Abstract
Long (27–29-bp dsRNA) Dicer-dependent substrates have been identified as potent mediators of RNAi-induced gene knockdown in HEK293 and HeLa cells. As the lengths of these molecules are reported to be below the threshold generally regarded as necessary for induction of the mammalian interferon (IFN) response, these long siRNA are being considered as RNAi substrates in both research and therapeutic settings. In this report, we demonstrate that >23-bp dsRNA can influence cell viability and induce a potent IFN response (highlighted by a strong up-regulation of the dsRNA receptor, Toll-like receptor 3) in a cell type-specific manner. This finding suggests that the length threshold for siRNA induction of the IFN response is not fixed but instead varies significantly among different cell types. Given the diversity of cell types that comprise whole organisms, these findings suggest great care should be taken when considering length variations of dsRNA molecules for RNAi experimentation, especially in therapeutic applications.
Keywords: siRNA, duplex length, interferon response, long dsRNA
INTRODUCTION
There is significant debate over the ability of siRNAs and longer dsRNAs to induce the innate immune response (Manche et al. 1992; Sledz et al. 2003; Kim et al. 2004). Though mammalian cells are exquisitely sensitive to the introduction of dsRNA, molecules less than 30 bp in length are generally believed to avoid induction of interferon (IFN) pathways (Manche et al. 1992; Kariko et al. 2004; Kim et al. 2004; Siolas et al. 2004). Recent studies have also suggested that long (27–29 bp) dsRNAs and shRNAs that enter the RNA interference (RNAi) pathway in a Dicer-dependent fashion provide more efficient gene silencing than shorter, Dicer-independent substrates (Kim et al. 2004; Siolas et al. 2004). These studies (which demonstrated potent and specific gene silencing by longer duplexes and hairpins) were performed in tumor cell lines that are often characterized by a host of genetic abnormalities including defects in the IFN response pathway (Stojdl et al. 2000; Khabar et al. 2004; Zughaier et al. 2005). In this work we have systematically examined the effects of dsRNA length on gene silencing, cell viability, and induction of the innate immune response in a range of cell lines. Our results show that different cell types manifest drastically different cell viability and IFN responses to long siRNAs, suggesting that the dsRNA length threshold varies considerably among cell types.
RESULTS
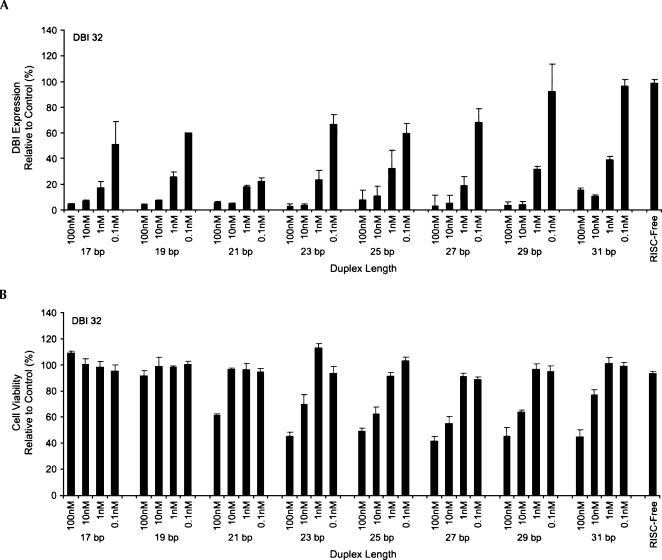
Initially, a panel of 24 dsRNAs having duplex lengths between 17 and 31 bp and targeting three different sites in the diazepam binding inhibitor gene (DBI; DBI 32, 34, and 35) was transfected into HeLa S3 cells at concentrations ranging from 0.1 to 100 nM. In all three cases, dsRNAs longer than 23 bp induced 20%–60% cell death at concentrations between 10 and 100 nM while shorter duplexes had no effect on cell viability (Fig. 1A,B; Supplementary Fig. 1A–D at http://www.dharmacon.com/tech/publications). The cell death phenotype was comparable for two different lipid delivery reagents (DharmaFECT 4 and Lipofectamine 2000) (Supplementary Fig. 1E at http://www.dharmacon.com/tech/publications). In addition, the effect on cell viability was target silencing-independent (shorter duplexes induced comparable levels of gene knockdown at equivalent or higher concentrations without cell death), a conclusion that was confirmed in a separate study that targeted three additional genes (EGFP, hnRNP H, and La) using sequences derived from the study by Kim et al. (2004; Supplementary Fig. 2A at http://www.dharmacon.com/tech/publications). It is worth noting that in two of the three cases studied, longer (>23 bp) duplexes exhibited greater potency than siRNAs (19 bp) at 1 nM concentrations, thus supporting previous claims by Kim et al. (2004) and Siolas et al. (2004) describing the heightened potency of longer molecules (Fig. 1A; Supplementary Fig. 1A,C at http://www.dharmacon.com/tech/publications). In the third case, target silencing by long and short duplexes was indistinguishable.
