Abstract
RNase P and RNase MRP are ribonucleoprotein enzymes required for 5′-end maturation of precursor tRNAs (pre-tRNAs) and processing of precursor ribosomal RNAs, respectively. In yeast, RNase P and MRP holoenzymes have eight protein subunits in common, with Pop1p being the largest at >100 kDa. Little is known about the functions of Pop1p, beyond the fact that it binds specifically to the RNase P RNA subunit, RPR1 RNA. In this study, we refined the previous Pop1 phylogenetic sequence alignment and found four conserved regions. Highly conserved amino acids in yeast Pop1p were mutagenized by randomization and conditionally defective mutations were obtained. Effects of the Pop1p mutations on pre-tRNA processing, pre-rRNA processing, and stability of the RNA subunits of RNase P and MRP were examined. In most cases, functional defects in RNase P and RNase MRP in vivo were consistent with assembly defects of the holoenzymes, although moderate kinetic defects in RNase P were also observed. Most mutations affected both pre-tRNA and pre-rRNA processing, but a few mutations preferentially interfered with only RNase P or only RNase MRP. In addition, one temperature-sensitive mutation had no effect on either tRNA or rRNA processing, consistent with an additional role for RNase P, RNase MRP, or Pop1p in some other form. This study shows that the Pop1p subunit plays multiple roles in the assembly and function of of RNases P and MRP, and that the functions can be differentiated through the mutations in conserved residues.
Keywords: site-directed random mutagenesis, pre-tRNA processing, 5.8S rRNA processing, ribonucleoprotein complex, steady-state activity
INTRODUCTION
Ribonuclease P (RNase P) is a ubiquitous endonuclease that removes 5′ leader sequences from precursor tRNAs (pre-tRNAs). RNase P activity has been found in Bacteria, Archaea, Eukarya, and organelles (Frank and Pace 1998; Xiao et al. 2002) and most forms of RNase P are ribonucleoprotein complexes, containing both protein and RNA subunits. Bacterial RNase P consists of an essential RNA subunit (∼130 kDa) and a small, basic protein subunit (∼15 kDa) (Frank and Pace 1998). Although both subunits are required for the in vivo RNase P function, the bacterial RNA subunit alone can catalyze pre-tRNA cleavage at high salt concentrations in vitro, and therefore is an RNA enzyme or “ribozyme” (Guerrier-Takada et al. 1983). Eukaryotic RNase P has a very similar RNA moiety, but the protein complement is far more complex than that of the bacterial enzyme. Nine- and 10-protein subunits have been identified in the nuclear RNase P from yeast and humans, respectively (Chamberlain et al. 1998; Jarrous 2002). The eukaryotic RNase P RNA appears to have lost the ability to cleave pre-tRNAs in the absence of the protein subunits.
In the yeast Saccharomyces cerevisiae, nuclear RNase P contains one RNA subunit, RPR1 RNA, and nine protein subunits: Pop1p, Pop3p, Pop4p, Pop5p, Pop6p, Pop7p, Pop8p, Rpp1p, and Rpr2p (Lee et al. 1991b; Chamberlain et al. 1998). All 10 subunits are essential for yeast growth and the in vivo RNase P activity (Chamberlain et al. 1998 and references therein). RPR1 RNA is synthesized as a precursor (pre-RPR1 RNA) that contains an 84-nucleotide (nt) 5′-leader sequence and a 3′-trailing sequence flanking the mature RPR1 sequence (Lee et al. 1991a). Pre-RPR1 RNA is assembled into a ribonucleoprotein complex with at least seven of the nine RNase P protein subunits before being processed to the mature form (Srisawat et al. 2002).
Eukaryotic RNase P is closely related to another ribonucleoprotein enzyme, RNase MRP, which has only been found in eukaryotes and appears to have evolved from RNase P to perform specialized functions. The demonstrated nuclear function of yeast RNase MRP is pre-rRNA in the maturation pathway of 5.8S rRNA (Lindahl and Zengel 1995; Reilly and Schmitt 1995; Tollervey 1995). Recently, an additional role for RNase MRP in cell cycle progression in yeast through selective mRNA turnover has been reported (Gill et al. 2004). The yeast RNase MRP holoenzyme consists of one RNA subunit, NME1 RNA, and at least 10 protein subunits (Schmitt and Clayton 1994; Chamberlain et al. 1998; Salinas et al. 2005). Eight proteins are the same as those in yeast RNase P: Pop1p, Pop3p, Pop4p, Pop5p, Pop6p, Pop7p, Pop8p, and Rpp1p (Chamberlain et al. 1998 and references therein). The protein subunit unique to RNase P is the Rpr2p, whereas Snm1p and Rmp1p are specific to RNase MRP (Schmitt and Clayton 1994; Chamberlain et al. 1998; Salinas et al. 2005). NME1 RNA and the RPR1 RNA conform to similar secondary structures and have several small patches of conserved sequence found in all RNase P RNAs (Forster and Altman 1990; Frank et al. 2000; Li et al. 2002). Thus, RNase MRP arose from eukaryotic nuclear RNase P through sequence divergence of a duplicated RNA subunit gene and substitution of one protein subunit with two others.
