in the FGFR2 extracellular domain were deleted to prevent receptor association. Minigenes containing frameshift mutations (IIIb-FRT and IIIc-FRT) were generated by adding a C nucleotide at position 119 in exon IIIb or position 113 in exon IIIc. Constructs were further modified by deletion of sequences from the middle of introns 7, 8, and 9. This reduced the size of intron 7 from 1076 to 414 nt, intron 8 from 1212 to 595 nt, and intron 9 from 1994 to 557 nt. These deletions were accompanied by insertion of EcoRI and BamHI sites in intron 7 and an XhoI site in intron 9, facilitating potential further modifications. These intron modifications resulted in no significant differences in splicing compared to those with the full intron sequences (data not shown). The bicistronic plasmids were derived from pIRESneo3, pIRESpuro3, and pIREShyg3 (Clontech). A synthetic intron located downstream of the multicloning site in pIRESneo3 and pIRESpuro3 was deleted, and the sequence between the AgeI and EcoRI sites was replaced with 5′-CCGGCGCGCCGCGACCATGGTCTCCGGA-3′ to yield vectors termed pIRESneo3ΔInt-AAPBE and pIRESpuro3ΔInt-AAPBE. The inserted sequence contained a unique PshAI site, and the sequences encoding the minigene-fluorescent protein fusions were inserted into the modified pIRESpuro3 and pIRESneo3 vectors using PshAI and NotI. Minigenes containing a hygromycin resistance cassette were generated by removing a NheI to NotI insert from the MCS of pIRESneo3 ΔInt-AAPBE and insertion into the respective sites in pIREShyg3. The resulting vector was further modified to eliminate an XhoI site by digestion, Pfu polymerase fill-in, and re-ligation to yield pIREShyg3ΔInt-AAPBE. This manipulation also eliminated a synthetic intron present in the parent plasmid. Minigene/fluorescent protein cassettes were then inserted after digestion from pIRESneo3ΔInt-AAPBE with AgeI and NotI and insertion into the same sites in pIREShyg3ΔInt-AAPBE. A bicistronic expression plasmid containing a blasticidin-resistance gene was created by first removing the neomycin-resistance cassette from pIRESneo3ΔInt-AAPBE with XmaI and XbaI. A coding sequence for blasticidin resistance (BSD) was amplified from the pCDNA6/V5-His plasmid (Invitrogen) using primers 5′-CCCCGGGATCCATGGCCAAGCCTTTGTCTCA-3′ and 5′-CTCAGATTAGCCCTCCCACACATAAC-3′. The PCR product was digested with XmaI and XbaI and inserted in place of the removed neomycin-resistance gene to generate pIRESblast2 ΔInt-AAPBE. cDNAs for RNA binding regulatory proteins were inserted in this vector for determination of their ability to change FGFR2 splicing. Fluorescent vectors containing a single artificial exon were created in several steps from the previously described PKC-neg-EGFP plasmid (Hovhannisyan et al. 2006). PKC-neg-33.51-EGFP was generated by removing the previously described exon 33.51 (and flanking splice sites) from pI-XN-33.51 with BamHI and XhoI and insertion into the same sites in the intron of PKC-neg-EGFP (Hovhannisyan and Carstens 2005). The 46-nt 33.51 exon was replaced with a 40-nt exon termed 40B, 5′-GGTTCACAACCATCGACCGCCTCAAGAAGATCTCTGACAG-3′. In addition, the last 8 nt of the 3′ splice site intron sequence (upstream of exon 33.51) was changed from 5′-TGGCTCAG-3′ to 5′-ACCCTTAG-3′ and the intronic portion of the 5′ splice site from 5′-GTAGTT-3′ to 5′-GTTGGT-3′. The resulting vector was termed PKC-neg-40B-EGFP. A related PKC-neg 40B-mRFP vector was created by replacing the coding sequence for EGFP with the mRFP coding sequence. Intron fragment 3 (IF3) is as previously described and was inserted into NotI and ClaI sites located in the intron downstream of exon 40B (Hovhannisyan and Carstens 2005). Separate versions of these minigenes were created using resistance genes for neomycin, puromycin, or hygromycin. Additional details regarding these plasmids, including sequences and maps, are available on request.
