Abstract
Pre-mRNA splicing is catalyzed by the spliceosome, a macromolecular machine dedicated to intron removal and exon ligation. Despite an abundance of in vitro information and a small number of in vivo studies, the pathway of yeast (Saccharomyces cerevisiae) in vivo spliceosome assembly remains uncertain. To address this situation, we combined in vivo depletions of U1, U2, or U5 snRNAs with chromatin immunoprecipitation (ChIP) analysis of other splicing snRNPs along an intron-containing gene. The data indicate that snRNP recruitment to nascent pre-mRNA predominantly proceeds via the canonical three-step assembly pathway: first U1, then U2, and finally the U4/U6•U5 tri-snRNP. Tandem affinity purification (TAP) using a U2 snRNP-tagged protein allowed the characterization of in vivo assembled higher-order splicing complexes. Consistent with an independent snRNP assembly pathway, we observed high levels of U1–U2 prespliceosomes under U5-depletion conditions, and we observed significant levels of a U2/U5/U6/Prp19-complex mature splicing complex under wild-type conditions. These complexes have implications for the steady-state distribution of snRNPs within nuclei and also reinforce the stepwise recruitment of U1, U2, and the tri-snRNP during in vivo spliceosome assembly.
Keywords: spliceosome, commitment complex, prespliceosome, genetic depletion, mass spectrometry
INTRODUCTION
Eukaryotic pre-mRNA processing includes the precise removal of noncoding introns, i.e., pre-mRNA splicing. Splicing is catalyzed by the spliceosome, a multi-megadalton complex whose recruitment and activity is orchestrated by cis sequence elements within each intron: the 5′ splice site (ss), branchpoint, and 3′ss. The spliceosome is remarkably complex and contains five small nuclear RNAs (U snRNAs), each of which is packaged as a U snRNP (small nuclear ribonucleoprotein particle) that functions in concert with numerous non-snRNP proteins (Nilsen 2003).
Based largely on in vitro experiments, spliceosome assembly is generally viewed as a stepwise process. It begins with the ATP-independent association of U1 snRNP with the 5′ss and the association of specific proteins with the branchpoint, called commitment complex in yeast and E complex in mammals. U2 snRNP then joins the commitment complex in an ATP-dependent step to form the prespliceosome. The preassembled U4/U6•U5 tri-snRNP then recognizes the prespliceosome to form the complete spliceosome. A number of structural rearrangements accompany or follow the addition of U2 snRNP and the tri-snRNP, including the release of U1 and U4 snRNPs coincident with the acquisition of the Prp19-complex (NTC), which is associated with the complete or catalytically active spliceosome (for reviews, see Burge et al. 1999; Villa et al. 2002; Jurica and Moore 2003). The net result is splicing: exon ligation with concomitant intron excision.
Biochemical purifications from yeast have been instrumental in defining the protein composition of spliceosome subcomplexes. These include the U1, U2, U5, U6, and U4/U6•U5 snRNPs, as well as the NTC (Neubauer et al. 1997; Gottschalk et al. 1998; Caspary et al. 1999; Gottschalk et al. 1999; Rigaut et al. 1999; Stevens and Abelson 1999; Bouveret et al. 2000; Stevens et al. 2001; Ohi et al. 2002; Wang et al. 2003). Although larger, more mature in vivo splicing complexes have not been characterized, the subcomplexes have been compared with the mass spectrometry analysis of more mature splicing complexes from HeLa nuclear extracts, purified after in vitro assembly (for review, see Jurica and Moore 2003).
Although these purifications have been essential in defining the composition of splicing complexes and subcomplexes, the pathway of in vivo spliceosome assembly and splicing is still poorly understood (Nilsen 2002). For example, the observation of a tetra-snRNP complex in yeast extracts (containing all splicing snRNPs except U1) suggests that a multi-snRNP complex rather than distinct U2 and U4/U6•U5 complexes may engage the pre-mRNA substrate (Gottschalk et al. 1999). Two purifications of the NTC from Saccharomyces cerevisiae and Schizosaccharomyces pombe reported the copurification of U2/U5/U6 snRNPs, suggesting significant levels of pre-/post-catalytic spliceosomes in vivo (Ohi et al. 2002; Wang et al. 2003). Furthermore, U2/U5/U6 complexes are the major U2 snRNP-containing complex in S. pombe extracts, suggesting a substantial difference in splicing complex dynamics or stability with S. cerevisiae (Huang et al. 2002). The most provocative report was the identification of a penta-snRNP harboring all five splicing snRNPs from budding yeast (Stevens et al. 2002). This complex was purified under salt conditions permissive for in vitro splicing, and there was evidence that the penta-snRNP is active without disassembly into subcomplexes.
In vitro studies have also revealed alternative snRNP complexes and challenged the canonical stepwise assembly pathway for higher eukaryotes. For example, stable interactions can form between a short 5′ss–containing oligo and a penta-snRNP (Malca et al. 2003). E complexes formed and purified under mild conditions contain U2 snRNP without the usual ATP or branchpoint requirement, and U2 snRNP components can influence stable U1 association with pre-mRNA (Das et al. 2000). U2 snRNA modifications are also required for efficient in vitro E complex formation (Donmez et al. 2004). Furthermore, the U5 snRNP component Prp8 makes ATP-dependent contacts with the 5′ss prior to the first chemical step of splicing; these are independent of U2 snRNP binding to the branchpoint (Wyatt et al. 1992; Maroney et al. 2000). Low ionic strength experiments may more accurately mimic the nuclear environment; e.g., a high concentration and large number of protein–protein contacts between splicing snRNPs may facilitate complex formation prior to spliceosome assembly. Aspects of the stepwise assembly pathway defined by native gel analysis would then reflect conformational changes rather than snRNP recruitment or assembly events per se. Nonetheless, it is unclear if these interactions reflect true in vivo complexes preassembled prior to association with the pre-mRNA substrate. Indeed, the failure of most in vitro assays to recapitulate cotranscriptional spliceosome assembly and splicing suggests that low salt purification and in vitro assembly/splicing assays may reflect poorly in vivo assembly pathways.
To bridge this gap, we and others have recently used chromatin immunoprecipitation (ChIP) to address in vivo cotranscriptional spliceosome assembly (Kotovic et al. 2003; Gornemann et al. 2005; Lacadie and Rosbash 2005). ChIP utilizes in vivo formaldehyde cross-linking and immunoprecipitation of fixed, sheared chromatin and can assay the cotranscriptional association of RNP proteins along a gene (Lei et al. 2001; Kotovic et al. 2003; Abruzzi et al. 2004). Our laboratory and the Neugebauer group showed that the association of U snRNPs with intron-containing genes requires splicing signals, and proposed that stepwise snRNP recruitment is the principal mechanism of spliceosome assembly. This suggestion was based primarily on the sequential ChIP signals of different snRNPs along the gene. However, this does not necessarily require sequential snRNP recruitment, as snRNP conformational changes might cause changes in epitope availability or “cross-linkability” (Lacadie and Rosbash 2005; Nilsen 2005).
