Abstract
RNA catalysis is important in the processing and translation of RNA molecules, yet the mechanisms of catalysis are still unclear in most cases. We have studied the role of nucleobase catalysis in the hairpin ribozyme, where the scissile phosphate is juxtaposed between guanine and adenine bases. We show that a modified ribozyme in which guanine 8 has been substituted by an imidazole base is active in both cleavage and ligation, with ligation rates 10-fold faster than cleavage. The rates of both reactions exhibit bell-shaped dependence on pH, with pKa values of 5.7 ± 0.1 and 7.7 ± 0.1 for cleavage and 6.1 ± 0.3 and 6.9 ± 0.3 for ligation. The data provide good evidence for general acid–base catalysis by the nucleobases.
Keywords: RNA catalysis, general acid–base, imidazole
INTRODUCTION
Compared with proteins, the catalytic resources of RNA are quite limited, consisting of proximity and orientational effects, hydrated metal ions, and nucleobases (Bevilacqua 2004; Lilley 2005). Despite the rather unpromising pKA values of the natural nucleobases in the free state, there is increasing evidence for their direct participation in RNA catalysis (Perrotta et al. 1999; Nakano et al. 2000; Lafontaine et al. 2002; Jones and Strobel 2003; Ke et al. 2004; Kuzmin et al. 2004).
The hairpin ribozyme comprises two formally unpaired loops in adjacent arms of a four-way junction that become intimately docked together to facilitate catalysis of site-specific cleavage and ligation (Fig. 1), leading to a rate enhancement of ≥105-fold. These reactions are brought about by transesterification reactions shown in Figure 2A. A degree of rate enhancement should be provided by orientation of the attacking nucleophile (Soukup and Breaker 1999) but the major effect is likely to derive from stabilization of the transition state and/or acid–base catalysis to generate a stronger nucleophile and a better leaving group. The crystal structure of a vanadate-substituted transition state analog of the ribozyme (Rupert et al. 2002; Fig. 2B) suggests the participation of two nucleobases. G8 is hydrogen-bonded to the 2′-O and the proS O of the scissile phosphate, while A38 forms hydrogen bonds to the 5′-O and the proR O. In addition to stabilizing the transition state, these bases are well placed to participate in general acid–base catalysis. In the cleavage reaction, G8 is positioned to deprotonate the 2′-O, while A38 could protonate the 5′-oxyanion leaving group. In the ligation reaction, G8 and A38 would act as general acid and base, respectively. G8U substitution does not impair global folding yet leads to 1000-fold reduction in cleavage activity (Wilson et al. 2001), as did removal of the nucleobase (Kuzmin et al. 2004). A 10,000-fold loss of activity resulted from an abasic site at position 38 (Kuzmin et al. 2005). While Bevilacqua (2003) has stressed the potential role of these nucleobases in general acid–base catalysis, Fedor has argued that the principal role is to provide electrostatic transition-state stabilization (Kuzmin et al. 2004, 2005). This was in part based on the observation that free bases restoring activity to ribozyme abasic at position 8 all possessed an exocyclic amine group.
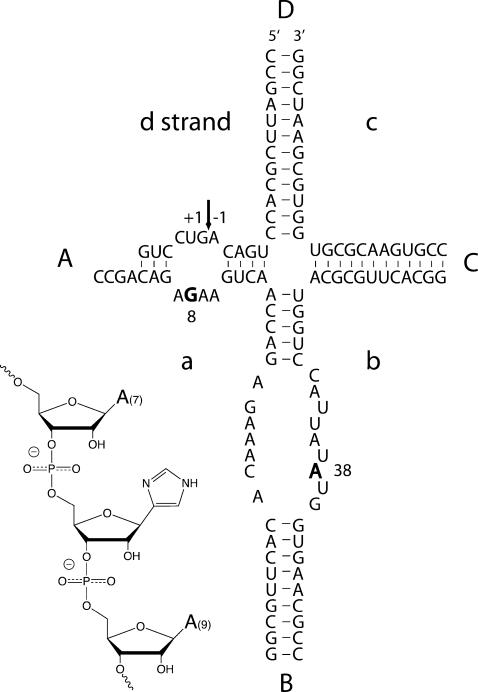
FIGURE 1.
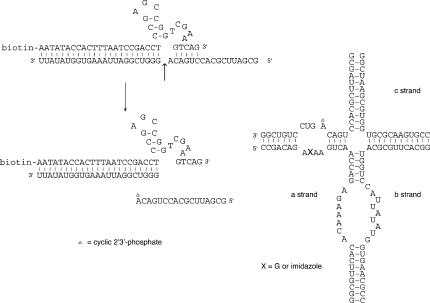
The hairpin ribozyme. The sequence of the hairpin ribozyme in the cleavage form used in these studies. The ribozyme comprises four strands (a–d). The cleavage site is marked with an arrow, and the locations of G8 and A38 are highlighted in bold. Coaxially stacked arms A and D are shown in black and C and D in gray to be consistent with the folding scheme shown in Figure 4A. The inset shows the local structure of the imidazole nucleotide (shown in its nonprotonated form) incorporated into the “a” strand at position 8.
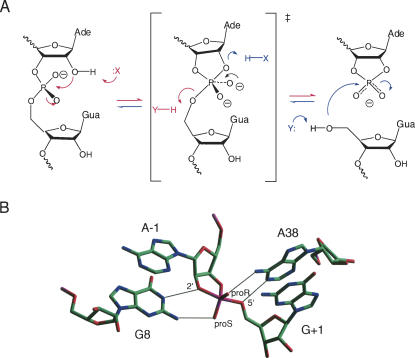
FIGURE 2.