Transfection of minigenes
Transfections of DT3 and AT3 cells were carried out as described (Muh et al. 2002). 293 Cells were transfected with Mirus Transit 293 transfection reagent according to the manufacturer's protocol. In transient transfection experiments, cells were harvested at 48 h for further analysis. Cotransfections were performed using equal amounts of the different plasmids. For stable selection, cells were transferred to selective media 24–48 h after transfection and selected for at least 2 wk prior to further analysis. The concentrations used were 1.0–1.5 μg/mL puromycin for DT3, C4-2, and 293 cells, and 15 μg/mL puromycin for AT3 cells. The G418 and hygromycin concentration used was 400–500 μg/mL. Blasticidin was used at 10–20 μg/mL.
Flow cytometric analysis (FCM) and FACS
Flow cytometric analysis (FCM) of EGFP alone was carried out using a FACScalibur (BD Biosciences) using FL1. For two-color fluorescent analysis, cells were analyzed using a BD LSRII with blue lasers configured at 610/20 for mRFP fluorescence and 530/30 for EGFP fluorescence. Compensation was performed in order to make sure that one fluorochrome didn't interfere with the other. Analysis was carried out using BD CellQuest Pro. Resulting mean fluorescence intensities (MFI) were determined as the computed mean of the fluorescence exhibited by a given population of cells on an arbitrary four-decade log scale. Isolation of single cell fluorescent clones was performed by FACS using a BD FACSVantage DIVA SE with blue lasers configured at 610/20 to detect mRFP fluorescence and 530/30 to detect EGFP fluorescence. To ensure that both green and red fluorescent proteins were expressed in sorted cells, parameters were established such that the top 25%–50% of cells based on fluorescence were single-cell-sorted into 96 well plates.
RNA isolation and RT-PCR analysis
Preparation of total RNA, reverse transcription, and PCR was performed essentially as previously described (Muh et al. 2002). For analysis of the splicing pattern from the endogenous transcript, the primers used were 5′-CCCGGGGAATTCACCACCATGCAGGCGATTAA-3′ and 5′-CCCGGGTCTAGATTTATAGTGATGCCCAGCCC-3′. Analysis of the fluorescent FGFR2 minigenes was carried out with forward primer 5′-GGCGCGCCGCGACCATGGTCTGGATCAAACATGTGGAAAA-3′ and either 5′-CTCGCCGGACACGCTGAACTTG-3′ (EGFP-R) or 5′-TTGGAGCCGTACTGGAACTGAGG-3′ (mRFP-R). For analysis of splicing using the PKC-neg.40B minigenes, we used forward primer 5′-CCAGATGCCCGCAGCCCCACAA-3′ and either EGFP-R or mRFP-R. Digestion with AvaI or HincII to evaluate exon IIIb or exon IIIc inclusion was performed as described (Muh et al. 2002). Quantification was done using a Molecular Dynamics PhosphorImager with correction for molar equivalents.
ACKNOWLEDGMENTS
We thank Arthur Tsai for comments on the manuscript and Lori Gostomski for technical support. We also thank Doug Black, Tom Cooper, Kunio Inoue, David Levens, Andrew Lieberman, David Ron, and Roger Tsien for providing plasmids used in this study. This work was supported by Public Health Service Grant CA093769 to R.P.C. W.L.M. is supported by PHS Grant CA59971.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.34906.
