Abstract
We describe a simple procedure for RNA 5′-adenylation using T4 DNA ligase. The 5′-monophosphorylated terminus of an RNA substrate is annealed to a complementary DNA strand that has a 3′-overhang of 10 nucleotides. Then, T4 DNA ligase and ATP are used to synthesize 5′-adenylated RNA (5′-AppRNA), which should find use in a variety of practical applications. In the absence of an acceptor nucleic acid strand, the two-step T4 DNA ligase mechanism is successfully interrupted after the adenylation step, providing 40%–80% yield of 5′-AppRNA after PAGE purification with few side products (the yield varies with RNA sequence). Optimized reaction conditions are described for 5′-adenylating RNA substrates of essentially any length including long and structured RNAs, without need for sequestration of the RNA 3′-terminus to avoid circularization. The new procedure is applicable on the preparative nanomole scale. This 5′-adenylation strategy using T4 DNA ligase is a substantial improvement over our recently reported adenylation method that uses T4 RNA ligase, which often leads to substantial amounts of side products and requires careful optimization for each RNA substrate. Efficient synthetic access to 5′-adenylated RNA will facilitate a range of applications by providing substrates for in vitro selection; by establishing a new protocol for RNA 5′-capping; and by providing an alternative approach for labeling RNA with 32P or biophysical probes at the 5′-terminus.
Keywords: RNA 5′-adenylation, T4 DNA ligase, in vitro selection, RNA capping, RNA labeling
INTRODUCTION
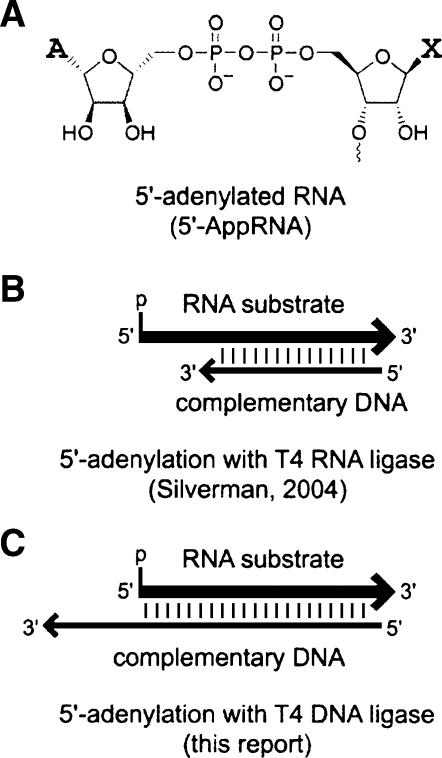
We recently described a practical approach for the synthesis of 5′-adenylated1 RNA (5′-AppRNA), which is RNA that has a 5′,5′-adenyl pyrophosphoryl cap structure (Fig. 1A; Silverman 2004). The 5′-AppRNA is desirable to investigate natural biochemical pathways in which such intermediates participate (for references, see Silverman 2004). The 5′-AppRNA will also be useful for in vitro selection procedures to identify nucleic acid enzymes that use 5′-AppRNA as a reactive RNA substrate (Hager and Szostak 1997; Coppins and Silverman 2004, 2005), as well as for applications such as RNA 5′-capping and 5′-labeling. In our previous report (Silverman 2004), the protein enzyme T4 RNA ligase was used along with a complementary DNA oligonucleotide (Fig. 1B) to achieve the net transfer of AMP onto the 5′-monophosphorylated terminus of the desired RNA substrate. This approach successfully interrupts the natural T4 RNA ligase mechanism, which normally does not stop at 5′-AppRNA but instead continues to join a 3′-hydroxyl group of a second RNA substrate to the first RNA substrate with loss of AMP and formation of ligated RNA. Although substantial quantities of 5′-AppRNA can indeed be generated using T4 RNA ligase by our previous strategy, the approach has certain drawbacks. Principal among these is that several side products are formed via undesired secondary reactions of the 5′-AppRNA. These side products are formed by RNA circularization or oligomerization, or they are formed by ligation to the RNA of a DNA “blocking oligonucleotide” that was included to suppress the first two side reactions. Ultimately, the formation of these side products means that the overall adenylation strategy using T4 RNA ligase must be optimized carefully for each individual substrate, and for certain RNA sequences the side products are formed in substantial amounts even after optimization. We therefore sought other approaches to prepare 5′-AppRNA that would not be subject to the same disadvantages.