The mechanism and active site of the hairpin ribozyme. (A) The cleavage and ligation reactions. Cleavage is achieved by a transesterification reaction involving SN2 attack of a 2′-O on the adjacent phosphate group, passing through a dianionic oxyphosphorane transition state in which the 2′- and 5′-O atoms are apical. Ligation is the reverse of this, involving attack of the 5′-O on the cyclic phosphate. X and Y are putative participants in general acid–base catalysis. In the cleavage reaction (red), X acts as a base to facilitate removal of the proton from the 2′-O, while Y protonates the 5′-O leaving group. In the ligation reaction (blue), the roles of X and Y reverse, so that X now acts as a general acid to protonate the 2′-O leaving group, while Y removes a proton from the 5′-O nucleophile. (B) The structure of the active site of the hairpin ribozyme in a crystal structure of a vanadate transition state analog (Rupert et al. 2002). G8 and A38 flank the scissile phosphate and form hydrogen bonds to the 2′- and 5′-O, respectively, as well as to the nonbridging O atoms.
We therefore tested whether imidazole could replace G8 with retention of catalytic activity. This is a good general base with a pKA close to neutrality that lacks an exocyclic amine group. It was convincingly used with genomic and anti-genomic HDV ribozyme variants, in which a cytosine proposed to participate in acid–base chemistry was substituted by uridine, where some cleavage activity was restored in the presence of high concentrations of imidazole in free solution (Perrotta et al. 1999; Nakano et al. 2000). However, the success of exogenous bases reactivating ribozyme activity may be impaired by lack of access to the active site in the correct orientation (Peracchi et al. 1998). Imidazole and its derivatives failed to restore activity to a position 8-abasic hairpin ribozyme (Kuzmin et al. 2004). We have recently developed a novel chemo-genetic strategy for investigating the role of nucleobases in ribozyme chemistry by means of the site-specific replacement of a conventional nucleotide base by imidazole (Harusawa et al. 1996; Araki et al. 2004). We demonstrated catalytic activity in a VS ribozyme, in which a critical adenosine was replaced by imidazole (Zhao et al. 2005). We have now explored the role of the nucleobase of G8 in hairpin ribozyme catalysis using the imidazole nucleoside.
RESULTS AND DISCUSSION
Cleavage activity of a G8Imidazole-substituted hairpin ribozyme
The hairpin ribozyme was constructed by the hybridization of four synthetic oligonucleotides as shown in Figure 1, and a version of strand a was synthesized containing the imidazole nucleoside in place of guanine at position 8. The substrate strand d was radioactively [5′-32P]-labeled to study the cleavage reaction.
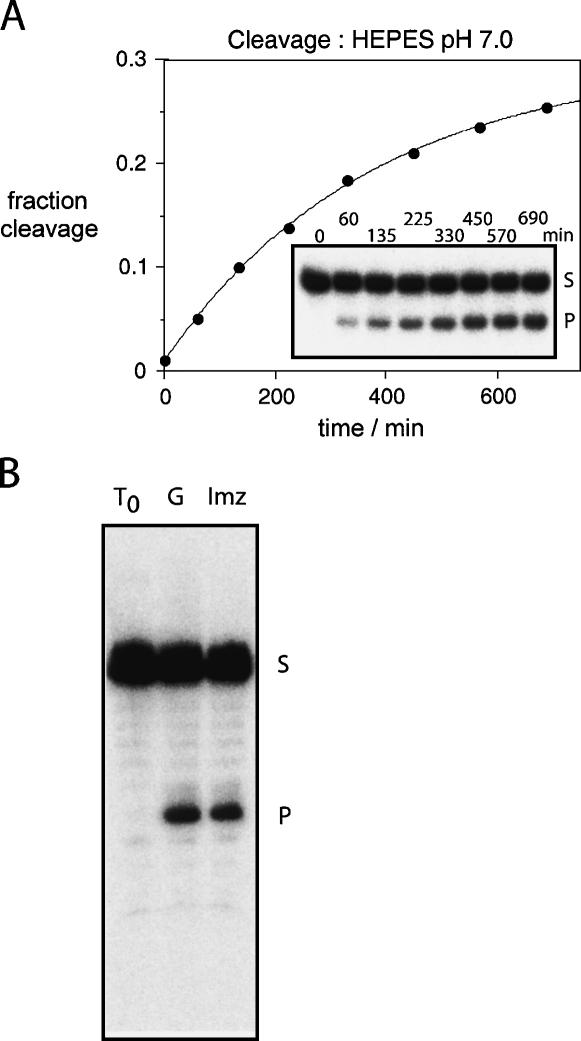
When the imidazole-substituted ribozyme was incubated at 25°C in the presence of 25 mM HEPES (pH 7.0), 10 mM MgCl2, 50 mM NaCl, product was generated at an observed rate of kobs = 0.0027 min−1 (Fig. 3A). This is substantially slower than the natural ribozyme under these conditions (kobs = 0.7 min−1) but is significantly faster than that for a G8U variant (kobs = 3 × 10−4 min−1). Furthermore, although the reaction is relatively slow, the extent of cleavage is significant, achieving a plateau value comparable to that obtained by the wild-type ribozyme under equivalent conditions. The reaction product has also been examined at higher electrophoretic resolution (Fig. 3B). Its rate of migration is indistinguishable from that generated by the natural ribozyme, showing that it is the 17-nt product expected for ribozyme cleavage.
FIGURE 3.