To further characterize in vivo spliceosome assembly and to circumvent technical caveats, we utilized in vivo depletion strategies in combination with ChIP assays to demonstrate that U1 and U2 snRNPs are fully capable of assembling individually into in vivo splicing complexes. The data support the canonical three-step spliceosome assembly pathway: U1, then U2, and finally U4/U6•U5 addition. We also purified in vivo splicing complexes via a slightly modified TAP (tandem affinity purification) approach. In combination with sucrose gradient analysis, the TAP protocol revealed high levels of U2/U5/U6/NTC complexes under nondepletion conditions and high levels of arrested U1–U2 prespliceosomes under U5-depletion conditions. This strategy of arresting and purifying in vivo complexes should be applicable to the biochemical study of multiple spliceosomal complexes.
RESULTS
Cotranscriptional snRNP recruitment to ACT1 after in vivo snRNP depletion
To test current models of in vivo spliceosome assembly, we utilized a genetic strategy to deplete cells of individual snRNPs (Patterson and Guthrie 1987; Seraphin and Rosbash 1989). Two strains were created with the U1 snRNA gene under GAL10 UAS transcriptional control (Gal-U1) as well as either TAP-tagged U2 snRNP or TAP-tagged U5 snRNP (Lea1p-TAP or Prp8p-TAP, respectively). Because U1 transcription is repressed in glucose, the stable U1 snRNP population is diluted twofold with each cell doubling. Since U1 snRNP is essential, growth slows and almost stops after 16 h in glucose. The U1 snRNP depletion also causes an inhibition of splicing; however, general cell physiology, including transcription and U snRNA levels, is relatively unaffected (Fig. 1B; Seraphin and Rosbash 1989). The two strains were also maintained in galactose as positive controls. Chromatin from the four samples was immunoprecipitated with IgG-Sepharose and bound DNA analyzed by real-time PCR. Primer sets spanning ACT1, which contains a canonical 5′ proximal intron, were used to monitor U snRNP association (Fig. 1A). The data were normalized to signal from PMA1, an intronless gene.
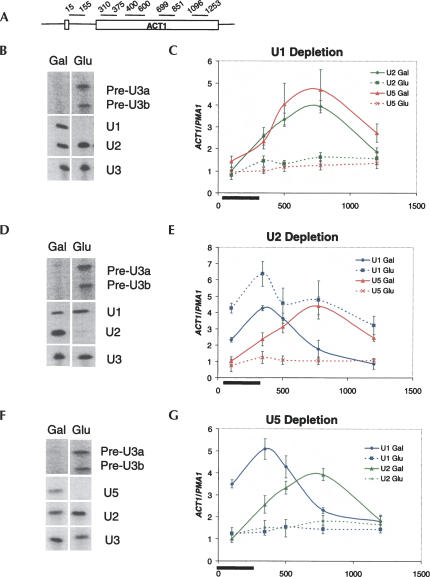
FIGURE 1.
U snRNA depletions affect cotranscriptional recruitment of U1, U2, and U5 snRNPs. TAP-tagged U snRNA depletion strains were grown in galactose or glucose prior to chromatin immunoprecipitations (ChIPs). Primer extensions confirming depletion of snRNAs and inhibition of splicing (as evident by accumulation of pre-U3a/b snRNA) are shown in the left panels (B,D,F). For ChIP (C,E,G), primer sets spanning the ACT1 gene (A) were used to monitor U snRNP association. The Y-axis reflects enrichment of U snRNPs to ACT1 relative to the intronless PMA1 gene to control for background binding. The X-axis represents distance from the ACT1 start codon. The black bar demarcates the location of the intron. The snRNP factors immunoprecipitated and found in graph legends are the following: U1 (U1C-TAP), U2 (Lea1p-TAP), and U5 (Prp8p-TAP). (B,C) U1 snRNA depletion, (D,E) U2 snRNA depletion, and (F,G) U5 snRNA depletion.
In galactose, signals for both U2 and U5 snRNP peak ∼800 bp after the start codon or ∼500 bp after the 3′ss, exactly as previously reported (Fig. 1C; Lacadie and Rosbash 2005). In glucose, however, there is little or no U2 or U5 signal. This indicates that U1 snRNP is required for U2 and tri-snRNP recruitment. The decrease in U2 and U5 signal is not due to a concomitant decrease in transcription, as Pol II signals are nearly identical from galactose or glucose-grown cells (data not shown).
The role of U2 snRNP in U1 and U5 snRNP recruitment was assayed similarly. A previously constructed Gal–U2 strain was used for TAP tagging of U1C (U1 snRNP) and Prp8p. After 16 h in glucose, U2 snRNP was depleted and splicing inhibited in both strains (Fig. 1D; Seraphin and Rosbash 1989).
In the positive control (Gal–U2 U1C-TAP grown in galactose), the peak U1 signal occurs at the second primer pair, near the ACT1 3′ss (Fig. 1E). U1 then decreases progressively along the gene as previously observed in wild-type strains (Kotovic et al. 2003; Gornemann et al. 2005; Lacadie and Rosbash 2005). The U1 pattern is similar after U2 snRNP depletion in glucose. However, U1 levels are modestly elevated relative to the galactose samples, suggesting an arrested U1 snRNP complex fails to undergo subsequent spliceosome assembly steps. The difference is not due to transcription levels since Pol II ChIP signals are nearly identical under the two growth conditions (data not shown). The Gal–U2 PRP8-TAP strain was assayed similarly. In galactose, the U5 signal along ACT1 is similar to that shown above for galactose-grown Gal–U1 PRP8-TAP, with a peak signal ∼500 bp after the 3′ss (Fig. 1E). In glucose, however, the U5 signal is absent from the ACT1 ORF (Fig. 1E), indicating that tri-snRNP recruitment is dependent upon U2 snRNP.
We next asked if the reverse relationship also exists, i.e., is U2 snRNP recruitment dependent on U5 snRNP or the tri-snRNP? To address this question, a Gal–U5 (Seraphin et al. 1991) strain expressing Lea1p-TAP was constructed. Based on the expectation that U1 snRNP recruitment should be independent of tri-snRNP activity (since U1 snRNP does not even require U2 snRNP [Fig. 1E]), we also constructed a positive control Gal–U5 strain with U1C-TAP (Fig. 1F).
Surprisingly, the U2 signal to ACT1 was absent from the Gal–U5 LEA1-TAP strain grown in glucose (Fig. 1G), consistent with an obligate tetra-snRNP recruitment model. However, U1 was also absent after U5 depletion (Fig. 1G), suggesting that U5 snRNP may also aid U1 recruitment in yeast. This recalls the cross-linking of U5 to a mammalian 5′ss in the absence of U2 snRNP binding to the branchpoint (Maroney et al. 2000). However, this interpretation appears incompatible with the results shown above, namely, that only U1 snRNP is recruited in the U2 snRNP depletion (Fig. 1E). One possibility is that in Gal–U2, U5, as well as U1, is recruited without U2 but that U5 is not captured by formaldehyde cross-linking. A related possibility is that U5 aids U1 recruitment in a more enigmatic fashion, i.e., without being stably recruited. A more testable explanation is that U5 depletion leads to high levels of blocked splicing complexes, which contain both U1 and U2 snRNP; U5 snRNP would be required to complete spliceosome assembly and splicing, which is required in turn for the recycling of U1 and U2 snRNP back to nascent transcription complexes.