FIGURE 1.
5′-Adenylated RNA and its synthesis using protein enzymes. (A) Structure of 5′-adenylated RNA (5′-AppRNA). X denotes the nucleobase at the 5′-terminus of the RNA strand that is 5′-adenylated. (B) Synthesis of 5′-AppRNA using T4 RNA ligase and a complementary DNA oligonucleotide that leaves an RNA 5′-overhang (Silverman 2004). (C) Synthesis of 5′-AppRNA using T4 DNA ligase and a complementary DNA oligonucleotide that leaves a recessed RNA 5′-terminus (DNA 3′-overhang), as reported here.
The impetus for our previous report (Silverman 2004) was the earlier finding by Chiuman and Li (2002) that 5′-adenylated DNA (5′-AppDNA) can be synthesized in a related fashion by using 5′-monophosphorylated DNA and T4 DNA ligase. Because T4 DNA ligase generally requires a double-stranded substrate whereas T4 RNA ligase requires a single-stranded substrate, certain key aspects of the RNA 5′-adenylation procedures are different. In preliminary studies for our previous report, we were unsuccessful in attempting to apply T4 DNA ligase to form 5′-AppRNA. Nevertheless, the disadvantages of T4 RNA ligase for 5′-AppRNA synthesis (as described above) led us to reevaluate the ability of T4 DNA ligase to perform this function. Here we report that T4 DNA ligase can indeed be used along with a complementary DNA oligonucleotide (Fig. 1C) to prepare 5′-AppRNA in good yield. Furthermore, the use of T4 DNA ligase avoids formation of most of the side products that are formed with T4 RNA ligase, and the need for careful optimization of reaction conditions for individual RNA substrates is substantially reduced. Therefore, T4 DNA ligase is superior to T4 RNA ligase for RNA adenylation. We anticipate that the establishment of this user-friendly synthetic approach will facilitate practical applications of 5′-AppRNA for in vitro selection, RNA 5′-capping, and RNA 5′-labeling with 32P or biophysical probes.
RESULTS
5′-Adenylation of RNA using T4 DNA ligase, ATP, and a complementary DNA oligonucleotide that provides a recessed RNA 5′-end
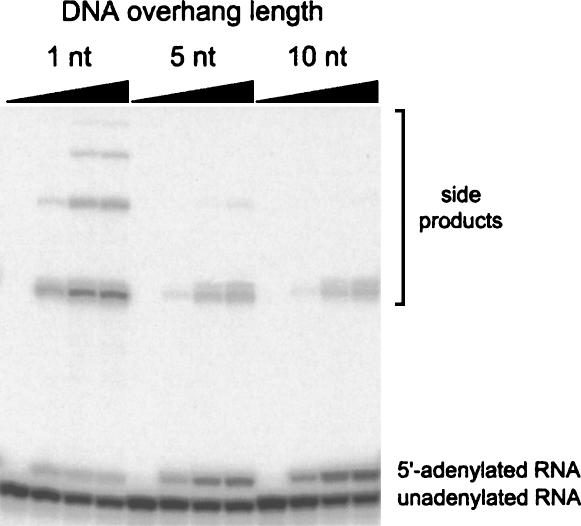
Our previously reported strategy for RNA 5′-adenylation using T4 RNA ligase subverts the second (ligation) step of the enzyme's natural mechanism by sequestering the 3′-terminus of the RNA substrate within an RNA:DNA duplex (Silverman 2004). However, the 5′-terminus of the RNA substrate remains single-stranded as a 5′-overhang, thereby permitting the first (adenylation) step of the mechanism (Fig. 1B). Because T4 DNA ligase instead requires a double-stranded substrate (Chiuman and Li 2002), the RNA adenylation substrate was rendered double-stranded by annealing it to a DNA oligonucleotide that creates a recessed RNA 5′-end (i.e., a DNA 3′-overhang; Fig. 1C). For T4 DNA ligase and a DNA substrate, this arrangement is sufficient to enable the adenylation step, but without an acceptor oligonucleotide also bound in the duplex, no further reaction occurs (Chiuman and Li 2002). Here we observe the same phenomenon for RNA 5′-adenylation with T4 DNA ligase, using 5′-phosphorylated 17-mer RNA substrate S1 as the test oligonucleotide. The length of the 3′-overhang on the complementary DNA oligonucleotide strongly affects the efficiency of adenylation; longer overhangs are better (Fig. 2). Because the rate and the yield were highest with 10 overhanging DNA nucleotides (of the lengths tested), we used DNA complements that provide a 10-nt DNA overhang for the remainder of our experiments. As a functional check on the structure of the 5′-AppRNA synthesized using T4 DNA ligase, adenylated RNA oligonucleotide 5′-App-S1 was shown to be fully competent for ligation to the 3′-terminus of a DNA strand, using an exactly complementary DNA oligonucleotide as a splint and T4 DNA ligase (∼95% ligation yield in <1 min without adding ATP; data not shown).