Cleavage reaction by the G8Imidazole hairpin ribozyme. (A) Progress curve for the cleavage reaction of the G8Imidazole hairpin ribozyme in 25 mM HEPES (pH 7.0), 50 mM NaCl, and 10 mM MgCl2 at 25°C. The data were fitted to a single exponential, yielding a cleavage rate of 0.0027 min−1. The corresponding electrophoretic separation of substrate (S) and product (P) is shown as an inset. Reaction time (minutes) is shown above each lane. (B) Analysis of the cleavage product at sequencing-gel resolution. The cleavage products for natural and G8Imidazole hairpin ribozymes were electrophoresed in a 25-cm 20% polyacrylamide gel containing 7 M urea. Reaction times were 1 min for the natural ribozyme (lane G) and 120 min for the G8Imidazole (lane Imz) ribozyme. A relatively short reaction time was used so that hydrolysis of the cyclic phosphate is negligible.
The results show that a functional ribozyme is generated when guanine 8 is replaced by imidazole, giving the correct cleavage product in good yield. This is consistent with a direct role for the nucleobase at position 8 in ribozyme catalysis.
Global folding of the G8Imidazole-substituted hairpin ribozyme is unaltered
A low rate of cleavage would be observed if substitution of imidazole for the guanine base at position 8 affected the loop–loop interaction that creates the active form of the ribozyme. We therefore investigated this possibility using fluorescence resonance energy transfer (FRET) between fluorescein and Cy3 fluorophores attached to the 5′-termini of the c and d strands, measured in the steady state. We have previously shown that this vector can be used to follow the metal ion-induced folding of the natural form of the ribozyme (Wilson and Lilley 2002), since the efficiency of energy transfer increases as the C–D end-to-end distance decreases as a consequence of the scissor-like rotation of the stacked helices that accompanies loop–loop interaction (Fig. 4A). For this purpose, we constructed a form of the ribozyme with a 2′-deoxyribose substitution at A-1 on the d strand to prevent cleavage activity. The same a and b strands were used to make this construct and the ribozymes used for analysis of ribozyme activity, and thus, the imidazole-containing strand was identical in both sets of experiments.
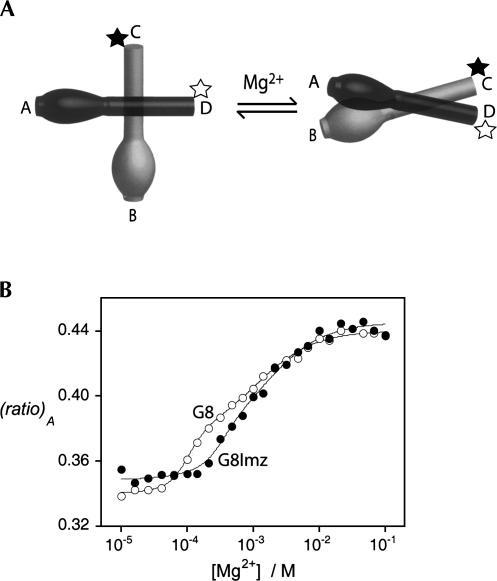
FIGURE 4.
Analysis of ion-induced folding of the natural and substituted hairpin ribozymes using fluorescence resonance energy transfer. (A) Schematic to show the analysis of folding by FRET. Fluorescein (black star) and Cy3 (open star) fluorophores were covalently linked to the 5′ termini of the C and D arms, respectively. Addition of magnesium ions induces a rotation of the stacked helices to allow an intimate interaction between the A and B loops, resulting in a shortening of the interfluorophore distance and hence an increase in FRET efficiency. (B) Plot of the extent of energy transfer, (ratio)A, as a function of Mg2+ concentration for the G8 (○) and G8Imidazole (●) hairpin ribozymes in 10 mM HEPES (pH 7.0), 50 mM NaCl at 4°C. The data were fitted to equation 2, giving estimated [Mg2+]1/2 values of (1.0 ± 0.2) × 10−4 M and (1.1 ± 0.3) × 10−3 M for the G8 ribozyme, and (3.5 ± 0.8) × 10−4 M and (1.5 ± 0.5) × 10−3 M for the G8Imidazole ribozyme.
We compared the extent of energy transfer as a function of Mg2+ ion concentration for the natural sequence and G8Imidazole ribozymes. The data are plotted as the normalized acceptor intensity, (ratio)A; this is directly proportional to FRET efficiency (Clegg 1992). The general shape of the profiles are similar for both species (Fig. 4B), with the G8Imidazole ribozyme requiring a higher concentration of Mg2+ to initiate folding. When fitted with equation 2, the natural ribozyme yields [Mg2+]1/2 values of (1.0 ± 0.2) × 10−4 M and (1.1 ± 0.3) × 10−3 M, in good agreement with previous results (Wilson and Lilley 2002). The apparent affinity for Mg2+ ions is threefold higher for the G8Imidazole ribozyme, indicating a weakening of the loop–loop interaction with the modification at position 8, with [Mg2+]1/2 values of (3.5 ± 0.8) × 10−4 M and (1.5 ± 0.5) × 10−3 M. Single-molecule studies have shown that cleavage-competent natural ribozymes are fully folded at 1 mM Mg2+, which correlates with the higher affinity cooperative Mg2+ ion binding (Nahas et al. 2004). Assuming this also applies to the G8Imidazole ribozyme, a [Mg2+]1/2 of 3.5 × 10−4 M indicates that the ribozyme will be fully folded in the presence of 10 mM Mg2+ ions as used for our activity measurements.