Functions of the individual protein subunits of yeast RNase P and RNase MRP are largely unknown, although several protein subunits can bind RNA. For example, Pop1p and Pop4p bind directly to the RPR1 RNA (Ziehler et al. 2001; Houser-Scott et al. 2002), whereas Snm1p can bind to the NME1 RNA directly (Schmitt and Clayton 1994). A separate study reported that Pop3p could bind to the RPR1 RNA, pre-tRNA, and single-stranded RNAs (Brusca et al. 2001), although the specificity of these interactions is not known. Recently, crystal structures and NMR structures of the archaeal homologs of Pop4p, Rpp1p, and Rpr2p have been solved (Boomershine et al. 2003; Sidote and Hoffman 2003; Numata et al. 2004; Sidote et al. 2004; Takagi et al. 2004; Kakuta et al. 2005), but structural information for the other protein subunits is not available. Information concerning the location of the various subunits in the holoenzyme complex comes from coprecipitation experiments and two-hybrid and three-hybrid studies (Jiang and Altman 2001; Houser-Scott et al. 2002; Hall and Brown 2004; Welting et al. 2004; Kifusa et al. 2005).
We mutagenized Pop1p (100 kDa), the largest common protein subunit of yeast RNase P and MRP, to investigate its contributions to RNases P and MRP. Pop1p is important for the assembly and cellular functions of RNase P and MRP (Lygerou et al. 1994; Srisawat et al. 2002), and is conserved from yeast to human (Lygerou et al. 1996b). Attempts to characterize the function of Pop1p in vitro have not been successful, largely because the individually expressed protein is not soluble and the yeast holoenzyme has not been reconstituted in vitro. Therefore, we mutagenized the evolutionarily conserved regions of Pop1p and examined the effects of the mutations on the in vivo functions and biogenesis of RNase P and RNase MRP. Alignment of the Pop1p sequences from yeast, worm, and human previously revealed three highly conserved regions (Lygerou et al. 1996b). In this study, we have found four conserved regions in the Pop1p family, including the three regions identified previously, by amino acid sequence alignment of additional putative Pop1p homologs. Mutated POP1 gene libraries were then created, in which highly conserved amino acids in the four conserved regions were randomized. Selections for viable sequence variations in POP1 were followed by screens for conditionally defective mutations. Subsequent characterization of the partially defective POP1 mutations showed that most defective mutations affect the assembly and in vivo functions of RNases P and MRP to various degrees. A few mutations that impair only RNase P function or only RNase MRP function were obtained, suggesting that Pop1p might participate differentially in the two enzymes. The collection of Pop1p mutations with distinct phenotypes is anticipated to be useful to further understand the biology of RNase P/MRP and RNA processing in the cell.
RESULTS
Alignment of Pop1p and homologs
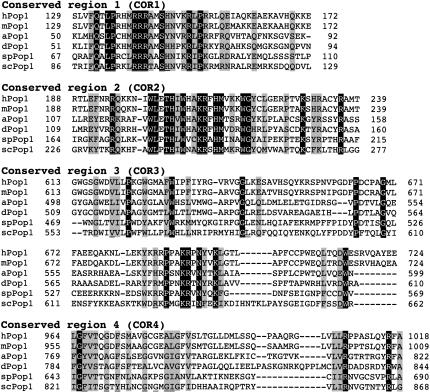
To identify putative homologs of the yeast Pop1p from other eukaryotes, we performed a BLAST search of the NCBI database with the sequence of Pop1p (http://www.ncbi.nlm.nih.gov/BLAST). The search revealed putative Pop1p homologs from Drosophila (NCBI accession no. NP_572236), Anopheles (NCBI accession no. XP_307517), mouse (NCBI accession no. NP_080616), and Schizosaccharomyces pombe (NCBI accession no. CAB61782). Sequences of the putative homologs were aligned with those of Pop1p from S. cerevisiae and human, using the algorithm Clustal W (http://us.expasy.org) with moderate manual adjustment (Fig. 1). The alignment confirmed the presence of three highly conserved regions identified previously (Lygerou et al. 1996b), COR1, -2, and -4, as well as an additional conserved sequence, COR3. Sequence conservation of the Pop1p family outside of the four conserved regions is low, with only 4% sequence identity.
FIGURE 1.