FIGURE 2.
Determining the DNA overhang requirement for RNA 5′-adenylation with T4 DNA ligase and ATP. These assays used RNA subtrate S1 (5′-GGAAGAGAUGGCGACGG-3′) and were performed in 40 mM Tris (pH 7.8), 10 mM MgCl2, 10 mM DTT, and 500 μM ATP at 37°C (5 μM RNA; t = 0 h, 2 h, 8 h, and 24 h). The yields of 5′-AppRNA for DNA overhang lengths of 1 nt, 5 nt, and 10 nt were 8%, 26%, and 37% under these conditions, indicating that the longer 10-nt overhang is preferred. A 20-nt DNA overhang led to approximately the same 5′-adenylation yield as a 10-nt overhang (data not shown). Additional studies defined optimal incubation conditions that differ in several ways from those used in this particular experiment (see text).
We optimized a number of additional variables for adenylation of S1: ATP concentration, reaction temperature, pH value, Mg2+ concentration, and enzyme concentration (data not shown). For ATP concentrations between 0.5 and 10 mM, the highest yield of adenylation product with the least amount of side products was achieved at the relatively high ATP concentration of 10 mM (Nilsson et al. 2001; higher concentrations were not tested). Of 16°C, 25°C, and 37°C, the highest ligation yield was observed at 25°C. Between pH 6.0 and 9.3, the optimal pH value was 7.0. Between 2 and 20 mM MgCl2, the optimal Mg2+ concentration was 10 mM. Finally, between 0.1 and 3 U/μL of T4 DNA ligase, the optimal enzyme concentration was 3 U/μL (higher concentrations were not tested).
Establishing generality of the approach using other RNA adenylation substrates
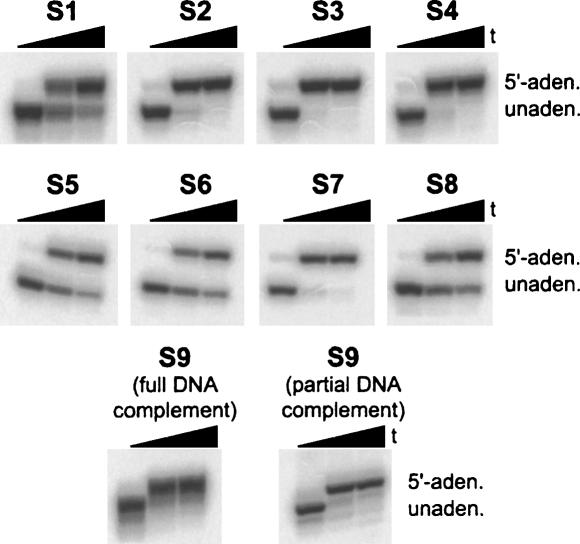
For general application of the new RNA 5′-adenylation strategy, a wide range of RNA substrates must be tolerated. As shown in Figure 3, generality was established using RNA oligonucleotide substrates S2–S9 (Silverman 2004). In each case, we used a complementary DNA oligonucleotide with a 10-nt overhang and the optimal reaction conditions determined above (40 mM MOPS at pH 7.0, 10 mM MgCl2, 10 mM DTT, and 10 mM ATP with 3 U/μL T4 DNA ligase at 25°C; Fig. 3). Oligonucleotides S2–S4 have the same sequence as S1 except with one of the other three nucleotides at the 5′-terminus; their successful adenylation demonstrates that all four RNA 5′-terminal nucleotides may be adenylated using T4 DNA ligase. Indeed, these assays showed that S1 was coincidentally adenylated less efficiently than the other three RNAs S2–S4. Substrates S5–S8 have considerably different sequence compositions and purine:pyrimidine ratios than S1, further establishing the generality of RNA 5′-adenylation with T4 DNA ligase.