Splicing complexes accumulate after U snRNP depletions
To verify this block-to-recycling model, we characterized these complexes by immunoprecipitation via the TAP-tagged snRNP proteins in different depletion strains. Specific association of an intron-containing gene with snRNPs provided a readout for the presence of in vivo splicing complexes. After RNA purification, DNase treatment, random priming, and real-time PCR with ACT1 and PMA1 primer sets, we estimated changes in the association of intron-containing RNAs with the three TAP-tagged splicing snRNPs due to the genetic depletions, i.e., in glucose versus in galactose.
We first examined two strains, Gal–U2 U1C-TAP and Gal–U2 PRP8-TAP, and normalized the ACT1 signal to the PMA1 signal to account for background binding. After U2 depletion, the U1 association with ACT1 pre-mRNA increased approximately fivefold (glucose vs. galactose), indicating stalled commitment complexes and consistent with potent cotranscriptional recruitment of U1 in the absence of U2 (Fig. 2A, bars 1,2). In contrast, U5 association with ACT1 decreased by about twofold to approximately background levels (Fig. 2A, bars 3,4), consistent with the notion that splicing complexes formed without U2 also lack U5, both cotranscriptionally and after release of the snRNPs from the transcription machinery.
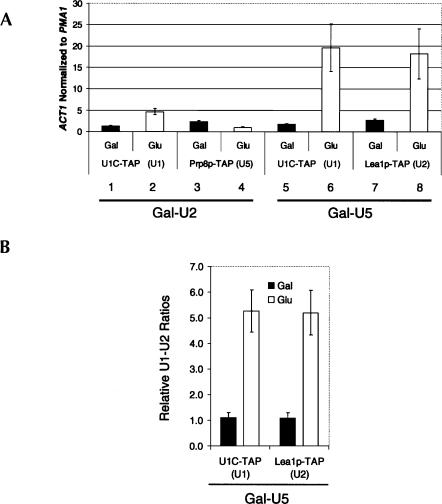
FIGURE 2.
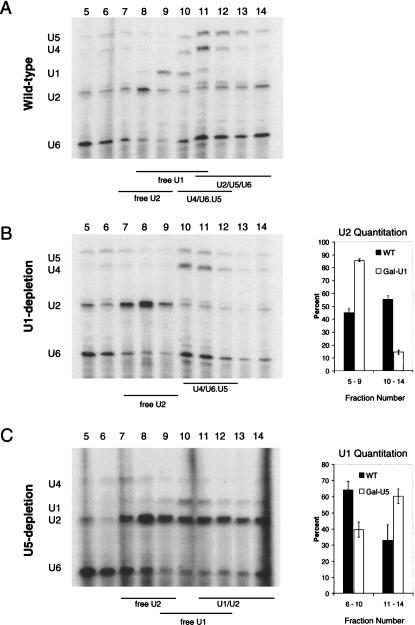
Post-transcriptional splicing complexes accumulate in snRNA depletions. (A) The Gal-U2 and Gal-U5 TAP tagged strains described in Figure 1 were used in a U snRNP immunoprecipitation experiment (see Materials and Methods). Values represent ACT1 levels normalized to intronless PMA1 to show enrichment over background. Black columns indicate strains grown in galactose (nondepletion), and white columns indicate those grown in glucose (depletion). ACT1 RNA is enriched on U1 snRNP (U1C-TAP) in the absence of U2 snRNP while U5 snRNP (Prp8p-TAP) levels decrease. Increased association of ACT1 with U1 indicates accumulation of commitment complexes. In U5-depletion strains, ACT1 is highly enriched on both U1 (U1C-TAP) and U2 snRNPs (Lea1p-TAP), indicative of prespliceosome formation. (B) U1 and U2 snRNAs from the same U snRNP IP experiments were analyzed by quantitative RT-PCR in Gal-U5 U1C-TAP and LEA1-TAP. Black columns represent the U1–U2 ratios when grown in galactose; white columns indicate the U1–U2 ratios in U5 depletion. In U1C-TAP, the ratio is U2:U1 and for LEA1-TAP, the ratio is U1:U2. U2 is more highly associated with U1 in Gal–U5 U1C-TAP and U1 more highly associated with U2 in Gal–U5 LEA1-TAP upon depletion. The reciprocal increased association between U1 and U2 suggests prespliceosomes form in the absence of U5.
We assayed the effects of U5 depletion on post-transcriptional complexes in a similar fashion by characterizing the U1C and Lea1p-TAP tagged Gal–U5 strains. Association of pre-mRNA with both U1 and U2 snRNPs increased ∼20-fold over background upon U5 depletion (Fig. 2A, bars 5–8), indicating that U1–U2–containing prespliceosomes form in vivo without U5 snRNP and supporting the canonical in vitro stepwise spliceosome assembly pathway. Interestingly, the ACT1 association with U1 snRNP is approximately fourfold higher in Gal–U5 depletion than in Gal–U2 depletion (Fig. 2A, cf. bars 6 and 2), suggesting higher in vivo steady-state levels of U1–U2 prespliceosomes without U5 than U1-only commitment complexes without U2. This also suggests that stalled prespliceosomes may be more stable than are commitment complexes.
An increase in prespliceosomes should also result in an increased association between U1 and U2 snRNPs. To this end, U1 and U2 snRNAs were analyzed by random priming and quantitative RT-PCR from the same immunoprecipitated samples mentioned above. As predicted, a fivefold increase in U1/U2 association accompanies the increase in snRNP-associated ACT1 mRNA in the U5-depleted sample; U1 becomes more U2-associated and U2 becomes more U1-associated (Fig. 2B). The high levels of stalled complexes without U5 strongly indicate that the loss of cotranscriptional assembly is due to a shift in a significant fraction of the U1 and U2 snRNP populations from assembly-competent to arrested in splicing complexes.
The stability of prespliceosomes and their resistance to extract preparation prompted us to analyze the complement of snRNAs associated with Lea1p-TAP after growth of the Gal-U5 LEA1-TAP strain under nondepletion (galactose) growth conditions. Surprisingly, we observed significant levels of U5 and U6 (Fig. 3A, lane 3). These associations are splicing dependent, as U5 and U6 association with Lea1p-TAP dramatically decreases upon U1 depletion, indicating that they are biologically meaningful interactions (Fig. 3B). Furthermore, we routinely observe lower levels of U5 and U6 snRNA associations with Lea1p-TAP with a more traditional extract protocol (Fig. 3C; see Materials and Methods). Although low levels of U5 and U6 have been previously observed to copurify with Lea1p-TAP (Dziembowski et al. 2004), this direct comparison suggests that these modest protocol modifications better reflect in vivo complex integrity. Consistent with the detection of abundant U2/U5/U6 splicing complexes, Prp8p-TAP also associates with U2 snRNP in addition to the expected U4, U5, and U6 snRNAs under the same growth and extract conditions (data not shown). Prp8p-TAP presumably purifies a mixture of U2/U5/U6 spliceosomes as well as tri-snRNP particles.
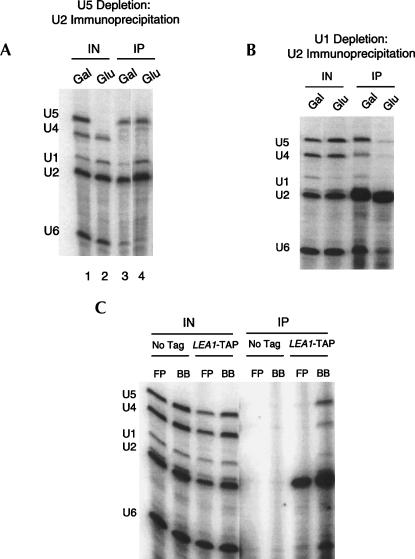
FIGURE 3.