FIGURE 1.
Duplexes greater than 23 bp in length induce toxicity in HeLa S3 cells. Bar graph showing the level of DBI silencing (A) and viability of HeLa S3 cells (B) transfected with DBI 32 dsRNA of varying duplex lengths (17–31 bp) at concentrations ranging from 0.1 to 100 nM. Experiments were performed in triplicate. Error bars represent standard deviations from the mean.
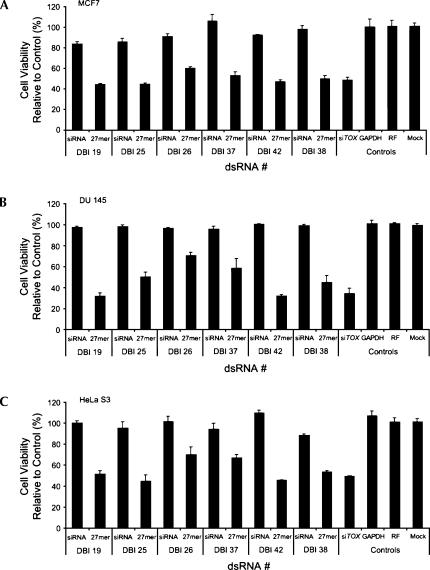
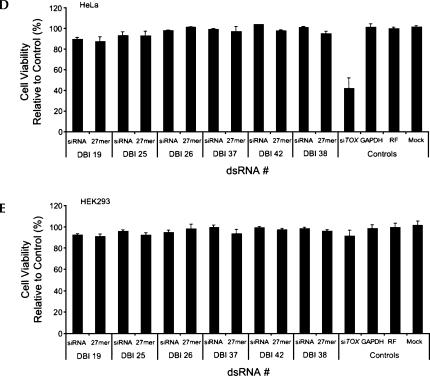
A side-by-side comparison between short (19 bp) and long (27 bp) dsRNA in DU 145, HEK293, HeLa, HeLa S3, and MCF7 cells identified varying degrees of sensitivity to dsRNA length. Transfection of long duplexes targeting DBI consistently induced 30%–60% cell death in DU 145, HeLa S3, and MCF7 cells (defined as “sensitive,” 100 nM) (Fig. 2A–C; Supplementary Fig. 2A at http://www.dharmacon.com/tech/publications). In contrast, the same duplexes failed to induce cell death in HeLa and HEK293 cells (defined as “insensitive”) (Fig. 2D,E; Supplementary Fig. 2B at http://www.dharmacon.com/tech/publications). These findings corroborate previous observations made by Kim et al. (2004) and Siolas et al. (2004) and imply that while long siRNA can induce potent and specific gene silencing without cell death in HeLa and HEK293 cells, this principle cannot be generalized. Nineteen base pair siRNAs did not induce significant changes in cell viability in any of the cell lines tested, indicating that the observed cell viability phenotype is target-independent, but siRNA length- and cell type–dependent.