FIGURE 3.
Establishing generality of the RNA 5′-adenylation strategy using RNA substrates S1–S9 and optimal reaction conditions of 40 mM MOPS (pH 7.0), 10 mM MgCl2, 10 mM DTT, and 10 mM ATP at 25°C (1 μM RNA; t = 0 h, 8 h, and 24 h). For S9, the full complement leaves a blunt RNA 3′-terminus, whereas the partial complement leaves 17 nt of RNA unpaired at the 3′-terminus. See Materials and Methods for RNA sequences. The gel images have been aligned vertically solely for presentation purposes. Unlike in the experiments of Figure 2, which were not performed under these optimal reaction conditions, essentially no side products were observed here (<2%; data not shown). We attribute this largely to the elevated ATP concentration (Nilsson et al. 2001).
Medium-length 34-mer RNA oligonucleotide S9 is sufficiently long such that a tightly bound 17-mer DNA oligonucleotide complementary to the 5′-end of S9 leaves the 3′-end of the RNA as a long (17-nt) single-stranded tail. For RNA adenylation using T4 RNA ligase, this arrangement led to rapid circularization immediately following adenylation, which necessitated the use of an additional complementary DNA oligonucleotide to block (sequester) the 3′-terminus (Silverman 2004). In contrast, here we found that RNA adenylation mediated by T4 DNA ligase does not require sequestration of the RNA 3′-terminus (Fig. 3). This substantially simplifies the planning and execution of the overall RNA 5′-adenylation strategy.
5′-Adenylation of a long and structured RNA substrate
We sought to 5′-adenylate a long, structured RNA substrate in addition to the short oligonucleotides S1–S8 and the medium-length S9. For this purpose we tested adenylation of P4–P6, a 160-nt independently folding domain of the Tetrahymena group I intron RNA (Murphy and Cech 1993; Cate et al. 1996; Juneau et al. 2001). The strategy of using a DNA oligonucleotide complementary to the 5′-terminus with a 10-nt DNA 3′-overhang was successful, as revealed by deoxyribozyme cleavage of the 5′-terminal portion of the RNA after adenylation (Fig. 4) In this experiment, it is notable that no special action was required with respect to the 3′-terminus of the P4–P6 RNA, which remained unmodified and unsequestered. This is in contrast to 5′-adenylation using T4 RNA ligase, for which the 3′-end of the long P4–P6 RNA substrate needed to be either covalently modified or otherwise blocked to avoid side reactions (Silverman 2004).
FIGURE 4.
5′-Adenylation of P4–P6, a long and structured RNA substrate. The 160-nt RNA substrate was 5′-32P-radiolabeled. Following 5′-adenylation (t = 0 h, 8 h, and 24 h using the optimal incubation conditions of Fig. 3), the 5′-terminal 14-nt portion of the RNA was cleaved using an 8–17 deoxyribozyme (see Materials and Methods), thereby allowing the 5′-adenylation to be detected by 20% PAGE. In all lanes, the band corresponding to the 146-nt 3′-portion of the P4–P6 RNA is present near the wells (not shown).
Preparative-scale RNA 5′-adenylation
Finally, we demonstrated that the T4 DNA ligase strategy can be applied for preparative-scale (nanomole) RNA 5′-adenylation. Using RNA substrates S1, S2, S6, and S7, 5′-adenylations were performed on the 1-nmol scale. The isolated yields of 5′-AppRNA after gel extraction and ethanol precipitation were 0.44–0.76 nmol (44%–76%). In all four cases, MALDI-MS confirmed the correct molecular weight within 0.1% for the 5′-AppRNA.
DISCUSSION
On the basis of an analogous approach to DNA 5′-adenylation using T4 DNA ligase (Chiuman and Li 2002) and our own procedure for RNA 5′-adenylation with T4 RNA ligase (Silverman 2004), here we describe a simple strategy for RNA 5′-adenylation using T4 DNA ligase, ATP, and a complementary DNA oligonucleotide (Fig. 1). The two methods for RNA and DNA 5′-adenylation differ considerably with respect to the required length of DNA 3′-overhang; a long 10-nt overhang is necessary for RNA adenylation (Fig. 2). Our new approach for RNA 5′-adenylation is experimentally much simpler than our previously reported approach that uses T4 RNA ligase for the same purpose (Silverman 2004). In particular, using T4 DNA ligase requires fewer variables to be optimized carefully because there are fewer concerns about secondary product formation. Furthermore, there is no need to sequester the 3′-terminus of RNA substrates that are longer than a short DNA complement (Figs. 3, 4), which is important in preparation of long 5′-AppRNAs (e.g., for in vitro selection experiments). Overall, the RNA 5′-adenylation strategy that uses T4 DNA ligase is currently our laboratory's favored approach for both small-scale reactions and for nanomole-scale preparation of 5′-AppRNA.