The pH dependence of the cleavage activity of a G8Imidazole-substituted hairpin ribozyme
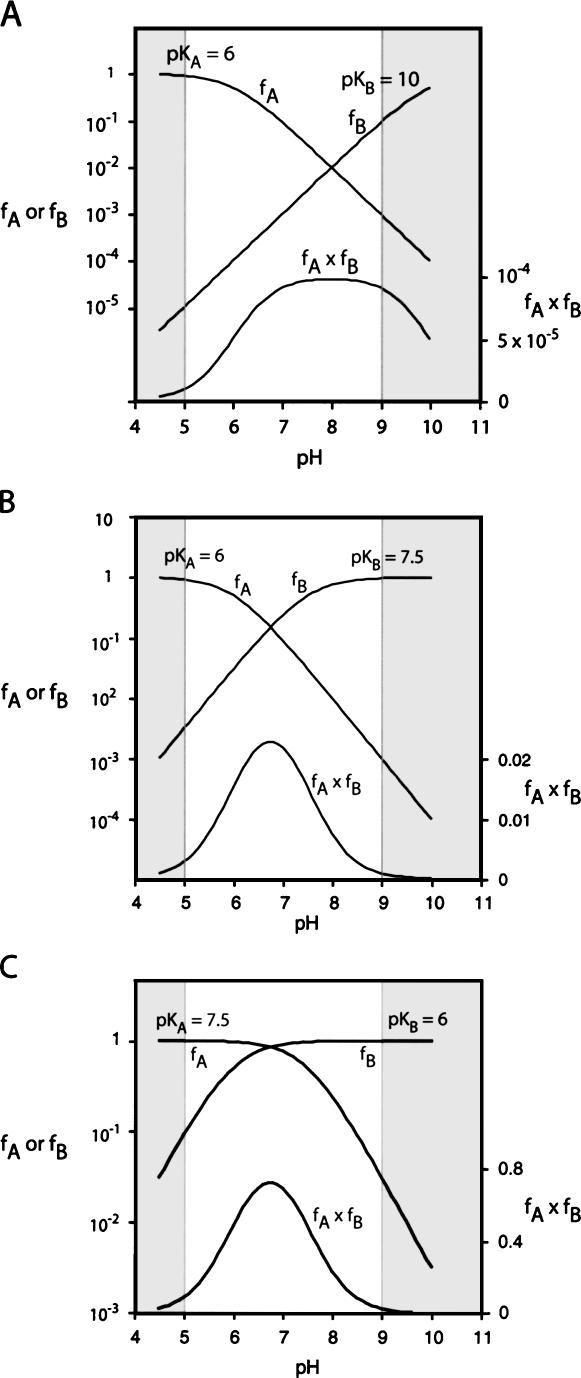
Cleavage and ligation rates of the natural hairpin ribozyme are pH-dependent. The rates of both reactions increase with pH up to neutrality, at which point they reach a plateau. Both correspond to the titration of a group with a pKA of close to 6.2 (Kuzmin et al. 2004; Nahas et al. 2004). This would be consistent with titration of A38, assuming an elevated pKA due to local environment as shown by simulation in Figure 5A. Removal of the base from A38 leads to a loss of activity, and perturbation of the pH profile, while a pKA close to 6.2 is retained under these conditions in corresponding G8-abasic derivatives despite a reduction in activity (Kuzmin et al. 2004, 2005). If A38 acts as a general acid to protonate the leaving group in the cleavage reaction, then G8 is likely to act as the general base, and thus is required to be in the deprotonated form. Since its pKA is expected to be >9, the concentration of the deprotonated form increases linearly with pH over the experimentally observable range (i.e., the unshaded region in Fig. 5A), and hence no reduction in rate is found since the deprotonation of A38 compensates so that the concentration of active ribozyme remains constant. However, if the role of this guanine is taken by imidazole with a lower pKA, the threshold for reduction in rate will occur at lower pH due to complete deprotonation of the imidazole around pH 8, leading to a bell-shaped pH profile (simulated in Fig. 5B). If A38 were to act as the base and the imidazole as the acid, as expected for the ligation reaction, then the variation of active fraction with pH has the same shape as that for the cleavage reaction (Fig. 5C). Although the ribozyme is now almost 100% in the required form for catalysis around neutrality, the intrinsic reactivities of this species are lower, since both acid and base are relatively weak.
FIGURE 5.
Calculated pH dependence of the cleavage reaction of the hairpin ribozyme as a function of base pKA, assuming general acid–base catalysis. The fractions of protonated acid fA, unprotonated base fB, and their product fA·fB have been calculated and plotted as a function of pH, following the approach of Bevilacqua (2003). The sections shaded in gray are the extreme values of pH not accessible to experimental study. Reaction rate should be proportional to the fraction of ribozyme in the appropriate state of protonation, i.e., fA·fB. Note that in these graphs fA and fB are plotted on a log10 scale (left), while fA·fB is plotted on a linear scale (right). (A) Plot for pKA values of 6 and 10 for the acid and the base, respectively. This corresponds to the natural ribozyme, assuming A38 is the acid and G8 is the base. Reaction rate increases with pH until reaching a plateau value close to neutrality. (B) Plot for pKA values of 6 and 7.5 for the acid and the base, respectively. This corresponds to that expected for a ribozyme in which G8 has been substituted by a base with a pKA of 7.5. A bell-shaped pH dependence is now expected. (C) Plot for pKA values of 7.5 and 6 for the acid and the base, respectively. This corresponds to that expected for a ribozyme in which G8 has been substituted by a base with a pKA of 7.5, now acting as the acid in the reverse reaction. Note that the shape of the pH profile is the same as that for the cleavage reaction.