REFERENCES
- Baraniak A.P., Lasda E.L., Wagner E.J., Garcia-Blanco M.A. A stem structure in fibroblast growth factor receptor 2 transcripts mediates cell-type-specific splicing by approximating intronic control elements. Mol. Cell. Biol. 2003;23:9327–9337. doi: 10.1128/MCB.23.24.9327-9337.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Bourgeois C.F., Lejeune F., Stevenin J. Broad specificity of SR (serine/arginine) proteins in the regulation of alternative splicing of pre-messenger RNA. Prog. Nucleic Acid Res. Mol. Biol. 2004;78:37–88. doi: 10.1016/S0079-6603(04)78002-2. [DOI] [PubMed] [Google Scholar]
- Brinkman B.M. Splice variants as cancer biomarkers. Clin. Biochem. 2004;37:584–594. doi: 10.1016/j.clinbiochem.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Campbell R.E., Tour O., Palmer A.E., Steinbach P.A., Baird G.S., Zacharias D.A., Tsien R.Y. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carstens R.P., McKeehan W.L., Garcia-Blanco M.A. An intronic sequence element mediates both activation and repression of rat fibroblast growth factor receptor 2 pre-mRNA splicing. Mol. Cell. Biol. 1998;18:2205–2217. doi: 10.1128/mcb.18.4.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Gatto-Konczak F., Bourgeois C.F., Le Guiner C., Kister L., Gesnel M.C., Stevenin J., Breathnach R. The RNA-binding protein TIA-1 is a novel mammalian splicing regulator acting through intron sequences adjacent to a 5′ splice site. Mol. Cell. Biol. 2000;20:6287–6299. doi: 10.1128/mcb.20.17.6287-6299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Moerlooze L., Spencer-Dene B., Revest J., Hajihosseini M., Rosewell I., Dickson C. An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal–epithelial signalling during mouse organogenesis. Development. 2000;127:483–492. doi: 10.1242/dev.127.3.483. [DOI] [PubMed] [Google Scholar]
- Ding J.H., Xu X., Yang D., Chu P.H., Dalton N.D., Ye Z., Yeakley J.M., Cheng H., Xiao R.P., Ross J., et al. Dilated cardiomyopathy caused by tissue-specific ablation of SC35 in the heart. EMBO J. 2004;23:885–896. doi: 10.1038/sj.emboj.7600054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis P.D., Smith C.W., Kemp P. Regulated tissue-specific alternative splicing of enhanced green fluorescent protein transgenes conferred by α-tropomyosin regulatory elements in transgenic mice. J. Biol. Chem. 2004;279:36660–36669. doi: 10.1074/jbc.M405380200. [DOI] [PubMed] [Google Scholar]
- Hovhannisyan R.H., Carstens R.P. A novel intronic cis element, ISE/ISS-3, regulates rat fibroblast growth factor receptor 2 splicing through activation of an upstream exon and repression of a downstream exon containing a noncanonical branch point sequence. Mol. Cell. Biol. 2005;25:250–263. doi: 10.1128/MCB.25.1.250-263.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovhannisyan R.H., Warzecha C.C., Carstens R.P. Characterization of sequences and mechanisms through which ISE/ISS-3 regulates FGFR2 splicing. Nucleic Acids Res. 2006;34:373–385. doi: 10.1093/nar/gkj407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui L., Zhang X., Wu X., Lin Z., Wang Q., Li Y., Hu G. Identification of alternatively spliced mRNA variants related to cancers by genome-wide ESTs alignment. Oncogene. 2004;23:3013–3023. doi: 10.1038/sj.onc.1207362. [DOI] [PubMed] [Google Scholar]
- Jin Y., Suzuki H., Maegawa S., Endo H., Sugano S., Hashimoto K., Yasuda K., Inoue K. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J. 2003;22:905–912. doi: 10.1093/emboj/cdg089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J.M., Castle J., Garrett-Engele P., Kan Z., Loerch P.M., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- Jones R.B., Wang F., Luo Y., Yu C., Jin C., Suzuki T., Kan M., McKeehan W.L. The nonsense-mediated decay pathway and mutually exclusive expression of alternatively spliced FGFR2IIIb and -IIIc mRNAs. J. Biol. Chem. 2001;276:4158–4167. doi: 10.1074/jbc.M006151200. [DOI] [PubMed] [Google Scholar]