Alignment of the four conserved regions (COR1–4) of Pop1p from S. cerevisiae (scPop1p) and its homologs from S. pombe (spPop1), Drosophila (dPop1), Anopheles (aPop1), mouse (mPop1), and human (hPop1). The sequences were aligned with the Clustal W algorithm. Identical amino acids are highlighted in black with white letters, whereas similar amino acids that occupy homologous positions are highlighted in gray. Less conserved positions that were also mutated are underlined. Positions of the amino acids in each Pop1 protein are indicated with numbers flanking the corresponding sequence. The alignment has confirmed the previously identified COR1, -2, and -4 (Lygerou et al. 1996b), and uncovered another conserved region, COR3.
In COR1, several Arg amino acids are conserved among the Pop1p family (positions 97, 98, 99, 107 in Fig. 1; Lygerou et al. 1996b) while COR2 is rich in Trp residues and basic amino acids. The basic and aromatic conserved amino acids in COR1 and COR2 could potentially contribute to the RNA binding by Pop1p, and these regions have been designated the “Pop1 domain,” signature sequences of the Pop1 family (Marchler-Bauer and Bryant 2004). Conserved positions in COR3 are more dispersed but are clearly identifiable. COR3 has been named as the “POPLD (NUC188) domain,” which is found in Pop1-like nucleolar proteins (Falquet et al. 2002; Staub et al. 2004). COR4 is a Gly-rich region, although not all the Gly residues can be aligned. The sequence of COR4 is similar to the consensus “G-patch” domain, a predicted globular domain found in several RNA-processing enzymes and RNA-binding proteins (Aravind and Koonin 1999). However, little is known about the role of the G-patch domain.
Site-directed randomization mutagenesis of Pop1p to obtain conditional defects
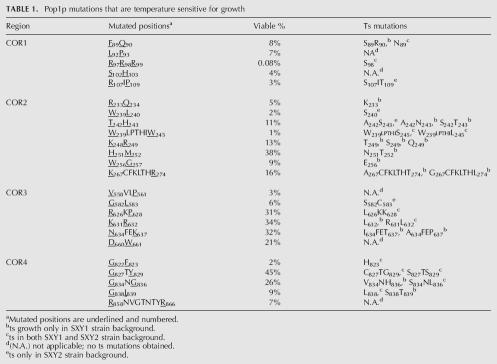
The roles of the conserved amino acids of the yeast Pop1p in assembly and function of RNase P and MRP were explored by mutagenesis of the conserved residues. Since Pop1p is required for yeast viability, we screened for conditionally defective phenotypes. Sequences encoding two or three conserved amino acids at a time were randomized by PCR mutagenesis (Materials and Methods), and viable variants were selected and screened for mutations that lead to temperature-sensitive (ts) or cold-sensitive (cs) growth phenotypes. The POP1 alleles in this study were tagged with triple haemagglutinin (3HA) epitopes to facilitate characterization.
Positions of the mutagenized amino acids are underlined and numbered in Table 1. Two conserved amino acids were mutated at a time, with the first two positions of both codons randomized. In theory this would generate a DNA library of 256 different sequence combinations for most of the libraries and 4096 DNA sequence variations for the three-position library at positions 97, 98, and 99. In total, 24 libraries of mutated pop1 genes were created for screening (Table 1).
TABLE 1.
Pop1p mutations that are temperature sensitive for growth
The percentage of viable colonies relative to the total number of transformants of each mutated pop1 library was determined to assess tolerance of the highly conserved positions in Pop1p for amino acid substitutions. The viable percentage varies from 0.08% to 45% for different mutated pop1 libraries (Table 1). It was unexpected that most of these absolutely conserved amino acids can be mutated to at least some alternative identities without causing severe functional defects.
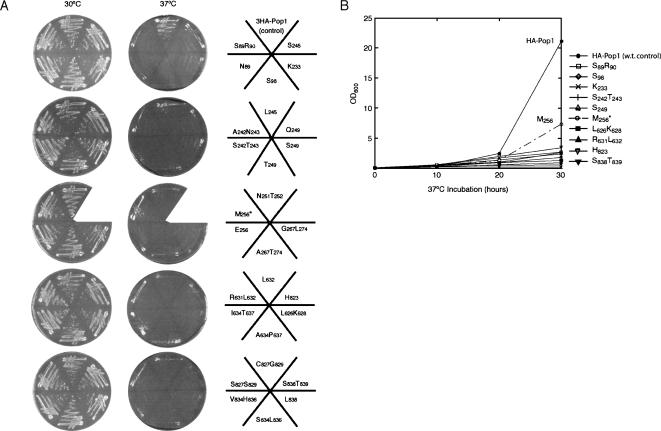
The strains with viable pop1 variants were grown at 30°C, 37°C, and 16°C to screen for ts and cs growth phenotypes, and the results are summarized in Table 1. No cs mutant strains were identified in this screen. Growth of the identified ts mutants at permissive (30°C) and nonpermissive (37°C) temperatures is shown in Figure 2A. In liquid cultures, the ts mutant strains grow substantially more slowly than the wild-type strain, except that the growth of the M256 mutant was only mildly impaired (Fig. 2B). The M256 mutant was therefore not analyzed in depth.