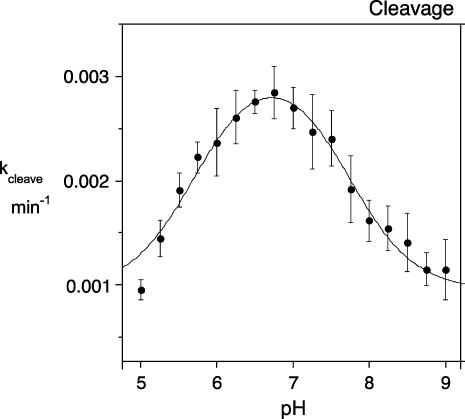
The experimentally observed dependence of cleavage rate with pH for the G8Imidazole-substituted ribozyme is indeed bell-shaped (Fig. 6). Fitting the data to a model based on two proton transfers gave estimated pKA values of 5.7 ± 0.1 and 7.7 ± 0.1. The lower pKA probably corresponds to that observed with the natural ribozyme, which is likely to be due to titration of A38. If so, then the pKA of A38 has become significantly lower in the modified ribozyme, indicating that the imidazole substitution has altered the environment of this nucleobase. This can be understood, since at the pH values at which the reaction rate is sensitive to the pKA of A38 a neutral guanine has been replaced by a cationic imidazole group within the active site of the ribozyme. This will tend to reduce the pKA of the nearby adenine. The higher pKA most probably is that of the imidazole; the value of 7.7 is near the upper end of the observed range for this base (Fersht 1999), and since the imidazole is close to a charged phosphate group in the active site, a high pKA would be expected. The value is also comparable to that of the equivalent histidine in RNaseA (Findlay et al. 1961).
FIGURE 6.
The pH dependence of the cleavage reaction of the G8Imidazole hairpin ribozyme. Plot of cleavage rate as a function of pH. Each point is the average and standard deviation of at least three independent experiments. The data were fitted to equation 1, giving estimated pKA values of 5.7 ± 0.1 and 7.7 ± 0.1.
The results show that the maximal cleavage activity of the G8Imidazole ribozyme is ∼200-fold lower than that of the natural ribozyme. Assuming an ∼200-fold increase in the concentration of the deprotonated base for the G8Imidazole ribozyme (i.e., a difference in pKA of 2.3 between imidazole and guanosine), this implies that the contribution of imidazole to the chemical step is ∼40,000-fold lower than that of guanosine. The low reactivity might be expected for two reasons. First, the ring nitrogen atoms of the imidazole are 2–4 Å from where N1 or N3 of guanine would be placed assuming that the ribose is not moved; participation of the imidazole in the reaction is therefore likely to require transient distortion of the active site. Similarly slow rates of cleavage were observed in the corresponding A756Imidazole VS ribozyme (Zhao et al. 2005) and in HDV C76U in the presence of exogenous imidazole (Perrotta et al. 1999). Second, the modified ribozyme will have lost some transition-state stabilization in the absence of hydrogen bonding to G8.
Ligation activity of a G8Imidazole-substituted hairpin ribozyme
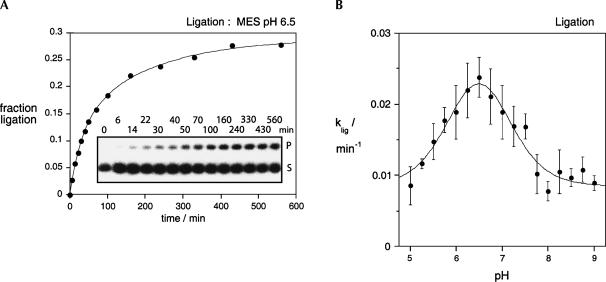
We have examined the ability of the G8Imidazole ribozyme to catalyze the ligation reaction. By the principle of microscopic reversibility, this reaction should pass through the same transition state as the cleavage reaction, and thus, if the imidazole is acting as a general base during cleavage, it should act as a general acid in ligation. The ribozyme was constructed using a substrate d strand in two sections, with a cyclic 2′3′ phosphate at A-1 created with the aid of a DNAzyme, and the 3′ substrate retained by a 7-bp helix (Fig. 7). The complete ribozyme was incubated with 20 mM MES (pH 6.5), 10 mM MgCl2, 50 mM NaCl, 0.6 mM HEPES, and the products analyzed by gel electrophoresis and quantified by PhosphorImaging (Fig. 8A). A discrete product of the correct size was generated in significant amounts over this timescale; the ultimate yield will be limited by the hydrolysis of the cyclic phosphate, which becomes important at longer times (Zhao et al. 2005). The observed reaction rate was kobs = 0.024 min−1. This is the rate of approach to equilibrium, i.e., the sum of ligation and cleavage rates. The rate of ligation is eightfold faster than the rate of cleavage by the modified ribozyme under the same conditions. A similar bias toward the ligation reaction has been measured in single-molecule and ensemble studies of the natural hairpin ribozyme (Fedor 1999; Nahas et al. 2004).
FIGURE 7.
Scheme for the generation of the ligation substrate. The 17-nt 5′ ligation substrate was generated using biotinylated 8–17 DNAzyme. This was hybridized together with the 3′ substrate fragment and strands a, b, and c in the presence of 1.5 mM HEPES (pH 7.5) to generate the ligation-competent hairpin ribozyme.
FIGURE 8.