Characterization of U2 snRNP-associated U snRNAs (A). U snRNAs from Gal–U5 LEA1-TAP were analyzed by primer extension following TAP purification from cells grown in galactose and glucose. In galactose, U2 is most highly associated with U5 and U6 snRNAs (low levels of U1 and U4 snRNA). In U5 depletion, U1, U5, and U6 snRNAs associate with U2 snRNP. (B) Lea1p-TAP immunoprecipitates predominantly U2 snRNA upon U1 depletion, indicating that U5 and U6 snRNP association is splicing dependent. (C) Extracts generated via traditional French press methods or our modified protocol were compared for association of snRNAs with Lea1p-TAP. FP, French press; BB, bead beating. LEA1-TAP or an untagged wild-type strain were used for immunoprecipitations. Lea1p-TAP specifically immunoprecipitates U2 snRNA when using traditional methods. However, with modified conditions, Lea1p-TAP interacts most highly with U2, U5, and U6 snRNAs with lower levels of U1 and U4.
Interestingly, there was still an association of U5 with U2 even after U5 depletion (Fig. 3A, lane 4; cf. lane 4 and input lane 2), suggesting that the residual U5 snRNA is predominantly associated with U2 snRNP. There was also the expected association of U1 with U2 under these conditions, i.e., a high level of prespliceosomes.
The surprising association of U5 with U2 inspired an estimate of in vivo splicing complex stability, which we assayed with a thiolutin immunoprecipitation assay. Thiolutin is a reversible RNA Pol II transcriptional inhibitor commonly used to measure mRNA half-life (Jimenez et al. 1973; Herrick et al. 1990). We reasoned that inhibiting transcription should terminate the pre-mRNA supply, resulting in substrate turnover and splicing complex disassociation. Immunoprecipitation with Lea1p-TAP and snRNA analysis would then show whether these complexes contain substrate or are stable entities. To this end, thiolutin was added to cells for 20 min prior to immunoprecipitation with Lea1p-TAP under U5-depletion conditions.
Analysis of U snRNAs showed a rapid and dramatic decrease in U1 levels associated with U2 snRNP after transcription inhibition under U5-depletion conditions (Fig. 4A, cf. lanes 5 and 6). There was much less effect on U5 or U6 levels, consistent with the nondepletion conditions (Fig. 4A; cf. lanes 3,4 and 5,6), in which the U2/U5/U6 complex is also stable. The data suggest there are two populations of U2-associated complexes upon U5 depletion: a relatively unstable U1–U2 prespliceosome pool and a stable U2/U5/U6 pool. Since most U5 snRNP is depleted and the remaining U5 is in stable U2/U5/U6 complexes, it is unlikely that this residual U5 influences the high levels of prespliceosomes that form and turnover. This also suggests that U5/tri-snRNP is normally in excess over U2 and that only a relatively small fraction of U5 is associated with U2 under wild-type conditions.
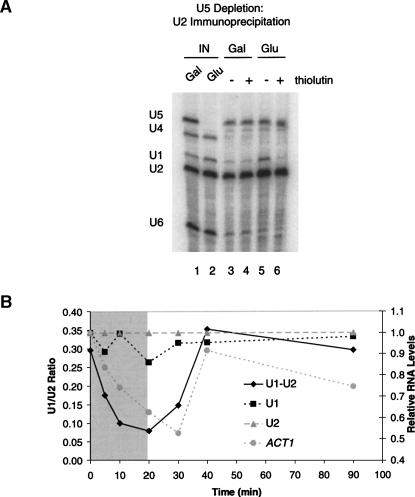
FIGURE 4.
Dynamics of prespliceosomes and U2/U5/U6 complexes during thiolutin treatment. (A) U snRNAs from Gal-U5 LEA1-TAP were analyzed by primer extension following TAP purification from cells grown in galactose and glucose with or without thiolutin (see Materials and Methods). Only U1–U2 prespliceosomes in U5 depletion turn over with thiolutin treatment, indicating that there are two separate complexes associated with U2 snRNP. (B) Prespliceosomes were monitored during thiolutin treatment and return to growth in Gal–U5 LEA1-TAP (see Materials and Methods). The gray section indicates thiolutin treatment; the white indicates time after thiolutin removal. The primary Y-axis represents U1–U2 ratios (prespliceosomes), and the secondary Y-axis represents U1, U2, and ACT1 mRNA all normalized to U2 snRNA levels and then to the first time point. Prespliceosomes decay with a half-life of ∼5 min and form with similar kinetics upon thiolutin removal and initiation of transcription. U1, U2, and ACT1 RNAs were analyzed from the same experiment to confirm effectiveness of thiolutin treatment. U1 and U2 snRNAs are stable during the thiolutin treatment, while ACT1 is labile and recovers with removal of thiolutin.
To extend these analyses, we took advantage of thiolutin reversibility to monitor de novo prespliceosome assembly. After transcriptional inhibition of a U5-depleted LEA1-TAP strain, cells were washed and returned to glucose-containing media. Aliquots were removed at the indicated times, cells were lysed, and complexes were immunoprecipitated. The association of U1 snRNA with U2 was measured by random priming and real-time PCR (U1:U2 ratio). Relative levels of U1, U2, and ACT1 were also assayed from total RNA to monitor the stabilities of these RNAs during transcriptional inhibition.
Prespliceosomes decayed remarkably quickly, with a half-life of ∼5 min (Fig. 4B). The decrease in prespliceosomes was accompanied by the expected behavior of U snRNA levels (stable) and ACT1 pre-mRNA levels (labile) in the total extract (Fig. 4B). Following the washout, prespliceosomes reform rapidly, coincident with a resupply of pre-mRNA. Despite the rapid disassociation/reassembly of prespliceosomes after thiolutin addition/washout, we presume that the stalled prespliceosomes are still sufficiently stable and abundant to trap most of the U1 snRNP pool and prevent detectable cotranscriptional prespliceosome assembly by ChIP.
High levels of endogenous U2 snRNP reside within splicing complexes
The significant association of U5 and U6 snRNPs with U2 snRNP under nondepletion conditions (Figs. 3A, 4A) suggests that a considerable fraction of yeast U2 snRNP is present in higher-order splicing complexes under exponential growth conditions. To address the distribution of these splicing snRNPs without potential biases from preferential immunoprecipitation of different complexes, we compared sucrose gradients from extracts derived from either U1-depleted cells or their wild-type counterparts.
Spliceosomal snRNP distributions from wild-type cells reflect previously reported particle sizes: U2 < U1 < U4/U6•U5 (Fig. 5). However, we observed significant levels of U2 extending into fractions containing higher molecular weight complexes, consistent with the association of a considerable fraction of U2 with other snRNPs (Fig. 5A). Importantly, U1 depletion significantly decreases the fraction of U2 in higher molecular weight complexes (Fig. 5B). We interpret these large U2 snRNP complexes to be the splicing-dependent U2/U5/U6 spliceosomes identified by immunoprecipitation (Fig. 3). Additionally, the distribution of U5 and U6 is shifted toward heavier complexes in the wild-type strain (Fig. 5A, fractions 12–14). This suggests that a significant fraction of these snRNPs is in a U2/U5/U6 complex, which is larger than the tri-snRNP. Quantitation of the U2 snRNA distribution confirms that U1 depletion causes a gross change in the snRNP distribution. It changes from 45:55 (fractions 5–9:10–14) in wild type to 85:15 after U1 depletion. Based on the gradient distribution and response to splicing inhibition (U1 depletion), we suggest that roughly half of U2 snRNP is in a U2/U5/U6/NTC complex at steady-state.