- Kanopka A., Muhlemann O., Petersen-Mahrt S., Estmer C., Ohrmalm C., Akusjarvi G. Regulation of adenovirus alternative RNA splicing by dephosphorylation of SR proteins. Nature. 1998;393:185–187. doi: 10.1038/30277. [DOI] [PubMed] [Google Scholar]
- Kolfschoten I.G., van Leeuwen B., Berns K., Mullenders J., Beijersbergen R.L., Bernards R., Voorhoeve P.M., Agami R. A genetic screen identifies PITX1 as a suppressor of RAS activity and tumorigenicity. Cell. 2005;121:849–858. doi: 10.1016/j.cell.2005.04.017. [DOI] [PubMed] [Google Scholar]
- Ladd A.N., Cooper T.A. Finding signals that regulate alternative splicing in the post-genomic era. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-11-reviews0008. reviews0008.1–0008.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladd A.N., Nguyen N.H., Malhotra K., Cooper T.A. CELF6, a member of the CELF family of RNA-binding proteins, regulates muscle-specific splicing enhancer-dependent alternative splicing. J. Biol. Chem. 2004;279:17756–17764. doi: 10.1074/jbc.M310687200. [DOI] [PubMed] [Google Scholar]
- Ladd A.N., Taffet G., Hartley C., Kearney D.L., Cooper T.A. Cardiac tissue-specific repression of CELF activity disrupts alternative splicing and causes cardiomyopathy. Mol. Cell. Biol. 2005;25:6267–6278. doi: 10.1128/MCB.25.14.6267-6278.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Roy M. Analysis of alternative splicing with microarrays: successes and challenges. Genome Biol. 2004;5:231. doi: 10.1186/gb-2004-5-7-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman A.P., Friedlich D.L., Harmison G., Howell B.W., Jordan C.L., Breedlove S.M., Fischbeck K.H. Androgens regulate the mammalian homologues of invertebrate sex determination genes tra-2 and fox-1. Biochem. Biophys. Res. Commun. 2001;282:499–506. doi: 10.1006/bbrc.2001.4617. [DOI] [PubMed] [Google Scholar]
- Lin S., Xiao R., Sun P., Xu X., Fu X.D. Dephosphorylation-dependent sorting of SR splicing factors during mRNP maturation. Mol. Cell. 2005;20:413–425. doi: 10.1016/j.molcel.2005.09.015. [DOI] [PubMed] [Google Scholar]
- Matlin A.J., Clark F., Smith C.W. Understanding alternative splicing: Towards a cellular code. Nat. Rev. Mol. Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- Matter N., Herrlich P., Konig H. Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature. 2002;420:691–695. doi: 10.1038/nature01153. [DOI] [PubMed] [Google Scholar]
- Modrek B., Lee C. A genomic view of alternative splicing. Nat. Genet. 2002;30:13–19. [Google Scholar]
- Muh S.J., Hovhannisyan R.H., Carstens R.P. A non-sequence-specific double-stranded RNA structural element regulates splicing of two mutually exclusive exons of fibroblast growth factor receptor 2 (FGFR2) J. Biol. Chem. 2002;277:50143–50154. doi: 10.1074/jbc.M207409200. [DOI] [PubMed] [Google Scholar]
- Pan Q., Shai O., Misquitta C., Zhang W., Saltzman A.L., Mohammad N., Babak T., Siu H., Hughes T.R., Morris Q.D., et al. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol. Cell. 2004;16:929–941. doi: 10.1016/j.molcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Patel N.A., Kaneko S., Apostolatos H.S., Bae S.S., Watson J.E., Davidowitz K., Chappell D.S., Birnbaum M.J., Cheng J.Q., Cooper D.R. Molecular and genetic studies imply Akt-mediated signaling promotes protein kinase CβII alternative splicing via phosphorylation of serine/arginine-rich splicing factor SRp40. J. Biol. Chem. 2005;280:14302–14309. doi: 10.1074/jbc.M411485200. [DOI] [PubMed] [Google Scholar]