Ligation reaction by the G8Imidazole hairpin ribozyme. (A) The ligation reaction at 25°C in 20 mM MES (pH 6.5) 10 mM MgCl2, 50 mM NaCl together with a residual 0.6 mM HEPES (pH 7.5) from the hybridization. The extent of product formation is plotted as a function of time and fitted to two exponential functions, the faster yielding an observed rate of 0.024 min−1. The corresponding electrophoretic separation of substrate (S) and product (P) is shown as an inset. Reaction time (minutes) is shown above each lane. (B) The pH dependence of ligation rate. Each point is the average and standard deviation of at least three independent experiments. The data were fitted to equation 1, giving estimated pKA values of 6.1 ± 0.3 and 6.9 ± 0.3.
We have measured the rate of the ligation reaction as a function of pH (Fig. 8B). A bell-shaped dependence was obtained once more, consistent with the participation of a group with a lower pKA compared with the natural ribozyme, as expected if imidazole were now acting as the general acid. Fitting the data to the model for two proton transfers gives pKA values of 6.1 ± 0.3 and 6.9 ± 0.3. The values differ from those obtained for the cleavage reaction, narrowing the bell shape of pH profile for the ligation reaction, which is largely due to a lowering of the higher (presumptive imidazole) pKA. This indicates that the local environment in the ground-state active sites for the two reactions are not identical for the modified ribozyme, an effect that might be accentuated by the substitution of imidazole for the guanine base. G8 is not hydrogen bonded to the phosphate group in the product of the cleavage reaction (Rupert et al. 2002), and therefore the imidazole may lie farther from the negative charge, thereby lowering the pKA in the state prior to the ligation reaction.
CONCLUSIONS
The cleavage and ligation activity of the imidazole-substituted hairpin ribozyme provide evidence for a direct role for the nucleobase at position 8 in catalysis. The pH dependence of both reactions are fully consistent with general acid–base catalysis, as are all available data on the pH dependence of the hairpin ribozyme either in its natural form or with substitutions at position 8 (Kuzmin et al. 2004). The imidazole group lacks the exocyclic amine previously found to be required in restoration of activity by exogenous bases with ribozyme abasic at position 8 (Kuzmin et al. 2004). Interaction with the amine (as in G8) may contribute to the transition state stabilization, but should not be required for general acid–base catalysis. The data show that electrostatic stabilization of the transition state by nucleobases is unable by itself to explain catalysis, since the charge of the active site in the modified ribozyme is net positive, compared with neutral for the natural situation. Thus, the natural ribozyme would be less appropriately charged for the stabilization of an dianionic transition state. While the chemistry of catalysis in the hairpin ribozyme is likely to include a number of different contributions including substrate orientation and transition-state stabilization, the present data provide good evidence that general acid–base catalysis is an important element.
MATERIALS AND METHODS
Synthesis of RNA
C4-linked imidazole ribonucleoside phosphoramidite with a pivaloyloxymethyl group was synthesized as described previously (Araki et al. 2004). This was incorporated into the “a” strand of the hairpin ribozyme by chemical synthesis using 2′-t-BDMS ribonucleoside β-cyanoethyl phosphoramidites as described previously (Zhao et al. 2005). The 5′ substrate strand used for ligation was transcribed in vitro using T7 RNA polymerase. All other RNA strands were synthesized using 2′-ACE-protected phosphoramidites (Scaringe 2000). Fully deprotected oligonucleotides were purified by electrophoresis in 20% polyacrylamide gels containing 7 M urea and recovered by electroelution.
Analysis of ribozyme cleavage reactions
Hairpin ribozyme strands a, b, and c (5 μM) were hybridized in 10 mM HEPES (pH 7.5), 50 mM NaCl by heating to 90°C, snap-cooled, left on ice for at least 20 min, and stored at –20°C. Ribozyme was diluted to 400 nM by addition of 50 mM NaCl containing radioactively [5′-32P]-labeled substrate strand (<20 nM) and incubated at 25°C for 15 min. A zero-time aliquot was removed, after which the cleavage reaction was initiated by addition of an equal volume of a solution comprising 50 mM buffer, 50 mM NaCl, and 20 mM MgCl2. All reactions were performed at 25°C under mineral oil to prevent evaporation. Aliquots (2 μL) were removed at various times, and the reaction terminated by addition to 8 μL of 20 mM EDTA in 95% formamide. Substrate and product were separated by electrophoresis in a 7-cm 20% polyacrylamide gel, and quantified by phosphorimaging. The buffers used were acetate, pH 5.0–5.5; MES, pH 5.5–6.75; HEPES, pH 6.75–8.0; and TAPS, pH 8.0–9.0. No significant difference in the rates of activity was observed for different buffers at the same pH.
Analysis of ribozyme ligation reactions
The strand containing the cyclic 2′3′ phosphate terminus for the ligation reaction was generated from a T7 RNA polymerase-transcribed RNA using a 5′-biotin-labeled 8–17 DNAzyme (Santoro and Joyce 1997). Cleavage of the RNA was performed immediately prior to use, after which reacted and unreacted DNAzyme was removed using streptavidin-coated magnetic beads. Ribozyme (100 nM) (strands a, b, and c) was hybridized with a trace of radioactively [5′-32P]-labeled 5′ substrate strand in 1.5 mM HEPES (pH 7.5), 5 mM MgCl2, 62.5 mM NaCl at 25°C for 20 min. A zero-time aliquot was removed, after which the ligation reaction was initiated by addition of 3′ substrate strand and appropriate buffer (using the range listed above), such that the resulting reaction solution comprised 20 mM buffer, 50 mM NaCl, 10 mM MgCl2, 1 μM 3′ substrate strand and a residual 0.6 mM HEPES (pH 7.5). Reactions were incubated at 25°C and 2 μL aliquots were removed at times 6–560 min, and the reaction was terminated by addition to 6 μL of 20 mM EDTA in formamide. Substrate and product were separated by electrophoresis in a 7-cm 20% polyacrylamide gel, and quantified by PhosphorImaging.