FIGURE 5.
High levels of U2 snRNP are in higher-order splicing complexes in vivo. Sucrose gradients of wild-type (A), U1-depleted (B), and U5-depleted (C) cells are shown. Extracts were layered onto 10%–30% sucrose gradients and fractions analyzed by primer extension. U snRNA distribution was quantitated from at least two independent sucrose gradients and is shown to the right of the corresponding gradient (see Materials and Methods). After U1 depletion (cf. B and A), a considerable fraction of U2 snRNP migrates as a mono-snRNP, indicating that under normal physiological conditions, a major fraction (∼50%) of U2 snRNP is in higher-order complexes. After U5 depletion (cf. C and A), U1 snRNA redistributes across the gradient toward larger complexes, consistent with accumulation of U1–U2 prespliceosomes.
Consistent with the above results, U5 depletion increases the size of U1-containing complexes (Fig. 5C). A similar quantitation indicates that the percentage distribution of U1 changes from ∼65:35 to 35:65 (fractions 6–10:11–14), consistent with an accumulation of U1–U2 prespliceosomes by immunoprecipitation (Figs. 2B, 3A). The data suggest that an approximate three- to fivefold change in free U1 snRNP is capable of abolishing cotranscriptional detection by ChIP (Fig. 1G).
Proteomic analysis of U2 snRNP complexes
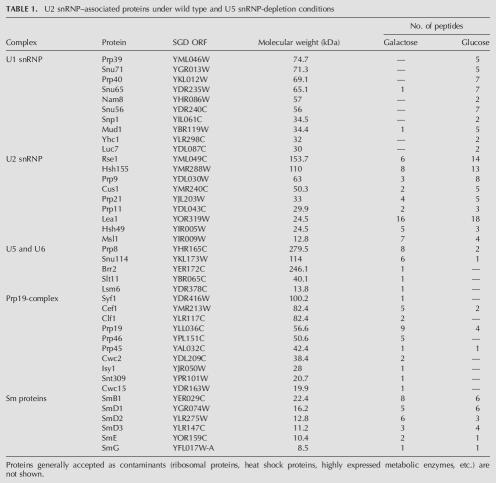
To characterize the composition of these stalled splicing complexes on a preparative scale, we purified U2 snRNP-associated factors from Gal-U5 LEA1-TAP and subjected the proteins to LC-MS/MS; proteins from cells grown in galactose were compared with cells depleted of U5 in glucose. Rather than compile an exhaustive list of proteins, we focused on proteins with the largest numbers of peptides. These are arranged by U snRNP or subsplicing complex (Table 1). Proteins generally accepted to be common contaminants, such as ribosomal, heat shock, and metabolic proteins, are not shown.
TABLE 1.
U2 snRNP–associated proteins under wild type and U5 snRNP-depletion conditions
The two data sets are striking in their differences and are entirely consistent with the U snRNA associations described above. Multiple known U2 snRNP proteins, including SF3a and SF3b components as well as the common set of Sm proteins, were identified in both purifications. Only from the galactose sample were large numbers of peptides identified from U5 and U6 snRNPs as well as the NTC complex. The association of the NTC with a U2/U5/U6 snRNP complex is consistent with previous reports of U2, U5, and U6 snRNA copurification with Clf1p and Cef1p (Ohi et al. 2002; Wang et al. 2003). Conversely, only from the glucose sample were all U1 snRNP proteins identified. The purification of this protein set is consistent with U1–U2 prespliceosomes and further confirms their tri-snRNP independent formation. As predicted from the snRNA analysis (Fig. 3A), a small number of U5 snRNP and NTC peptides were still observed in glucose, consistent with the presence of some residual U2/U5/U6/NTC stable complex. The data further indicate that a substantial fraction of U2 snRNP is present in higher-order splicing complexes under wild-type conditions and that U5 depletion causes tri-snRNP–independent formation of prespliceosomes. They also suggest that our slightly modified protocol reflects well in vivo complexes and has implications for future proteomic analyses of other in vivo splicing complexes.
DISCUSSION
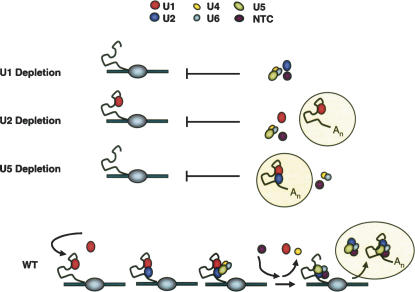
The study of spliceosome assembly has suffered from a lack of in vivo assays. Two groups recently reported a novel ChIP-based approach to study cotranscriptional spliceosome assembly in budding yeast (Kotovic et al. 2003; Gornemann et al. 2005; Lacadie and Rosbash 2005). Here, we further address the in vivo spliceosome assembly pathway by combining in vivo snRNP depletions with ChIP and splicing complex immunoprecipitation. We also extend current approaches to study the protein composition of in vivo formed splicing complexes. Our analyses support the canonical in vitro stepwise spliceosome assembly model. U1 is required for recruitment of all subsequent splicing snRNPs. U1–pre-mRNA complex (commitment complex) formation is independent of U2 snRNP and the tri-snRNP, and U1–U2 prespliceosomes form in the absence of tri-snRNP. We also show that stalled splicing complexes have a dramatic impact on cotranscriptional events and that a major fraction of U2 snRNP resides within higher-order splicing complexes. Importantly, these complexes can be readily purified for proteomic analyses. These results are summarized in a model (Fig. 6).
FIGURE 6.
Spliceosome assembly in U snRNA depletion strains. Spliceosome assembly pathways are shown in Gal–U1, Gal–U2, Gal–U5, and wild-type (WT) strains. Cotranscriptional and post-transcriptional complexes (circled) are depicted. No spliceosome assembly occurs in the absence of U1 snRNP (Gal–U1). However, commitment complex forms cotranscriptionally upon U2 snRNP depletion, while tri-snRNP addition is inhibited (Gal–U2). In the absence of U5 snRNP (tri-snRNP), prespliceosomes are only observed post-transcriptionally because free U1 and U2 are titrated away from the site of transcription (Gal–U5). Our data suggest that spliceosome assembly proceeds in a stepwise fashion in vivo and does not require higher-order snRNP complexes (tetra-snRNP, penta-snRNP, etc.).
Although the stepwise recruitment of U snRNPs during spliceosome assembly was generally accepted, the purification and characterization of a biologically active penta-snRNP complex under splicing permissive conditions challenged this view. Indeed, a preformed complex has theoretical benefits over a stepwise assembly pathway. For example, the local concentration of snRNPs should be much higher in a preformed penta-snRNP and lead to an increase in splicing efficiency. However, the recent ChIP analyses suggested that the conventional in vitro stepwise recruitment pathway is the main in vivo mechanism of assembly (Gornemann et al. 2005). This conclusion was based primarily on the absence of a ChIP signal for U5 snRNP in a strain lacking the nuclear cap binding complex, i.e., a strong U2 snRNP signal without a U5 snRNP signal. This interpretation was subsequently challenged on the basis of inherent limitations of the ChIP procedure (Nilsen 2005).