FIGURE 2.
(Continued on next page)
FIGURE 2.
Nonspecific dsRNA-mediated cytotoxicity induced by long siRNA is unique to specific cell types. The effect of six different long (27 bp) dsRNAs and siRNAs (19 bp) targeting DBI on cell viability in MCF7 (A), DU 145 (B), HeLa S3 (C), HeLa (D), and HEK293 (E) cells at 100 nM concentrations. Experiments were all performed in triplicate. Error bars represent standard deviations from the mean. RF represents RISC-free control.
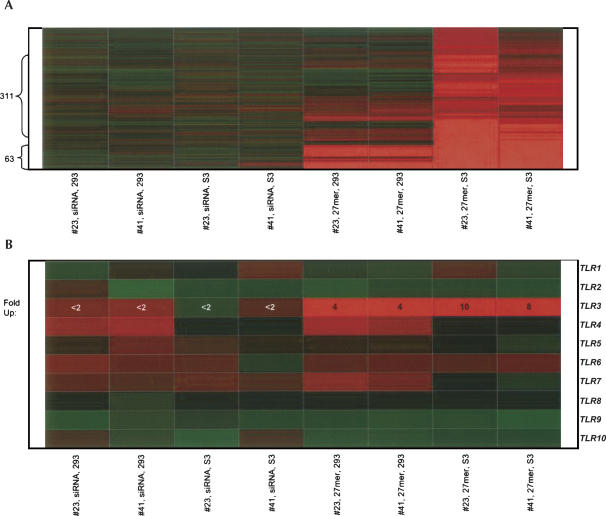
Microarray analysis performed on “sensitive” (HeLa S3) and “insensitive” (HEK293) cell lines transfected with siRNA (19 bp) and long dsRNA (27 bp) offers a possible explanation underlying the dsRNA-induced reduction in cell viability. Specifically, gene expression signature analysis revealed that while both cell types responded to the presence of long dsRNA by inducing expression of genes associated with the innate immune reaction, the response by the sensitive cell line was more widespread than that observed in the insensitive cell line. Introduction of long duplexes into HEK293 (insensitive) cells (100 nM) led to a greater than twofold induction of 63 genes, 34 of which were interferons or IFN-inducible genes (Fig. 3A; Supplementary Table 1 at http://www.dharmacon.com/tech/publications). This finding reveals that long siRNAs do in fact induce an IFN-like gene expression profile in cell lines that otherwise exhibit little or no overt signs of cell stress (e.g., cell death). Transfection of long (27 bp) siRNA into the sensitive cell line, HeLa S3, induced expression of this original cluster (Fig. 3A) plus an additional set of >300 genes that (again) included transcripts associated with IFN and apoptotic signaling pathways. The IFN induction was concentration-dependent with varying levels of the innate immune signature being clearly present at both 10 and 100 nM (Supplementary Fig. 3, Table 2 at http://www.dharmacon.com/tech/publications). As (1) no IFN-like expression profile was observed at 1 nM and (2) a fraction of the long siRNA retained functionality at this concentration, these results indicate that, under in vitro conditions, a narrow range of concentrations can be identified where potent and nontoxic gene silencing can be achieved with a subset of long (>23 bp) siRNA. In all cases tested, a 10-fold increase over the minimal functional concentration elicited an induction of genes possibly associated with IFN response and a reduction in cell viability. In contrast, siRNAs (19 bp) did not alter cell viability or induce the IFN-like gene expression profile, even at concentrations as high as 100-fold over the minimal functional concentration (Fig. 3A; 100 nM, Supplementary Fig. 3 at http://www.dharmacon.com/tech/publications). Thus, under conditions where concentrations cannot be controlled precisely and multiple cell types with varying dsRNA length sensitivities exist (e.g., in vivo), induction of nonspecific responses by long siRNAs is likely unavoidable.