Analysis of pH dependence of reaction rate
The pH dependence of cleavage and ligation rates were fitted to a model assuming a requirement for a general acid and base in protonated and deprotonated forms, respectively, and a background rate of reaction not requiring acid–base catalysis (knonAB), i.e.,
where kAB is the intrinsic rate of general acid–base catalyzed reaction and pKAacid and pKAbase are the acid dissociation constants of the general acid and base, respectively.
Analysis of ion-induced folding using FRET
Hairpin ribozyme species were constructed by incubating stoichiometric quantities of unlabeled a and b strands, fluorescein-labeled c strand, and Cy3-labeled d strand at 90°C for 2 min, followed by snap-cooling on ice. The hybridized species were purified, their fluorescence spectra recorded at 4°C in 10 mM HEPES (pH 7.0), 50 mM NaCl, and the extent of energy transfer determined as described previously (Wilson and Lilley 2002). The reported parameter (ratio)A is directly proportional to the efficiency of energy transfer. Conversion to an absolute measure of energy transfer was not possible since the low yield of fluorescent G8Imidazole hairpin ribozyme prevented an accurate measurement of an absorption spectra for this species. Ion-induced folding by the hairpin ribozyme requires a complex model to describe the observed binding over a broad range of Mg2+ and Na+ concentrations (Wilson and Lilley 2002). Single-molecule studies have demonstrated that this complexity arises from a heterogeneous ribozyme population, with all molecules exhibiting simple two-state folding (Tan et al. 2003). Therefore, data were fitted to a model allowing for two independent populations, using the Hill coefficients of 3 and 1 determined previously (Wilson and Lilley 2002):
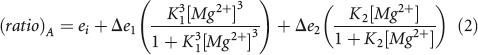 |
where ei is the energy transfer in the absence of magnesium ions, Δe 1 and Δe 2 are changes in energy transfer on addition of magnesium ions, and K 1 and K 2 are apparent association constants for magnesium ions. These are reported as the corresponding magnesium ion concentration at which a transition is half-complete, [Mg2+]1/2 = 1/K1 or 1/K2(Wilson and Lilley 2002). The quoted uncertainties are the asymptotic standard deviations generated by the nonlinear curve fitting routine.
RNA sequences
The unmodified ribozyme comprised strands a–c (all written 5′ to 3′):
a: CCGACAGAGAAGUCAACCAGAGAAACACACUUGCGG;
b: CCGCAAGUGGUAUAUUACCUGGUACGCGUUCACGG;
c: CCGUGAACGCGUGGUGCGAAUCGG.
The imidazole-containing aIm strand was
aIm: CCGACAGAImAAGUCAACCAGAGAAACACACUUGCGG
where Im denotes the imidazole nucleoside.
The G8U-containing a strand was
a: CCGACAGAUAAGUCAACCAGAGAAACACACUUGCGG.
For the cleavage reaction, the substrate d strand was
d: CCGAUUCGCACCUGACAGUCCUG.
The template for synthesis of the RNA strand used to construct the ligation-competent ribozyme by T7 RNA polymerase comprised the following oligonucleotides:
AATATACCACTTTAATCCGACCCTGTCAGGTGCGAATCGCTATAGTGAGTCGTATTACC
and GGTAATACGACTCACTATAGCG.
The 40-nt transcript was annealed to the 8–17 DNAzyme:
biotin-AATATACCACTTTAATCCGACCTCCGAGCCGGTCGAAGTCAG.
The sequence of the 3′ substrate strand for ligation was
GUCCUGUCGG.
The additional strands used to assemble the species used for the FRET analysis of folding had the sequence
c: Fluorescein-CCGUGAACGCGUGGUGCGAAUCGG;
d: Cy3-CCGAUUCGCACCUGACAGUCCUG.
ACKNOWLEDGMENTS
We thank the Cancer Research-UK (Dundee) and “High-Tech Research Center” Project for Private Universities with matching fund subsidy from MEXT in 2002-2006 (Osaka) for financial support.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.11706.