A disadvantage of our new 5′-adenylation approach is that T4 DNA ligase is relatively expensive in comparison with T4 RNA ligase. For example, a 1-nmol scale adenylation with T4 DNA ligase uses ∼$14 of commercial enzyme, whereas T4 RNA ligase procedure (Silverman 2004) uses as little as ∼$1.50 of enzyme. Fortunately, we have found that an overexpressed His6-tagged version of T4 DNA ligase successfully 5′-adenylates RNA with efficiency comparable to that of the commercial enzyme (data not shown). This indicates that large-scale RNA 5′-adenylation reactions are not prohibitively expensive using T4 DNA ligase.
A range of applications should be facilitated by the straightforward synthetic access to 5′-AppRNA that is described here. Our own in vitro selection efforts using 5′-triphosphate RNA substrates (Wang and Silverman 2003a; Purtha et al. 2005) can now be extended to RNAs for which 5′-triphosphates are difficult to prepare and 5′-adenylates serve as close functional surrogates (Hager and Szostak 1997; Coppins and Silverman 2004, 2005). Because 5′-adenylated RNA should be resistant to exonuclease degradation from the 5′-terminus, 5′-capping of RNA by adenylation should be a useful RNA modification, perhaps for in vivo applications. Because the α-phosphate of ATP is transferred to the adenylate portion of the 5′-AppRNA (Fig. 1), RNA 5′-adenylation using T4 DNA ligase and α-32P-ATP is an alternative approach to RNA 5′-radiolabeling that may be of practical utility. Finally, because the adenylate moiety has an ribose 2′,3′-diol group that may be oxidized with sodium periodate to aldehydes, RNA 5′-adenylation enables labeling of the 5′-terminus by subsequent reductive amination or other coupling reactions using a variety of amine reagents combined with biophysical probes (Wells and Cantor 1977; Odom et al. 1980; Proudnikov and Mirzabekov 1996). This overall 5′-labeling approach will be particularly useful when a method for RNA labeling is sought that does not require incorporation by solid-phase synthesis of special 5′-terminal RNA nucleotides and the RNA 3′-terminus cannot be used as a modification site, e.g., when it is blocked as a 2′,3′-cyclic phosphate.
MATERIALS AND METHODS
Oligonucleotide preparation and sequences
RNA substrate oligonucleotides S1–S9 each terminating in a 2′,3′-diol were prepared by solid-phase synthesis (Dharmacon, Inc. or HHMI-Keck Laboratory, Yale University). Their nucleotide sequences are as follows: S1, 5′-GGAAGAGAUGGCGACGG-3′; S2, S3, and S4 are the same as S1 except the first nucleotide was changed to A, U, and C, respectively; S5, 5′-AAUUGCGGGAAAG-3′; S6, 5′-CUUUCCCGCAAUUCC-3′; S7, 5′-UAAUACGACUCACUAUA-3′. S8, 5′-UCGUGCGGCUCACUGUA-3′; S9, 5′-UAAUACGACUCACUAUAGGAAGAGAUGGCGACGG-3′. Note that the sequence of S9 is the sequence of S7 plus 17 additional nt (which correspond to the sequence of S1). DNA oligonucleotides were obtained from IDT. All RNA and DNA oligonucleotides were purified by 20% denaturing PAGE followed by ethanol precipitation and quantified by absorbance (A260). The RNA substrates were 5′-phosphorylated with γ-32P-ATP (Perkin-Elmer) and T4 PNK (Fermentas) and purified by PAGE. For the preparative-scale adenylation reactions, the RNA substrates were 5′-phosphorylated in 2-nmol portions (1 mM ATP, 10 U T4 PNK from Fermentas, 1× PNK buffer, 50-μL reaction volume, 37°C, 2–4 h), purified by PAGE, and requantified by absorbance.