FIGURE 2.
Growth of the 3HA-Pop1p mutants on synthetic minimum medium. The strain background for this test was SXY1. (A) Growth at 30°C and 37°C on solid medium. Position of each strain is illustrated in the right panel. Strain SXY1/M256, indicated with an asterisk, is ts for growth on plates, but the growth in liquid medium at 37°C is only mildly slow compared to the wild-type control strain (shown in B). (B) Growth curves of representative strains at 37°C in liquid medium. The growth curves of strains 3HA-Pop1 (control) and M256 are indicated on the plot.
Pre-tRNA processing in pop1 mutants
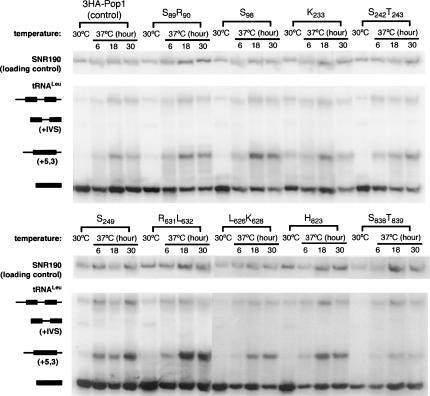
Since Pop1p is required for RNase P function, effects of the ts Pop1p mutations on in vivo pre-tRNALeu processing were examined at 30°C and at 6, 18, and 30 h after the shift to 37°C (Fig. 3). Previous studies have shown that in the normal maturation pathway of tRNALeu, the three major RNA species are the primary transcript; an intermediate containing the intervening sequence but not the 5′ and 3′ extensions (“+IVS” in Fig. 3); and mature tRNALeu, which is spliced and contains the mature 5′ and 3′ ends (Lee et al. 1991b). The amount of the primary transcript and that of the “+IVS” species are normally low, with the vast majority of the tRNALeu in the mature form. If RNase P activity is deficient, an aberrant, diagnostic intermediate with 5′ and 3′ extensions (“+5,3”) and the primary transcript accumulate (Lee et al. 1991b; Xiao et al. 2005). In this study, the normal processing profile of tRNALeu is observed at 30°C in the control strain containing wild-type 3HA-Pop1p (Fig. 3). However at 37°C, the “+5,3” species is increased even in the control strain, possibly reflecting an effect on the rate of processing events. Although terminal processing normally precedes splicing, the order is not obligatory (Lee et al. 1991b), and the “+5,3” form of pre-tRNALeu is a good indicator of decreased activity in RNase P-catalyzed cleavage.
FIGURE 3.
Northern blot analysis of pre-tRNALeu processing in representative 3HA-pop1 mutants at 30°C and 37°C. Incubation time of the strains at 37°C is indicated above the corresponding lanes, and the balance of pre-tRNALeu forms in total RNA was isolated by Northern blot. Four major species of tRNALeu are the primary transcript, an intermediate containing the intron (“+IVS”), an alternative intermediate containing 5′ and 3′ extensions (“+5,3”), and the mature tRNALeu (Lee et al. 1991b). Identities and schematic diagrams of tRNALeu species are shown to the left of the blots. Black bars indicate mature tRNA sequences, and lines represent the 5′ leader, the 3′ trailer, and the intervening sequences. The signals of the primary transcript and the intermediate “+5,3” were separately normalized to that of the loading control, SNR190 RNA. The resulting ratios in strain 3HA-Pop1 were set at 1 and were used for comparison with the ratios from other mutant strains. The normalized amounts of primary transcript and “+5,3” intermediate in 3HA-pop1 mutants at 30°C and 37°C (6 h) are listed in Table 2.
After 6 h of incubation at 37°C, accumulation of the primary transcript ranges from 0.88-fold (mutant S827S829) to 4.4-fold (mutant S249) over the wild-type strain (Table 2). Most mutations also result in accumulation of “+5,3” tRNALeu by two- to threefold at 37°C (Table 2). Mutations that display the most pronounced pre-tRNA accumulation occur at position 249 (T249, S249, Q249) and at position 823 (H823) as indicated by the increase in the amounts of both the primary transcript and the “+5,3” species (Table 2). Three mutations, S89R90, N89, and S827S829, do not cause significant accumulation of the primary transcript or the “+5,3” species (≤twofold accumulation, Fig. 3; Table 2). The data suggest that the conserved amino acids R249 and F823 in wild-type Pop1p are important for in vivo pre-tRNA processing while the contribution of F89, Q90, G827, or Y829 to RNase P function is small, if any. Effects of the other Pop1p mutations on RNase P function are intermediate.