FIGURE 3.
Gene expression signatures identify length sensitive induction of genes associated with the innate immune response. (A) Heat map of genes induced in HEK293 (insensitive cell line, lanes 1,2) and HeLa S3 (sensitive cell line, lanes 3,4) cells transfected with one of two 19-bp siRNA (#23 or #41) targeting DBI. Lanes 5–8 represent HEK293 (lanes 5,6) and HeLa S3 cells (lanes 7,8) transfected with long (27 bp) RNA duplexes (comparable to siRNA #23 and #41). The Y-axis represents the number of genes that are up-regulated (red) by twofold or more. (B) Heat map of Toll-like receptors (TLR)1–10 in insensitive (HEK293) and sensitive (HeLa S3) cell lines transfected with long (27 bp) dsRNA and short (19 bp) siRNA.
The expression of two prominent genes associated with dsRNA induction of the innate immune response, Toll-like receptor 3 (TLR3) and the RNA-dependent protein kinase (PRKR/EIF2AK2), are strongly correlated with the phenotypes observed in sensitive and insensitive cell lines. Long dsRNAs induced a two- and a fourfold increase in PRKR expression in HEK293 and HeLa S3 cells, respectively (100 nM; data not shown). Similarly, while expression levels of TLRs 1, 2, 4, 5, 6, 7, 8, 9, and 10 remained constant after transfection of long siRNA, 27-bp dsRNA induced fourfold and 8- to 10-fold changes in TLR3 expression in insensitive and sensitive cells, respectively (100 nM, Fig. 3B; 10 nM, Supplementary Fig. 4 at http://www.dharmacon.com/tech/publications). None of the documented PRKR or TLR3 responses were observed when siRNA (19 bp) were transfected into either cell line at 100 nM concentrations.
DISCUSSION
The work presented above demonstrates that sensitivity to long siRNA (as measured by changes in viability and IFN induction) is cell type-dependent. Though our results in insensitive cell lines concur with previous studies by Kim et al. (2004) and Siolas et al. (2004), investigation of an expanded collection of cell types shows that the conclusions generated from studies on HEK293 and HeLa are not broadly applicable. While these studies failed to identify expression profiles consistent with an IFN response using shorter siRNA (e.g., 19 bp), it is conceivable that under particular sets of conditions (i.e., unique sets of cell lines and delivery reagents) small duplexes might also generate an IFN response. That said, these studies demonstrate that minimal structural variations in dsRNA length can result in drastic increases in cellular stress and suggest that the utility of long (>23 bp) siRNAs in basic research and/or therapeutic applications (where multiple cell types must be considered) may be limited.
None of the sequences employed in this study contain the recently reported GUCCUUCAA (Hornung et al. 2005) or GU-rich (Judge et al. 2005) motifs that are purported to induce the IFN pathway (possibly through TLR7). Thus, our results intimate the presence of an exquisitely sensitive, differentially regulated, length-dependent sensor of dsRNA. One possible candidate for such a detector is the highly up-regulated TLR3 gene that together with TLR7 and/or TLR8 has been implicated in the IFN response to viral and synthetic RNAs (Alexopoulou et al. 2001; Takeda et al. 2003; Beutler 2004). Previous work by Kariko et al. (2004) showed that the degree of IFN induction (specifically IL-8 and IFN-β) by siRNA could be modulated by the level of TLR3 expression. Here we provide further support for the involvement of TLR3 in dsRNA-mediated IFN induction by showing that TLR3 expression is modulated by the presence of dsRNA and that the degree of up-regulation is dependent upon both the length of the transfected dsRNA and the cell type under investigation. Together with preliminary studies that show pretreatment of the HeLa S3 cells with bafilomycin (an inhibitor of endosomal maturation and TLR signal transduction) eliminates cell death associated with 27-bp duplexes (see Supplementary Fig. 5 at http://www.dharmacon.com/tech/publications), these data suggest a central role for TLR3 in the contrasting phenotypic outcomes observed in sensitive and insensitive cells.