Our depletion experiments imply that there is little snRNP interdependence during assembly, especially between U1 snRNP and the rest of the splicing snRNPs. The abundance of U1–U2 prespliceosomes in the absence of U5 snRNP also argues against an obligate U1-tetra-snRNP two-step pathway. Although the snRNA depletion strategy makes it possible that a two-step pathway, as well as the traditional three-step pathway, normally operate in vivo, we conclude that higher-order splicing complexes are not required for U1 or U2 snRNP recruitment by pre-mRNA.
A lack of snRNP dependence in yeast suggests potential differences with the mammalian spliceosome assembly pathway. For example, decreases in U2 snRNP activity negatively impact U1 snRNP interactions with a 5′ss in mammalian in vitro systems (Das et al. 2000; Donmez et al. 2004). Nonetheless, we observe no effect of U2 depletion on U1 recruitment in vivo. Furthermore, U5 snRNP can make ATP-dependent contacts with the 5′ss in the absence of U2 snRNP in higher eukaryotes (Maroney et al. 2000), which we were unable to detect in vivo by ChIP or by snRNP immunoprecipitation (Figs. 1E, 2A). These assembly pathway differences may reflect pre-mRNA organizational differences, namely, single 5′ proximal introns of regular length in yeast and multiple large introns with more degenerate splicing signals in metazoans. Furthermore, SR proteins and other metazoan-specific features may make more significant contributions.
Our purifications of U2 snRNP-associated complexes also suggest that there are substantial in vivo quantities of pre-/post-catalytic splicing complexes. This is because they contain significant levels of NTC proteins and lack U1 and U4 snRNA. These observations further suggest that spliceosome disassembly may be partially rate limiting in snRNP recycling and homeostasis. Similar conclusions were based on a Clf1p-TAP (S. cerevisiae NTC component) purification that identified substoichiometric levels of U2/U5/U6; this interaction was lost in response to temperature-sensitive (ts) splicing mutants (Wang et al. 2003). In contrast, Cdc5/Cef1 (S. pombe NTC component) that copurified with U2/U5/U6 was not sensitive to ts splicing mutants (Ohi et al. 2002). This discrepancy may reflect differences between S. cerevisiae and S. pombe. Some of our results are more consistent with the former conclusion, since the integrity of the U2/U5/U6 complex is compromised by U1 depletion (Figs. 3B, 5B). However, this requires a 16-h incubation in glucose, and our thiolutin experiments suggest that precatalytic spliceosomes (including prespliceosomes) are converted into other complexes or are degraded much more rapidly than the U2/U5/U6/NTC complex. The latter complex appears stable during the 20 min of transcriptional shutoff (Fig. 4A), suggesting that it may contain a slowly turning over lariat product but does not contain pre-mRNA substrate or intermediates, i.e., that it is post-catalytic.
Although differences in the mass spectrometry approaches preclude an accurate comparison with the published literature (B. Seraphin, pers. comm.), we reproducibly observed better preservation of complex integrity with these slight modifications to the standard TAP protocol (Fig. 3C; Puig et al. 2001; see Materials and Methods). They were designed to mirror the U snRNP immunoprecipitation experiments (Figs. 2–4), which represent conditions that are well-controlled and biologically relevant. Potential differences may reflect growth conditions (cells in log phase, i.e., OD600 ≤ 1.0) and the procedure for extract preparation. We routinely lyse cells (never frozen before lysis) by bead beating rather than other means of lysis such as French press or liquid nitrogen/grinding. Additionally, we lyse cells directly in immunoprecipitation buffer, and the extract is applied immediately to IgG-Sepharose rather than dialyzing the extract for several hours. The latter may encourage complex rearrangements or dissociation (see Materials and Methods). Our approach also exploited a single genetic or physiological (glucose vs. galactose) change between two samples, and the results show that specific in vivo complexes can be manipulated in vivo and then purified in vitro. In another example, Snu71p-TAP (a U1 snRNP protein) purifications from otherwise wild-type strains contain significant levels of the nuclear cap binding complex protein Cbp80p, evident by silver staining as well as mass spectrometry (data not shown); previous purifications with the standard methods did not report Cbp80p (Neubauer et al. 1997; Gottschalk et al. 1998; Rigaut et al. 1999). Taken together with other data in this article, these results indicate that our modified TAP purification protocol should complement the predominant strategy of purifying in vitro assembled complexes on synthetic pre-mRNAs and allow the characterization of other in vivo RNP complexes, including other in vivo arrested splicing complexes.
Based on the prevalence of U2/U5/U6/NTC complexes in our purifications and the data suggesting they are under physiological regulation in vivo, it seems unlikely that most of the snRNP population in vivo is in a tetra-/penta-snRNP complex poised for splicing (Stevens et al. 2002). Although a modest in vivo quantity of these higher-order complexes is possible, our results suggest that the snRNP distribution within S. cerevisiae may more closely resemble that in S. pombe, in which a substantial fraction of U2 snRNP is present in U2/U5/U6 complexes (Huang et al. 2002).
A relative paucity of free U2 snRNP may contribute to the largest apparent difference in cotranscriptional spliceosome assembly dynamics, namely, early-rapid U1 snRNP addition but later-slower U2 snRNP and U4/U6•U5 addition. Interestingly, a different picture emerges from in vitro spliceosome assembly: Commitment complex is only observed when U2 snRNP is inhibited or ATP absent, suggesting a kinetic preference for higher-order spliceosomes (Seraphin and Rosbash 1989; Liao et al. 1992). Although this in vivo–in vitro difference may reflect the delayed transcription of the branchpoint sequence relative to the 5′ss and/or limiting quantities of branchpoint binding proteins (Mud2p and BBP) in vivo, it is equally likely that free U2 snRNP is limiting in vivo but not in vitro, e.g., U2/U5/U6 complex disassociates during splicing extract preparation. Future experiments should be able to further explore the relationship between in vivo snRNP pools and cotranscriptional spliceosome assembly.
MATERIALS AND METHODS
Yeast strains and growth conditions
Yeast strains and plasmids used in this study are listed in Table 2. Yeast strains were manipulated according to standard procedures (Guthrie and Fink 1991). For U snRNA depletion experiments, log phase cells growing in 2% galactose were diluted into 2% glucose-containing media to an OD600 of 0.01 and grown for 16 h to an OD600 of ∼0.6. Gal–U1 (BS-Y82), Gal–U2 (BS-Y88), and Gal–U5 (BS-Y119) were parent strains for TAP tagging. Genomic DNAs from U1C, LEA1, and PRP8 TAP-tagged strains (Open Biosystems) were used as templates for PCR integration of the TAP tag into depletion strains. Primers used for strain construction are available upon request. Transformants were confirmed by PCR and Western blotting.
TABLE 2.
Yeast strains and plasmids
Chromatin immunoprecipitations
ChIPs and quantitative PCR were performed as previously described (Lacadie and Rosbash 2005). PCRs were performed on a Rotor-Gene 3000 PCR machine (Corbett Research). ACT1 ChIP signals are expressed as the IP-to-IN ratio for ACT1 over the IP-to-IN ratio for the intronless PMA1 gene. Reported ACT1/PMA1 values are the averages of at least three independent experiments, and error bars represent ±1 SD. Sequences of oligos used for PCR are available upon request.