TABLE 2.
Quantification of the RNA processing in 3HA-pop1 mutants
Maturation of the RPR1 RNA in pop1 mutants
In order to investigate whether the deficient pre-tRNA processing in the pop1 mutants was due to an assembly defect of the RNase P holoenzyme, maturation of the RPR1 RNA was examined. Pre-RPR1 RNA is assembled into a ribonucleoprotein complex with at least seven protein subunits of RNase P, including Pop1p, before being processed to the mature RNA (Srisawat et al. 2002). Mutations in RPR1 RNA that compromise its structure or prevent protein association also slow maturation of the RPR1 RNA, possibly by blocking the formation of the correct RNP complex required for processing. Accumulation of pre-RPR1 RNA is therefore diagnostic for an assembly problem in RNase P holoenzyme.
When wild-type yeast is grown to mid-log phase at 30°C, the ratio of mature to precursor RPR1 RNA is ∼5:1. In this study, the ratio in the control strain carrying the wild-type 3HA-Pop1p is 2.8 at 30°C and 2.4 at 37°C, slightly lower than normal (Table 2; Fig. 4), suggesting that 3HA tagging of Pop1p or growth at high temperature might slow assembly of RNase P or maturation of the RPR1 RNA, although not enough to slow growth or substantially alter tRNA processing at 30°C.
FIGURE 4.
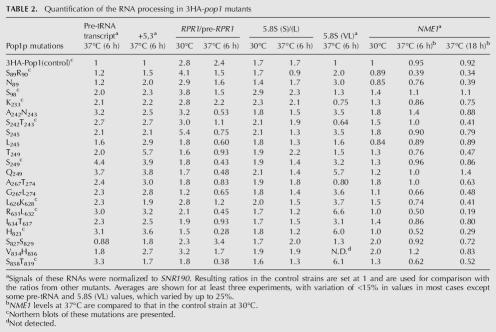
Northern blot analysis of precursor and mature RPR1 RNA in representative 3HA-pop1 mutants at 30°C and 37°C. Incubation time of the strains at 37°C is indicated. Precursor and mature RPR1 RNA are detected by Northern blotting, with SNR190 RNA probed as a loading control. Ratio of the mature to precursor RPR1 RNA in each mutant strain at 30°C and 37°C (6 h) is shown in Table 2.
Most defective Pop1p mutations affect RPR1 RNA maturation at 37°C (Fig. 4). In 14 mutant strains containing mutations in COR2–4, the ratio of mature to precursor RPR1 RNA is decreased by at least twofold after 6 h of incubation at 37°C (Table 2, column 5). These mutations are A242N243, S242T243, S245, L245, T249, S249, Q249, A267T274, G267L274, L626K628, R631L632, I634T637, H823, and S838T839. Several of these mutations (for example, H823) alter the ratio of the pre-RPR1 RNA to the mature form even at 30°C (Table 2; Fig. 4). Additionally, cellular levels of both the precursor and mature RPR1 RNAs in these 14 strains decline rapidly at 37°C compared to the control and normalized to SNR190 snRNA (Fig. 5; data not shown), suggesting that the altered RNase P complexes might not be as stable as the wild-type enzyme. The other Pop1p mutations, including all the mutations obtained in COR1, do not significantly affect maturation of the RPR1 RNA, although slow decreases in the level of the RPR1 RNAs are observed at 37°C (Table 2; Fig. 4; data not shown).
FIGURE 5.
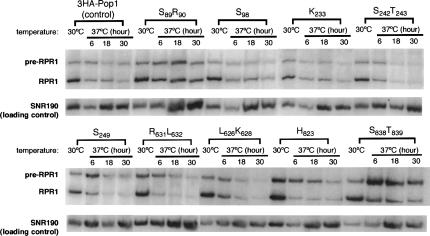
Northern blot analysis of 5.8S rRNA processing in representative 3HA-pop1 mutants at 30°C and 37°C. Incubation time of the strains at 37°C is indicated above the corresponding lanes. Three species of 5.8S rRNA are detected by Northern blotting. These include the long form (“5.8S (L)”), the short form (“5.8 (S)”), and the very long form (“5.8S (VL)”) (Shuai and Warner 1991; Lindahl et al. 1992; Schmitt and Clayton 1993; Chu et al. 1994). The SNR190 RNA is probed as a loading control. Ratio of the short to long forms of 5.8S rRNA in the mutants at 30°C and 37°C (6 h) is summarized in Table 2. Amount of 5.8S (VL) rRNA was normalized to that of SNR190 RNA. The resulting ratio in the control strain 3HA-Pop1 at 37°C was set at 1 and was used for comparison with the ratio from other mutant strains. The normalized amounts of 5.8S (VL) rRNA at 37°C are shown in Table 2.