MATERIALS AND METHODS
RNAs, genes, and cell lines
Double-stranded RNAs (dsRNAs) targeting the diazepam binding inhibitor (DBI, NM_020548) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH, NM_002046), and all other genes were synthesized using 2′-ACE chemistry (Scaringe 2000) at Dharmacon, Inc. For the experiments showing dsRNAs of different lengths, the dsRNA molecules contained a duplex region (e.g., 19, 21, 23, 25, 27, 29, or 31 bp) and two nucleotide 3′ overhangs on their sense strands. The duplexes were designed by extending sequence from the 5′ end of the sense strand, thus preserving the thermodynamic stability and targeting region (or motif) of the 5′ AS end of the molecule. For all remaining experiments, siRNAs were 19 bp with two nucleotide 3′ overhangs on both strands, and the 27-mer duplexes contained 27 bp and were blunt-ended. Sequences targeting DBI, GAPDH, and additional genes (e.g., EGFP, U55762; hnRNP H, L22009; and La, NM_003142.2) are provided in Supplementary Table 3 at http://www.dharmacon.com/tech/publications.
All five cell lines used in this study—DU 145, HEK293, HeLa, HeLa S3, and MCF7—were obtained from ATCC and cultured under prescribed media conditions at 37°C, 5% CO2.
Transfection, quantifying gene silencing, and assessing toxicity
Cells were plated in 96-well plates (1–2 × 104 cells/well) for 18–24 h before transfection. On the day of transfection, RNA-lipid complexes were introduced into each well of cells (0.1–100 nM RNA, for HEK293 and DU 145, 0.4 μL/well DharmaFECT 1; for HeLa, 0.2 μL/well DharmaFECT 1; for MCF7, 0.2 μL/well DharmaFECT 4; for HeLa S3, 0.4 μL/well DharmaFECT 4). Twenty-four to 72 h post-transfection, the level of target knockdown was assessed using a branched DNA assay (Genospectra) specific for the targets of interest. Cell viability was determined using an alamarBlue assay (BioSource Intl, Inc.) according to manufacturer's instructions. Where indicated, “siTOX,” representing a toxic duplex (siCONTROL TOX Non Targeting siRNA), is used as an indicator of the success of transfection. “GAPDH” represents a siRNA (siCONTROL GAPDH siRNA) targeting the GAPDH housekeeping gene. “RISC Free” represents a nontargeting duplex (RISC-Free siRNA control) carrying chemical modifications that prevent interaction with the RNA Induced Silencing Complex. “Mock” represents lipid-only treated cells.
Gene profiling experiments
HEK293 and HeLa S3 cells were plated in 96-well plates (1 × 104 cells/well) and cultured for 24 h (37°C, 5% CO2). All duplexes were transfected at 100, 10, or 1 nM using DharmaFECT 1 (HEK293 cells, 0.4 μL/well) or DharmaFECT 4 (HeLa S3 cells, 0.4 μL/well) siRNA transfection reagents. Total RNA purification was performed using RNeasy columns with on-column DNase digestion (Qiagen). RNA integrity was analyzed with the RNA 6000 Nano LabChip on a 2100 Bioanalyzer (Agilent).
For each sample, RNA was amplified and Cy3- or Cy5-labeled using a Low Input RNA Fluorescent Linear Amplification Kit (Agilent). Hybridizations were performed using Human 1A (V2) Oligo Microarrays (Agilent) with Cy3-labeled reference RNA derived from mock- or untransfected cells. Arrays were scanned on a Model G2505B Scanner (Agilent), and the raw image was processed using Feature Extraction (v7.5.1), Spotfire Decision Site 8.1, and the Spotfire Functional Genomics Modules. A cutoff was applied (the log of combined Cy3 and Cy5 channel signal >2.8) to eliminate noise associated with low signal data points. In addition, a twofold cutoff (log ratio of >0.3 or < −0.3) was applied to up- or down-regulated genes used in comparative analysis. TLR3 was not identified by these procedures due to the low level of TLR3 expression in HeLa S3 and HEK293 cells. To assess the effects of long and short siRNA on TLRs 1–10, the combined channel cutoff was disregarded.