U snRNP immunoprecipitations
U snRNP immunoprecipitation experiments were performed in genetic depletion strains grown in 2% glucose or 2% galactose. Cultures (100 mL) were grown for 16 h to an OD600 ∼0.4–0.8, centrifuged, and washed in 5 mL IPP150 buffer (10 mM Tris-Cl at pH 8.0, 150 mM NaCl, 0.1% NP-40, 1.5 mM MgCl2). Pellets were resuspended in 1 mL IPP150, supplemented with RNasin (40 U/mL; Promega) and PMSF (1 mM), and lysed by bead beating three times for 1 min with 1 min on ice in between in a Mini Bead Beater 8 (Biospec Products). Lysates were centrifuged for 5 min at 5000 rpm and supernatants applied to 10 μL of prewashed IgG-Sepharose beads (Amersham Biosciences). Following 1 h at 4°C, bound complexes were washed three times for 5 min with 1 mL IPP150. Input and IP fractions were phenol-chloroform extracted and ethanol precipitated, and samples were treated with DNase I (Promega). cDNA synthesis of RNA samples was primed with random hexamers (NEB) and reverse transcription performed with Superscript II (Invitrogen) according to the manufacturer's protocol. Quantitative real-time PCR was performed on a Rotor-Gene 3000 PCR machine (Corbett Research). Reported ACT1/PMA1 values are the averages of at least three independent experiments, and error bars represent ±1 SD.
Thiolutin experiments were performed by adding thiolutin (Sigma Aldrich) to a final concentration of 5 μg/mL. For time course experiments, aliquots were removed and immediately placed on ice. Cells were then treated as described in U snRNP immunoprecipitations. In the return to growth experiment, after 20 min of thiolutin treatment, cells were washed three times with room-temperature water and resuspended in glucose-containing media (Jimenez et al. 1973).
Primer extensions of spliceosomal snRNAs from U snRNP IP experiments were performed as described previously (Pikielny and Rosbash 1986). Sequences of oligos are available upon request.
Sucrose gradients
Sucrose gradients were performed by layering 100 μL of extract on top of 2 mL linear 10%–30% gradients made in IP buffer (see U snRNP Immunoprecipitations). Gradients were centrifuged at 50,000 rpm for 4 h at 4°C in a Beckman TLS-55 rotor. Fractions (150 μL) were collected manually from the top of the gradient. Samples were phenolchloroform–extracted and ethanol-precipitated, and U snRNAs were analyzed by primer extension. For U2 snRNA analysis in wild type and Gal-U1 (Fig. 5B), the percentage of each fraction was pooled from fractions 5–9 and 10–14. For U1 snRNA analysis in wild type and Gal-U5 (Fig. 5C), the percentage of each fraction was pooled from fractions 6–10 and 11–14. The pooled percent of total was averaged and error reported as ±1SD.
Spliceosome purifications
Purification of U2 snRNP-containing complexes were performed with Gal-U5 LEA1-TAP. Four liters of cells were harvested at an OD600 ∼1.0 for both glucose and galactose cultures after 16 h of growth. Purification was performed essentially as described with the following modifications (Puig et al. 2001): Cells were centrifuged and washed with ice-cold IPP150 buffer. Cells were resuspended in IPP150 supplemented with RNasin (Promega) and protease inhibitors (Roche). Extracts were prepared by bead beating three times for 1 min with 1 min on ice in between. Glass beads were removed and lysates centrifuged at 4°C in an SW40 rotor at 20,000 rpm for 30 min. The supernatant was centrifuged at 4°C in an SW40 rotor at 33,500 rpm for 75 min. The lysate was added directly to IgG-agarose beads (Sigma), and TAP purification protocol followed as described (Puig et al. 2001).
Mass spectrometry
Mass spectrometry was performed by the Taplin Biological Mass Spectrometry Facility at Harvard Medical School with an LTQ linear ion-trap mass spectrometer (ThermoFinnigan). Excised gel bands were subjected to a modified in-gel trypsin digestion procedure (Shevchenko et al. 1996). Peptides were subjected to electrospray ionization prior to entering an LTQ linear ion-trap mass spectrometer (ThermoFinnigan). Eluting peptides were detected, isolated, and fragmented to produce a tandem mass spectrum of specific fragment ions for each peptide. Peptide sequences and protein identity were determined by matching protein or translated nucleotide databases with the acquired fragmentation pattern by the software program, Sequest (ThermoFinnigan) (Eng et al. 1994).
ACKNOWLEDGMENTS
We thank our local colleagues, especially S. Lacadie and K. Abruzzi, for helpful discussions and comments on the manuscript. Thanks also to B. Seraphin for communicating unpublished results as well as manuscript suggestions and to K. Neugebauer, B. Rymond, T. Nilsen, D. Libri, and D. Belostotsky for manuscript comments. Mass spectrometry was performed at the Taplin Biological Mass Spectrometry Facility (Harvard Medical School, Department of Cell Biology, laboratory of Dr. S. Gygi). This work was supported in part by a grant to M.R. from the NIH (R01 GM23549) and the NIH Genetics Training Grant (5T32 GM07122).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.50506.