The 14 mutations that substantially affect RPR1 RNA maturation also cause significant defects in pre-tRNA processing at 37°C (Fig. 3; Table 2). Moreover, in mutants S89R90, N89, and S827S829, where the RPR1 RNA processing is not affected by the mutations, the pre-tRNA profile is similar to that of the control strain (Fig. 3; Table 2). Thus, maturation and stability of RNase P holoenzyme often correlates with in vivo pre-tRNA processing and pre-tRNA processing defects in these mutants could be mainly attributable to the lack of RNase P. However, there are situations where a correlation is missing. Mutations S98, K233, and V834H836 affect pre-tRNA processing, but the maturation of the RPR1 RNA is hardly changed by the mutations. Thus, these amino acids might contribute to other functions, such as recognition of substrate pre-tRNA, catalysis, or subcellular localization of the enzyme.
5.8S rRNA processing in pop1 mutants
Because Pop1p is also an integral subunit of RNase MRP, we examined the 5.8S rRNA maturation in pop1 mutants. Under normal growth conditions two forms of 5.8S rRNA can be detected by Northern blots. They are the short form “5.8S (S)” and the long form “5.8S (L),” with 5.8S (S) normally being two- to threefold more abundant (Li et al. 2004; Xiao et al. 2005). In an RNase MRP deficient strain, the ratio of the 5.8S (S) to 5.8S (L) is reduced, and a very long form of 5.8S rRNA “5.8S (VL)” accumulates (Shuai and Warner 1991; Lindahl et al. 1992; Schmitt and Clayton 1993; Chu et al. 1994).
The ratio of 5.8S (S) to 5.8S (L) is similar in the control strain and the pop1 mutant strains at 30°C, ranging from 1.5 to 3 (Fig. 5; Table 2). After switching to 37°C, mutant S89R90 displays the most severe 5.8S rRNA processing defect. The amount of 5.8S (S) decreased rapidly at 37°C (Fig. 6), resulting in a twofold reduction in the ratio of 5.8S (S) to 5.8S (L) rRNA after only 6 h (Table 2). The temperature dependence of the 5.8S rRNA processing correlates well with the ts growth phenotype of the strain. This mutation also results in the accumulation of the longer 5.8S (VL) RNA at both 30°C and 37°C (Fig. 5; Table 2). Conversely, the pre-tRNA processing in this mutant is not notably affected (Fig. 4; Table 2), suggesting F89 and Q90 in Pop1p are required for the function of RNase MRP but not for RNase P. Characterization of the N89 mutation at this position further supports this idea (Table 2).
FIGURE 6.
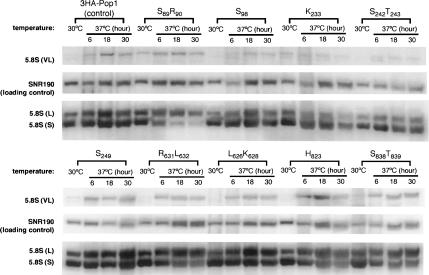
Northern blot analysis on levels of the NME1 RNA in representative 3HA-pop1 mutants at 30°C and 37°C. Incubation time of the strains at 37°C is indicated. Amount of the NME1 RNA is normalized to SNR190 RNA. The resulting ratio in the control strain 3HA-Pop1 at 30°C was set at 1 and was used for comparison with the ratio from other mutants. The normalized amount of NME1 RNA in each strain at 30°C and 37°C (6 h and 18 h) is listed in Table 2.
Mutations A242N243, S245, S249, Q249, G267L274, L626K628, R631L632, I634T637, H823, and S838T839 also affect RNase MRP function as indicated by the substantial (>threefold) accumulation of the 5.8S (VL) RNA at 37°C (Table 2). However, unlike mutant S89R90, the ratios of short to long forms of 5.8S in these strains do not decrease substantially until after 18 h of incubation at 37°C (Fig. 5; data not shown). These mutations also affect the in vivo RNase P function to various extents.
There are also examples of mutations that affect pre-tRNA processing but not 5.8S rRNA ratios. The 5.8S rRNA processing in ts mutant strains S98, K233, S242T243, A267T274, and V834H836 appears normal (Fig. 5; Table 2), but each strain has a moderate defect in pre-tRNA processing. One mutation, S827S829, is particularly noteworthy in that it does not have any obvious defect in either tRNA or 5.8S rRNA processing at 37°C, yet the strain does not grow at 37°C (Fig. 2). Possible causes of this defect will be addressed in the Discussion.
It is worth noting that different amino acid identities at the same conserved positions in Pop1p can have different effects on 5.8S rRNA processing. For instance, mutation A242N243 accumulates the 5.8S (VL) RNA, while another mutation at the same position (S242T243) does not. The same observations also apply to positions K267 and R274 (Table 2; data not shown). It is not surprising that different amino acid substitutions would be differentially tolerated, as seen by the observation that only a subset of amino acid combinations are tolerated at all at most of the conserved positions (Table 1).