The DNA oligonucleotides for use with S1–S8 were complementary to the above RNA sequences and including the desired length of DNA 3′-overhang. The overhang sequence that provides up to 10 nt of 3′-overhang was all or part of … TATAGTGAGT-3′ except for S7 and S9, for which the overhang was all or part of … CGCGACAGAC-3′ (to avoid improper secondary structure formation with the RNA). The DNA oligonucleotide for 5′-adenylation of S9 was either fully complementary to S9 or complementary to only the S7 portion of the RNA sequence (5′-portion; 17 out of 34 nt), in both cases with a 10-nt DNA 3′-overhang.
The P4–P6 RNA (Silverman and Cech 1999) was prepared by T7 RNA polymerase transcription from a linearized plasmid DNA template. The transcription solution included excess GMP (10 mM GMP, 0.5 mM GTP) to provide >95% 5′-monophosphate RNA. The corresponding DNA oligo was a 60-mer complementary to nt 102–151 of P4–P6 with a 10-nt DNA 3′-overhang. The 8–17 deoxyribozyme and the procedure used to cleave P4–P6 for the experiment of Figure 4 were described previously (Silverman 2004).
RNA 5′-adenylation procedure
The optimal analytical-scale 5′-adenylation procedure (Fig. 3) was as follows: A sample containing 0.25 pmol of 5′-32P-radiolabeled RNA substrate, 10 pmol of unradiolabeled 5′-monophosphorylated RNA substrate, and 30 pmol of DNA oligonucleotide in 8 μL of 5 mM Tris (pH 7.5), 15 mM NaCl, and 0.1 mM EDTA was annealed by heating to 95°C for 3 min and cooling on ice for 5 min. The solution was brought to 10-μL total volume containing 40 mM MOPS (pH 7.0), 10 mM MgCl2, 10 mM DTT, 10 mM ATP, and 30 Weiss units of T4 DNA ligase (MBI Fermentas). Final concentrations were 1 μM RNA substrate, 3 μM DNA oligonucleotide, and 3 U/μL T4 DNA ligase. The sample was fully immersed in a 37°C water bath. At appropriate time points, 1.5-μL aliquots were quenched into 8 μL of stop solution (80% formamide, 1× TB, 50 mM EDTA, and 0.25% each bromophenol blue and xylene cyanol, where 1× TB contains 89 mM each Tris and boric acid at pH 8.3). Quenched aliquots were electrophoresed on 20% denaturing PAGE and imaged with a PhosphorImager (Figs. 2, 3). Use of the 8–17 deoxyribozyme to determine 5′-adenylation of the P4–P6 RNA (Fig. 4) was described previously (Silverman 2004).
For the preparative-scale 5′-adenylation reactions, 1 nmol of 5′-phosphorylated RNA was annealed in a 60-μL volume and incubated in 100-μL total volume (10 μM RNA) at 25°C for 24 h using the procedure described above. Samples were separated on 20% denaturing PAGE and extracted as described (Wang and Silverman 2003b). The yields of 5′-adenylated S1, S2, S6, and S7 were 0.58, 0.76, 0.44, and 0.71 nmol, which parallel the yields in the analytical-scale assay of Figure 3. Because of the 10-nt DNA 3′-overhang, the RNA and DNA were readily separated. In other preparative 5′-adenylation reactions using an RNA substrate synthesized by in vitro transcription from a double-stranded DNA template (Milligan et al. 1987), we have successfully used the antisense DNA template strand as the DNA oligonucleotide for 5′-adenylation (data not shown).
MALDI-TOF mass spectrometry of the 5′-adenylated RNA
MALDI-TOF mass spectrometry was used to confirm the identities of the 5′-adenylated RNAs synthesized on the 1-nmol scale. The observed (calculated) mass values for 5′-AppRNA were as follows: S1, 6017 (6017); S2, 6006 (6001); S6, 5014 (5018); S7, 5747 (5748). Mass spectra were obtained on an Applied Biosystems Voyager DE-STR instrument in the Mass Spectrometry Laboratory of the UIUC School of Chemical Sciences.
ACKNOWLEDGMENTS
This research was supported by the Burroughs Wellcome Fund (New Investigator Award in the Basic Pharmacological Sciences), the March of Dimes Birth Defects Foundation (5-FY02-271), the NIH (GM-65966), and the UIUC Department of Chemistry. Acknowledgement is made to the donors of The Petroleum Research Fund, administered by the American Chemical Society, for partial support of this research (38803-G4). S.K.S. is the recipient of a fellowship from The David and Lucile Packard Foundation. We thank Lisa Cooper, James Heeres, and Chao Quan for technical assistance during the initial phase of this study.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.33106.