REFERENCES
- Abruzzi K.C., Lacadie S., Rosbash M. Biochemical analysis of TREX complex recruitment to intronless and intron-containing yeast genes. EMBO J. 2004;23:2620–2631. doi: 10.1038/sj.emboj.7600261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouveret E., Rigaut G., Shevchenko A., Wilm M., Seraphin B. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 2000;19:1661–1671. doi: 10.1093/emboj/19.7.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C.B., Tuschl T., Sharp P.A. Splicing of precursors to mRNAs by the spliceosome. In: Gesteland R.R., et al., editors. The RNA world. 2d ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1999. pp. 525–560. [Google Scholar]
- Caspary F., Shevchenko A., Wilm M., Seraphin B. Partial purification of the yeast U2 snRNP reveals a novel yeast pre- mRNA splicing factor required for pre-spliceosome assembly. EMBO J. 1999;18:3463–3474. doi: 10.1093/emboj/18.12.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R., Zhou Z., Reed R. Functional association of U2 snRNP with the ATP-independent spliceosomal complex E. Mol. Cell. 2000;5:779–787. doi: 10.1016/s1097-2765(00)80318-4. [DOI] [PubMed] [Google Scholar]
- Donmez G., Hartmuth K., Luhrmann R. Modified nucleotides at the 5′ end of human U2 snRNA are required for spliceosomal E-complex formation. RNA. 2004;10:1925–1933. doi: 10.1261/rna.7186504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziembowski A., Ventura A.P., Rutz B., Caspary F., Faux C., Halgand F., Laprevote O., Seraphin B. Proteomic analysis identifies a new complex required for nuclear pre-mRNA retention and splicing. EMBO J. 2004;23:4847–4856. doi: 10.1038/sj.emboj.7600482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng J.K., McCormack A.L., Yates J.R.I. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Gornemann J., Kotovic K.M., Hujer K., Neugebauer K.M. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol. Cell. 2005;19:53–63. doi: 10.1016/j.molcel.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Gottschalk A., Tang J., Puig O., Salgado J., Neubauer G., Colot H.V., Mann M., Seraphin B., Rosbash M., Luhrmann R., et al. A comprehensive biochemical and genetic analysis of the yeast U1 snRNP reveals five novel proteins. RNA. 1998;4:374–393. [PMC free article] [PubMed] [Google Scholar]
- Gottschalk A., Neubauer G., Banroques J., Mann M., Luhrmann R., Fabrizio P. Identification by mass spectrometry and functional analysis of novel proteins of the yeast [U4/U6.U5] tri-snRNP. EMBO J. 1999;18:4535–4548. doi: 10.1093/emboj/18.16.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C., Fink G.R. Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:389–398. [PubMed] [Google Scholar]
- Herrick D., Parker R., Jacobson A. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae . Mol. Cell. Biol. 1990;10:2269–2284. doi: 10.1128/mcb.10.5.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T., Vilardell J., Query C.C. Pre-spliceosome formation in S. pombe requires a stable complex of SF1–U2AF(59)-U2AF(23) EMBO J. 2002;21:5516–5526. doi: 10.1093/emboj/cdf555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez A., Tipper D.J., Davies J. Mode of action of thiolutin, an inhibitor of macromolecular synthesis in Saccharomyces cerevisiae . Antimicrob. Agents Chemother. 1973;3:729–738. doi: 10.1128/aac.3.6.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica M.S., Moore M.J. Pre-mRNA splicing: Awash in a sea of proteins. Mol. Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- Kotovic K.M., Lockshon D., Boric L., Neugebauer K.M. Cotranscriptional recruitment of the U1 snRNP to intron-containing genes in yeast. Mol. Cell. Biol. 2003;23:5768–5779. doi: 10.1128/MCB.23.16.5768-5779.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacadie S.A., Rosbash M. Cotranscriptional spliceosome assembly dynamics and the role of U1 snRNA: 5′ss base pairing in yeast. Mol. Cell. 2005;19:65–75. doi: 10.1016/j.molcel.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Lei E.P., Krebber H., Silver P.A. Messenger RNAs are recruited for nuclear export during transcription. Genes & Dev. 2001;15:1771–1782. doi: 10.1101/gad.892401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X.C., Colot H.V., Wang Y., Rosbash M. Requirements for U2 snRNP addition to yeast pre-mRNA. Nucleic Acids Res. 1992;20:4237–4245. doi: 10.1093/nar/20.16.4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malca H., Shomron N., Ast G. The U1 snRNP base pairs with the 5′ splice site within a penta-snRNP complex. Mol. Cell. Biol. 2003;23:3442–3455. doi: 10.1128/MCB.23.10.3442-3455.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroney P.A., Romfo C.M., Nilsen T.W. Functional recognition of 5′ splice site by U4/U6.U5 tri-snRNP defines a novel ATP-dependent step in early spliceosome assembly. Mol. Cell. 2000;6:317–328. doi: 10.1016/s1097-2765(00)00032-0. [DOI] [PubMed] [Google Scholar]
- Neubauer G., Gottschalk A., Fabrizio P., Séraphin B., Luhrmann R., Mann M. Identification of the proteins of the yeast U1 small nuclear ribonucleoprotein complex by mass spectrometry. Proc. Natl. Acad. Sci. 1997;94:385–390. doi: 10.1073/pnas.94.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen T.W. The spliceosome: No assembly required? Mol. Cell. 2002;9:8–9. doi: 10.1016/s1097-2765(02)00430-6. [DOI] [PubMed] [Google Scholar]
- Nilsen T.W. The spliceosome: The most complex macromolecular machine in the cell? Bioessays. 2003;25:1147–1149. doi: 10.1002/bies.10394. [DOI] [PubMed] [Google Scholar]
- Nilsen T.W. Spliceosome assembly in yeast: One ChIP at a time? Nat. Struct. Mol. Biol. 2005;12:571–573. doi: 10.1038/nsmb0705-571. [DOI] [PubMed] [Google Scholar]
- Ohi M.D., Link A.J., Ren L., Jennings J.L., McDonald W.H., Gould K.L. Proteomics analysis reveals stable multiprotein complexes in both fission and budding yeasts containing Myb-related Cdc5p/Cef1p, novel pre-mRNA splicing factors, and snRNAs. Mol. Cell. Biol. 2002;22:2011–2024. doi: 10.1128/MCB.22.7.2011-2024.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson B., Guthrie C. An essential yeast snRNA with a U5-like domain is required for splicing in vivo. Cell. 1987;49:613–624. doi: 10.1016/0092-8674(87)90537-x. [DOI] [PubMed] [Google Scholar]
- Pikielny C.W., Rosbash M. Specific small nuclear RNAs are associated with yeast spliceosomes. Cell. 1986;45:869–877. doi: 10.1016/0092-8674(86)90561-1. [DOI] [PubMed] [Google Scholar]
- Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., Seraphin B. The tandem affinity purification (TAP) method: A general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- Seraphin B., Rosbash M. Identification of functional U1 snRNA-pre-mRNA complexes committed to spliceosome assembly and splicing. Cell. 1989;59:349–358. doi: 10.1016/0092-8674(89)90296-1. [DOI] [PubMed] [Google Scholar]
- Seraphin B., Abovich N., Rosbash M. Genetic depletion indicates a late role for U5 snRNP during in vitro spliceosome assembly. Nucleic Acids Res. 1991;19:3857–3860. doi: 10.1093/nar/19.14.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Wilm M., Vorm O., Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Stevens S.W., Abelson J. Purification of the yeast U4/U6.U5 small nuclear ribonucleoprotein particle and identification of its proteins. Proc. Natl. Acad. Sci. 1999;96:7226–7231. doi: 10.1073/pnas.96.13.7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens S.W., Barta I., Ge H.Y., Moore R.E., Young M.K., Lee T.D., Abelson J. Biochemical and genetic analyses of the U5, U6, and U4/U6 × U5 small nuclear ribonucleoproteins from Saccharomyces cerevisiae . RNA. 2001;7:1543–1553. [PMC free article] [PubMed] [Google Scholar]
- Stevens S.W., Ryan D.E., Ge H.Y., Moore R.E., Young M.K., Lee T.D., Abelson J. Composition and functional characterization of the yeast spliceosomal penta-snRNP. Mol. Cell. 2002;9:31–44. doi: 10.1016/s1097-2765(02)00436-7. [DOI] [PubMed] [Google Scholar]
- Villa T., Pleiss J.A., Guthrie C. Spliceosomal snRNAs: Mg2+-dependent chemistry at the catalytic core? Cell. 2002;109:149–152. doi: 10.1016/s0092-8674(02)00726-2. [DOI] [PubMed] [Google Scholar]
- Wang Q., Hobbs K., Lynn B., Rymond B.C. The Clf1p splicing factor promotes spliceosome assembly through N-terminal tetratricopeptide repeat contacts. J. Biol. Chem. 2003;278:7875–7883. doi: 10.1074/jbc.M210839200. [DOI] [PubMed] [Google Scholar]
- Wyatt J.R., Sontheimer E.J., Steitz J.A. Site-specific cross-linking of mammalian U5 snRNP to the 5′ splice site before the first step of pre-mRNA splicing. Genes & Dev. 1992;6:2542–2553. doi: 10.1101/gad.6.12b.2542. [DOI] [PubMed] [Google Scholar]