Cellular level of the NME1 RNA in pop1 mutants
We examined the assembly or stability of RNase MRP in the pop1 mutant strains by probing the cellular level of the NME1 RNA. Levels of NME1 RNA in the control and 3HA-pop1 mutant strains do not change much at 30°C (<twofold; Fig. 6; Table 2). Most mutant strains contain slightly more cellular NME1 RNA than the control strain. At 37°C, the NME1 levels are considerably decreased after 18 h of incubation in the following nine mutant strains: S89R90, N89, S242T243, T249, G267L274, R631L632, L626K628, H823, and S838T839 (≥twofold decrease; Table 2; Fig. 6). In particular, mutations S89R90, R631L632, and H823 significantly reduce the amount of NME1 RNA early in the 37°C incubation (6 h; Table 2). Therefore, assembly or stability of the RNase MRP holoenzyme is likely affected by these Pop1p mutations. In several other 3HA-pop1 mutant strains (S98, K233, A242N243, S245, L245, S249, Q249, I634T637, S827S829, and V834H836), the level of the NME1 RNA is comparable to that in the control strain at 37°C (Table 2; Fig. 6; data not shown), suggesting that the mutations have little effect on RNase MRP levels.
Most of the mutations that affect RNase MRP levels also alter the 5.8S rRNA maturation (Table 2). For example, the three mutations (S89R90, R631L632, and H823) that severely reduce the level of NME1 RNA cause drastic defects in 5.8S rRNA processing at 37°C (Table 2; Figs. 5, 6). The shortage of RNase MRP in these mutants could account for the defects in 5.8S rRNA maturation to some extent. On the other hand, mutations S98, K233, S827S829, and V834H836 do not notably decrease the level of the NME1 RNA or the maturation of 5.8S rRNA (Table 2). These observations present a correlation between assembly or stability and cellular function of RNase MRP.
The relationship between stability and function of RNase MRP is not absolute in some strains with Pop1p mutations. For example, mutation S242T243 lowers the RNase MRP level but does not appear to affect the processing of 5.8S rRNA (Figs. 4, 6; Table 2). In contrast, mutations A242N243, S245, S249, Q249, and I634T637 accumulate the very long form of 5.8S rRNA while the level of RNase MRP complex in these mutants is not notably affected, indicative of RNase MRP functional defects. Thus, the defects in Pop1p caused by these mutations affect MRP function directly, rather that indirectly through stability effects.
Association of mutated Pop1p with the RPR1 and NME1 RNAs
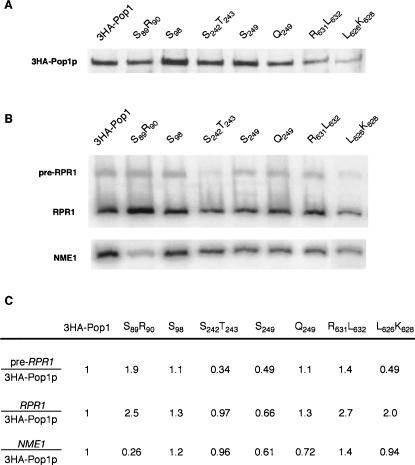
The processing profiles of the RPR1 RNA and the cellular levels of the NME1 RNA indicate that assembly or stability of the RNase P and MRP complexes are impaired to various degrees by the pop1 mutations. To examine the effects of the pop1 mutations on the association with RNA subunits, we performed coimmunoprecipitation (co-IP) of the RPR1 and NME1 RNAs with 3HA-Pop1p. The ratios of the copurified RNA subunits to 3HA-Pop1p are used to access how well the subunits associate, and the results are shown in Figure 7.
FIGURE 7.
Association of the mutated Pop1p with the RPR1 and NME1 RNAs in vivo. (A) Western blotting of 3HA-Pop1p protein in the HA immunoprecipitates. (B) Northern blotting of the NME1 RNA, and precursor and mature RPR1 RNAs copurified with the mutated 3HA-Pop1p. (C) Quantitation of the amounts of 3HA-Pop1p and RNA species in the immunoprecipitates. Signals of the NME1 RNA and the precursor and mature RPR1 RNAs on the Northern blot were divided by the signals of 3HA-Pop1p on the Western blot to account for the difference in the immunoprecipitation efficiency among samples. The resulting ratio in the 3HA-Pop1p control strain was arbitrarily set at 1 and was used for comparison with ratios from the mutant strains.