REFERENCES
- Cate J.H., Gooding A.R., Podell E., Zhou K., Golden B.L., Kundrot C.E., Cech T.R., Doudna J.A. Crystal structure of a group I ribozyme domain: Principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- Chiuman W., Li Y. Making AppDNA using T4 DNA ligase. Bioorg. Chem. 2002;30:332–349. doi: 10.1016/s0045-2068(02)00018-4. [DOI] [PubMed] [Google Scholar]
- Coppins R.L., Silverman S.K. A DNA enzyme that mimics the first step of RNA splicing. Nat. Struct. Mol. Biol. 2004;11:270–274. doi: 10.1038/nsmb727. [DOI] [PubMed] [Google Scholar]
- Coppins R.L., Silverman S.K. A deoxyribozyme that forms a three-helix-junction complex with its RNA substrates and has general RNA branch-forming activity. J. Am. Chem. Soc. 2005;127:2900–2907. doi: 10.1021/ja044881b. [DOI] [PubMed] [Google Scholar]
- Hager A.J., Szostak J.W. Isolation of novel ribozymes that ligate AMP-activated RNA substrates. Chem. Biol. 1997;4:607–617. doi: 10.1016/s1074-5521(97)90246-5. [DOI] [PubMed] [Google Scholar]
- Juneau K., Podell E., Harrington D.J., Cech T.R. Structural basis of the enhanced stability of a mutant ribozyme domain and a detailed view of RNA–solvent interactions. Structure. 2001;9:221–231. doi: 10.1016/s0969-2126(01)00579-2. [DOI] [PubMed] [Google Scholar]
- Milligan J.F., Groebe D.R., Witherell G.W., Uhlenbeck O.C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy F.L., Cech T.R. An independently folding domain of RNA tertiary structure within the Tetrahymena ribozyme. Biochemistry. 1993;32:5291–5300. doi: 10.1021/bi00071a003. [DOI] [PubMed] [Google Scholar]
- Nilsson M., Antson D.O., Barbany G., Landegren U. RNA-templated DNA ligation for transcript analysis. Nucleic Acids Res. 2001;29:578–581. doi: 10.1093/nar/29.2.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom O.W., Jr., Robbins D.J., Lynch J., Dottavio-Martin D., Kramer G., Hardesty B. Distances between 3′ ends of ribosomal ribonucleic acids reassembled into Escherichia coli ribosomes. Biochemistry. 1980;19:5947–5954. doi: 10.1021/bi00567a001. [DOI] [PubMed] [Google Scholar]
- Proudnikov D., Mirzabekov A. Chemical methods of DNA and RNA fluorescent labeling. Nucleic Acids Res. 1996;24:4535–4542. doi: 10.1093/nar/24.22.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purtha W.E., Coppins R.L., Smalley M.K., Silverman S.K. General deoxyribozyme-catalyzed synthesis of native 3′–5′ RNA linkages. J. Am. Chem. Soc. 2005;127:13124–13125. doi: 10.1021/ja0533702. [DOI] [PubMed] [Google Scholar]
- Silverman S.K. Practical and general synthesis of 5′-adenylated RNA (5′-AppRNA) RNA. 2004;10:731–746. doi: 10.1261/rna.5247704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman S.K., Cech T.R. Energetics and cooperativity of tertiary hydrogen bonds in RNA structure. Biochemistry. 1999;38:8691–8702. doi: 10.1021/bi9906118. [DOI] [PubMed] [Google Scholar]
- Wang Y., Silverman S.K. Deoxyribozymes that synthesize branched and lariat RNA. J. Am. Chem. Soc. 2003a;125:6880–6881. doi: 10.1021/ja035150z. [DOI] [PubMed] [Google Scholar]
- Wang Y., Silverman S.K. Characterization of deoxyribozymes that synthesize branched RNA. Biochemistry. 2003b;42:15252–15263. doi: 10.1021/bi0355847. [DOI] [PubMed] [Google Scholar]
- Wells B.D., Cantor C.R. A strong ethidium binding site in the acceptor stem of most or all transfer RNAs. Nucleic Acids Res. 1977;4:1667–1680. doi: 10.1093/nar/4.5.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]