- Pozzoli U., Sironi M. Silencers regulate both constitutive and alternative splicing events in mammals. Cell. Mol. Life Sci. 2005;62:1579–1604. doi: 10.1007/s00018-005-5030-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relogio A., Ben-Dov C., Baum M., Ruggiu M., Gemund C., Benes V., Darnell R.B., Valcarcel J. Alternative splicing microarrays reveal functional expression of neuron-specific regulators in Hodgkin lymphoma cells. J. Biol. Chem. 2005;280:4779–4784. doi: 10.1074/jbc.M411976200. [DOI] [PubMed] [Google Scholar]
- Sheives P., Lynch K.W. Identification of cells deficient in signaling-induced alternative splicing by use of somatic cell genetics. RNA. 2002;8:1473–1481. [PMC free article] [PubMed] [Google Scholar]
- Sorek R., Ast G. Intronic sequences flanking alternatively spliced exons are conserved between human and mouse. Genome Res. 2003;13:1631–1637. doi: 10.1101/gr.1208803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan K., Shiue L., Hayes J.D., Centers R., Fitzwater S., Loewen R., Edmondson L.R., Bryant J., Smith M., Rommelfanger C., et al. Detection and measurement of alternative splicing using splicing-sensitive microarrays. Methods. 2005;37:345–359. doi: 10.1016/j.ymeth.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Stark G.R., Gudkov A.V. Forward genetics in mammalian cells: Functional approaches to gene discovery. Hum. Mol. Genet. 1999;8:1925–1938. doi: 10.1093/hmg/8.10.1925. [DOI] [PubMed] [Google Scholar]
- Stoecklin G., Ming X.F., Looser R., Moroni C. Somatic mRNA turnover mutants implicate tristetraprolin in the interleukin-3 mRNA degradation pathway. Mol. Cell. Biol. 2000;20:3753–3763. doi: 10.1128/mcb.20.11.3753-3763.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin G., Colombi M., Raineri I., Leuenberger S., Mallaun M., Schmidlin M., Gross B., Lu M., Kitamura T., Moroni C. Functional cloning of BRF1, a regulator of ARE-dependent mRNA turnover. EMBO J. 2002;21:4709–4718. doi: 10.1093/emboj/cdf444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery J.P., Chopin D. Epithelial cell plasticity in development and tumor progression. Cancer Metastasis Rev. 1999;18:31–42. doi: 10.1023/a:1006256219004. [DOI] [PubMed] [Google Scholar]
- Toba G., Qui J., Koushika S.P., White K. Ectopic expression of Drosophila ELAV and human HuD in Drosophila wing disc cells reveals functional distinctions and similarities. J. Cell Sci. 2002;115:2413–2421. doi: 10.1242/jcs.115.11.2413. [DOI] [PubMed] [Google Scholar]
- Ule J., Jensen K.B., Ruggiu M., Mele A., Ule A., Darnell R.B. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- Ule J., Ule A., Spencer J., Williams A., Hu J.S., Cline M., Wang H., Clark T., Fraser C., Ruggiu M., et al. Nova regulates brain-specific splicing to shape the synapse. Nat. Genet. 2005;37:844–852. doi: 10.1038/ng1610. [DOI] [PubMed] [Google Scholar]
- Underwood J.G., Boutz P.L., Dougherty J.D., Stoilov P., Black D.L. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol. Cell. Biol. 2005;25:10005–10016. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables J.P. Aberrant and alternative splicing in cancer. Cancer Res. 2004;64:7647–7654. doi: 10.1158/0008-5472.CAN-04-1910. [DOI] [PubMed] [Google Scholar]
- Wagner E.J., Baines A., Albrecht T., Brazas R.M., Garcia-Blanco M.A. Imaging alternative splicing in living cells. Methods Mol. Biol. 2004;257:29–46. doi: 10.1385/1-59259-750-5:029. [DOI] [PubMed] [Google Scholar]
- Wang F., Kan M., McKeehan K., Jang J.H., Feng S., McKeehan W.L. A homeo-interaction sequence in the ectodomain of the fibroblast growth factor receptor. J. Biol. Chem. 1997;272:23887–23895. doi: 10.1074/jbc.272.38.23887. [DOI] [PubMed] [Google Scholar]