REFERENCES
- Araki L., Harusawa S., Yamaguchi M., Yonezawa S., Taniguchi N., Lilley D.M.J., Zhao Z., Kurihara T. Synthesis of C4-linked imidazole ribonucleoside phosphoramidite with pivaloyloxymethyl (POM) group. Tetrahedron Lett. 2004;45:2657–2661. [Google Scholar]
- Bevilacqua P.C. Mechanistic considerations for general acid-base catalysis by RNA: Revisiting the mechanism of the hairpin ribozyme. Biochemistry. 2003;42:2259–2265. doi: 10.1021/bi027273m. [DOI] [PubMed] [Google Scholar]
- Bevilacqua P.C. Mechanism of catalytic RNA. Biopolymers. 2004;73:69–70. doi: 10.1002/bip.10522. [DOI] [PubMed] [Google Scholar]
- Clegg R.M. Fluorescence resonance energy transfer and nucleic acids. Methods Enzymol. 1992;211:353–388. doi: 10.1016/0076-6879(92)11020-j. [DOI] [PubMed] [Google Scholar]
- Fedor M.J. Tertiary structure stabilization promotes hairpin ribozyme ligation. Biochemistry. 1999;38:11040–11050. doi: 10.1021/bi991069q. [DOI] [PubMed] [Google Scholar]
- Fersht A. Structure and mechanism in protein science: A guide to enzyme catalysis and protein folding. W.H. Freeman and Co; New York: 1999. [Google Scholar]
- Findlay D., Herries D.G., Mathias A.P., Rabin B.R., Ross C.A. The active site and mechanism of action of bovine pancreatic ribonuclease. Nature. 1961;190:781–784. doi: 10.1038/190781a0. [DOI] [PubMed] [Google Scholar]
- Harusawa S., Murai Y., Moriyama H., Imazu T., Ohishi H., Yoneda R., Kurihara T. Efficient and β-stereoselective synthesis of 4(5)-(β-D-ribofuranosyl)- and 4(5)-(2-deoxyribofuranosyl)imidazoles. J. Org. Chem. 1996;61:4405–4411. doi: 10.1021/jo952136z. [DOI] [PubMed] [Google Scholar]
- Jones F.D., Strobel S.A. Ionization of a critical adenosine residue in the Neurospora Varkud Satellite ribozyme active site. Biochemistry. 2003;42:4265–4276. doi: 10.1021/bi020707t. [DOI] [PubMed] [Google Scholar]
- Ke A., Zhou K., Ding F., Cate J.H., Doudna J.A. A conformational switch controls hepatitis delta virus ribozyme catalysis. Nature. 2004;429:201–205. doi: 10.1038/nature02522. [DOI] [PubMed] [Google Scholar]
- Kuzmin Y.I., Da Costa C.P., Fedor M.J. Role of an active site guanine in hairpin ribozyme catalysis probed by exogenous nucleobase rescue. J. Mol. Biol. 2004;340:233–251. doi: 10.1016/j.jmb.2004.04.067. [DOI] [PubMed] [Google Scholar]
- Kuzmin Y.I., Da Costa C.P., Cottrell J.W., Fedor M.J. Role of an active site adenine in hairpin ribozyme catalysis. J. Mol. Biol. 2005;349:989–1010. doi: 10.1016/j.jmb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Lafontaine D.A., Wilson T.J., Zhao Z.-Y., Lilley D.M.J. Functional group requirements in the probable active site of the VS ribozyme. J. Mol. Biol. 2002;323:23–34. doi: 10.1016/s0022-2836(02)00910-5. [DOI] [PubMed] [Google Scholar]
- Lilley D.M. Structure, folding and mechanisms of ribozymes. Curr. Opin. Struct. Biol. 2005;15:313–323. doi: 10.1016/j.sbi.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Nahas M.K., Wilson T.J., Hohng S., Jarvie K., Lilley D.M.J., Ha T. Observation of internal cleavage and ligation reactions of a ribozyme. Nat. Struct. Mol. Biol. 2004;11:1107–1113. doi: 10.1038/nsmb842. [DOI] [PubMed] [Google Scholar]
- Nakano S., Chadalavada D.M., Bevilacqua P.C. General acid-base catalysis in the mechanism of a hepatitis delta virus ribozyme. Science. 2000;287:1493–1497. doi: 10.1126/science.287.5457.1493. [DOI] [PubMed] [Google Scholar]
- Peracchi A., Karpeisky A., Maloney L., Beigelman L., Herschlag D. A core folding model for catalysis by the hammerhead ribozyme accounts for its extraordinary sensitivity to abasic mutations. Biochemistry. 1998;37:14765–14775. doi: 10.1021/bi980867y. [DOI] [PubMed] [Google Scholar]
- Perrotta A.T., Shih I., Been M.D. Imidazole rescue of a cytosine mutation in a self-cleaving ribozyme. Science. 1999;286:123–126. doi: 10.1126/science.286.5437.123. [DOI] [PubMed] [Google Scholar]
- Rupert P.B., Massey A.P., Sigurdsson S.T., Ferré-D'Amaré A.R. Transition state stabilization by a catalytic RNA. Science. 2002;298:1421–1424. doi: 10.1126/science.1076093. [DOI] [PubMed] [Google Scholar]
- Santoro S.W., Joyce G.F. A general-purpose RNA-cleaving DNA enzyme. Proc. Natl. Acad. Sci. 1997;94:4262–4266. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaringe S.A. Advanced 5′-silyl-2′-orthoester approach to RNA oligonucleotide synthesis. Methods Enzymol. 2000;317:3–18. doi: 10.1016/s0076-6879(00)17003-x. [DOI] [PubMed] [Google Scholar]
- Soukup G.A., Breaker R.R. Relationship between internucleotide linkage geometry and the stability of RNA. RNA. 1999;5:1308–1325. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan E., Wilson T.J., Nahas M.K., Clegg R.M., Lilley D.M.J., Ha T. A four-way junction accelerates hairpin ribozyme folding via a discrete intermediate. Proc. Natl. Acad. Sci. 2003;100:9308–9313. doi: 10.1073/pnas.1233536100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson T.J., Lilley D.M.J. Metal ion binding and the folding of the hairpin ribozyme. RNA. 2002;8:587–600. doi: 10.1017/s1355838202020514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson T.J., Zhao Z.-Y., Maxwell K., Kontogiannis L., Lilley D.M.J. Importance of specific nucleotides in the folding of the natural form of the hairpin ribozyme. Biochemistry. 2001;40:2291–2302. doi: 10.1021/bi002644p. [DOI] [PubMed] [Google Scholar]
- Zhao Z., McLeod A., Harusawa S., Araki L., Yamaguchi M., Kurihara T., Lilley D.M.J. Nucleobase participation in ribozyme catalysis. J. Am. Chem. Soc. 2005;127:5026–5027. doi: 10.1021/ja0502775. [DOI] [PubMed] [Google Scholar]