SUPPLEMENTAL DATA
Supplemental materials are available online at http://www.dharmacon.com/tech/publications/.
ACKNOWLEDGMENTS
We thank Queta Boese for helpful comments on the manuscript. Julia Kendall and Cara Kotas were instrumental in manuscript preparation. Special thanks to the Dharmacon Production Team for siRNA and RNA synthesis.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2340906.
REFERENCES
- Alexopoulou L., Holt A., Medzhitov R., Flavell R. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- Beutler B. Inferences, questions and possibilities in Toll-like receptor signalling. Nature. 2004;430:257–263. doi: 10.1038/nature02761. [DOI] [PubMed] [Google Scholar]
- Hornung V., Guenthner-Biller M., Bourquin C., Ablasser A., Schlee M., Uematsu S., Noronha A., Manoharan M., Akira S., de Fougerolles A., et al. Sequence-specific potent induction of IFN-α by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat. Med. 2005;11:263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- Judge A.D., Sood V., Shaw J.R., Fang D., McClintock K., MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat. Biotechnol. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- Kariko K., Bhuyan P., Capodici J., Weissman D. Small interfering RNAs mediate sequence-independent gene suppression and induce immune activation by signaling through Toll-like receptor 3. J. Immunol. 2004;172:6545–6549. doi: 10.4049/jimmunol.172.11.6545. [DOI] [PubMed] [Google Scholar]
- Khabar K., Al-Haj L., Al-Zoghaibi F., Marie M., Dhalla M., Polyak S., Williams B. Expressed gene clusters associated with cellular sensitivity and resistance towards anti-viral and anti-proliferative actions of interferon. J. Mol. Biol. 2004;342:833–846. doi: 10.1016/j.jmb.2004.07.065. [DOI] [PubMed] [Google Scholar]
- Kim D.-H., Behlke M.A., Rose S.D., Chang M.-S., Choi S., Rossi J.J. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat. Biotechnol. 2004;23:222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- Manche L., Green S.R., Schmedt C., Mathews M.B. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol. Cell. Biol. 1992;12:5238–5248. doi: 10.1128/mcb.12.11.5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaringe S.A. Advanced 5′-silyl-2′-orthoester approach to RNA oligonucleotide synthesis. Methods Enzymol. 2000;317:3–18. doi: 10.1016/s0076-6879(00)17003-x. [DOI] [PubMed] [Google Scholar]
- Siolas D., Lerner C., Burchard J., Ge W., Linsley P.S., Paddison P.J., Hannon G.J., Cleary M.A. Synthetic shRNAs as potent RNAi triggers. Nat. Biotechnol. 2004;23:227–231. doi: 10.1038/nbt1052. [DOI] [PubMed] [Google Scholar]
- Sledz C., Holko M., de Veer M., Silverman R., Williams B. Activation of the interferon system by short-interfering RNAs. Nat. Cell Biol. 2003;5:834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- Stojdl D., Lichty B., Knowles S., Marius R., Atkins H., Sonenberg N., Bell J. Exploiting tumor-specific defects in the interferon pathway with a previously unknown oncolytic virus. Nat. Med. 2000;6:821–825. doi: 10.1038/77558. [DOI] [PubMed] [Google Scholar]
- Takeda K., Kaisho T., Akira S. Toll-like receptors. Annu. Rev. Immunol. 2003;21:335–376. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- Zughaier S.M., Zimmer S.M., Datta A., Carlson R.W., Stephens D.S. Differential induction of the Toll-like receptor 4-MyD88-dependent and -independent signaling pathways by endotoxins. Infect. Immun. 2005;73:2940–2950. doi: 10.1128/IAI.73.5.2940-2950.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]