- Wang Z., Rolish M.E., Yeo G., Tung V., Mawson M., Burge C.B. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119:831–845. doi: 10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Westbrook T.F., Martin E.S., Schlabach M.R., Leng Y., Liang A.C., Feng B., Zhao J.J., Roberts T.M., Mandel G., Hannon G.J., et al. A genetic screen for candidate tumor suppressors identifies REST. Cell. 2005;121:837–848. doi: 10.1016/j.cell.2005.03.033. [DOI] [PubMed] [Google Scholar]
- Wu H.C., Hsieh J.T., Gleave M.E., Brown N.M., Pathak S., Chung L.W. Derivation of androgen-independent human LNCaP prostatic cancer cell sublines: Role of bone stromal cells. Int. J. Cancer. 1994;57:406–412. doi: 10.1002/ijc.2910570319. [DOI] [PubMed] [Google Scholar]
- Xu X., Fu X.D. Conditional knockout mice to study alternative splicing in vivo. Methods. 2005;37:387–392. doi: 10.1016/j.ymeth.2005.07.019. [DOI] [PubMed] [Google Scholar]
- Xu Q., Lee C. Discovery of novel splice forms and functional analysis of cancer-specific alternative splicing in human expressed sequences. Nucleic Acids Res. 2003;31:5635–5643. doi: 10.1093/nar/gkg786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Weinstein M., Li C., Naski M., Cohen R.I., Ornitz D.M., Leder P., Deng C. Fibroblast growth factor receptor 2 (FGFR2)-mediated reciprocal regulation loop between FGF8 and FGF10 is essential for limb induction. Development. 1998;125:753–765. doi: 10.1242/dev.125.4.753. [DOI] [PubMed] [Google Scholar]
- Xu X., Yang D., Ding J.H., Wang W., Chu P.H., Dalton N.D., Wang H.Y., Bermingham J.R., Jr., Ye Z., Liu F., et al. ASF/SF2-regulated CaMKIIδ alternative splicing temporally reprograms excitation–contraction coupling in cardiac muscle. Cell. 2005;120:59–72. doi: 10.1016/j.cell.2004.11.036. [DOI] [PubMed] [Google Scholar]
- Yan G., Fukabori Y., McBride G., Nikolaropolous S., McKeehan W.L. Exon switching and activation of stromal and embryonic fibroblast growth factor (FGF)-FGF receptor genes in prostate epithelial cells accompany stromal independence and malignancy. Mol. Cell. Biol. 1993;13:4513–4522. doi: 10.1128/mcb.13.8.4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasumoto H., Matsubara A., Mutaguchi K., Usui T., McKeehan W.L. Restoration of fibroblast growth factor receptor2 suppresses growth and tumorigenicity of malignant human prostate carcinoma PC-3 cells. Prostate. 2004;61:236–242. doi: 10.1002/pros.20093. [DOI] [PubMed] [Google Scholar]
- Yeakley J.M., Fan J.B., Doucet D., Luo L., Wickham E., Ye Z., Chee M.S., Fu X.D. Profiling alternative splicing on fiber-optic arrays. Nat. Biotechnol. 2002;20:353–358. doi: 10.1038/nbt0402-353. [DOI] [PubMed] [Google Scholar]
- Yeo G.W., Van Nostrand E., Holste D., Poggio T., Burge C.B. Identification and analysis of alternative splicing events conserved in human and mouse. Proc. Natl. Acad. Sci. 2005;102:2850–2855. doi: 10.1073/pnas.0409742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.H., Kangsamaksin T., Chao M.S., Banerjee J.K., Chasin L.A. Exon inclusion is dependent on predictable exonic splicing enhancers. Mol. Cell. Biol. 2005a;25:7323–7332. doi: 10.1128/MCB.25.16.7323-7332.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.H., Leslie C.S., Chasin L.A. Computational searches for splicing signals. Methods. 2005b;37:292–305. doi: 10.1016/j.ymeth.2005.07.011. [DOI] [PubMed] [Google Scholar]
- Zheng Z.M. Regulation of alternative RNA splicing by exon definition and exon sequences in viral and mammalian gene expression. J. Biomed. Sci. 2004;11:278–294. doi: 10.1159/000077096. [DOI] [PMC free article] [PubMed] [Google Scholar]