Representative pop1 mutations that have differential effects on RNase P and MRP assembly, as suggested by Northern analyses on RNA processing (Table 2), were chosen for the co-IP experiment. In particular, mutation S89R90 affects the cellular level of RNase MRP RNA, but not RNase P RNA. Mutations S242T243, S249, Q249, and L626K628 affect the maturation of the RNase P RNA but barely change the RNase MRP RNA level at early times after a shift to 37°C. Mutation R631L632 lowers levels of both RNase P and MRP RNAs, while neither the RNase P nor MRP RNA levels are affected by mutation S98. Because the mutant strains normally stopped growing by 10 h at 37°C, cultures were grown at 37°C for 6 h before harvest.
The amounts of 3HA-Pop1p and the RNA subunits that coimmunoprecipitated were determined by Western (Fig. 7A) and Northern (Fig. 7B) blots. The NME1 RNA and the precursor and mature RPR1 RNAs are found to be associated with all the mutated Pop1p proteins tested, although the ratio of the RNA subunits to Pop1p varies in different strains. Consistent with the Northern analysis, mutation S89R90 significantly reduces the NME1 RNA that is bound to Pop1p (∼fourfold reduction compared to the control), without decreasing the binding of the RPR1 RNAs to Pop1p (Fig. 7B,C). The reason for the increased association of RPR1 RNA with Pop1p is currently not known but could be due to decreased competition from NME1 RNA. This result is consistent with the suggestion that amino acids F89 and Q90 in wild-type Pop1p are required for the assembly of only RNase MRP.
In mutants S242T243 and L626K628, less pre-RPR1 RNA is associated with Pop1p compared to the wild-type strain, while the amount of mature RPR1 RNA bound is not decreased (Fig. 7B,C). The data suggest that these two mutations affect the initial assembly of the precursor RNase P complex but that the mature RNase P holoenzyme seems to be stable once formed. The association of Pop1p with NME1 RNA is not affected by these two mutations, as reflected by the amount of NME1 RNA copurified. This is consistent with the observation that the cellular level of the NME1 RNA in these mutants remains stable after short incubation at 37°C (6 h; Table 2), although stability of the NME1 RNA is decreased after longer incubation at 37°C (Fig. 6; Table 2). It is not clear whether the effects of these two mutations on the amount of the NME1 RNA in vivo are direct or indirect.
The binding of Pop1p to the NME1 RNA, mature RPR1 RNA, and pre-RPR1 RNA is reproducibly reduced in mutant S249 (Fig. 7C). This suggests that the assembly or stability of RNases P and MRP complexes is impaired by the mutation. In contrast, S98 does not decrease the ratio of Pop1p to the RNA subunits, in agreement with the presence of wild-type levels of the RPR1 and NME1 RNAs in this strain, even though the in vivo pre-tRNA processing is compromised. It is possible that this mutation affects other steps in the maturation pathway of pre-tRNA, such as the nucleolar localization of RNase P or catalysis of the pre-tRNA cleavage reaction.
It is surprising that mutation R631L632 does not reduce the association of Pop1p with either the RPR1 RNA or the NME1 RNA, although this mutation significantly decreases the levels of both RNA subunits at 37°C (Table 2). It is possible that both the Pop1p and the RNA subunits are turning over in these mutants, thus maintaining a constant ratio of protein to RNA. Mutation Q249 presents similar phenotypes in that it drastically reduces the ratio of mature to pre-RPR1 RNA in the cell, which suggests a stability problem of the RNase P holoenzyme. However, the Q249 Pop1p associates with RPR1 RNA at the same ratio as the wild-type control. This mutation only mildly affects the binding of Pop1p to the NME1 RNA.
A summary of the effects of Pop1p amino acid changes on RNases P and MRP is provided in Figure 8.
FIGURE 8.
Summary of the effects of the Pop1p mutations on RNA processing at 37°C. Amino acid sequences of the four conserved regions (CORs) in Pop1p from S. cerevisiae are listed, with positions provided on both sides of the sequences. Effects of the mutations at the highlighted positions on tRNA processing, maturation of the RPR1 RNA, processing of the 5.8S rRNA, and stability of the NME1 RNA are summarized. (+) The mutation causes defects in RNA processing/stability. (−) No obvious defects are detected when the position is mutated. (+/−) The mutated position either impairs RNA processing/stability or does not cause significant defects, depending on the nature of the mutations. Brackets denote that the listed phenotypes of RNA processing are observed when the two indicated positions are mutated at the same time.
Steady-state kinetics of the pre-tRNA cleavage reaction
To examine the effects of these mutations on the enzymatic activity of RNase P, steady-state kinetics for both the wild-type and RNase P holoenzyme with POP1 mutations were determined using the yeast pre-tRNATyr as previous described (Ziehler et al. 2000; Xiao et al. 2005) at both 30°C and 37°C to test for temperature sensitivity of the enzyme. Ideally it would be useful to also characterize the kinetic behavior of the RNase MRP activities, but we are unable to reproduce in vitro RNase MRP assays (Lygerou et al. 1996a) with sufficient reliability for kinetics.
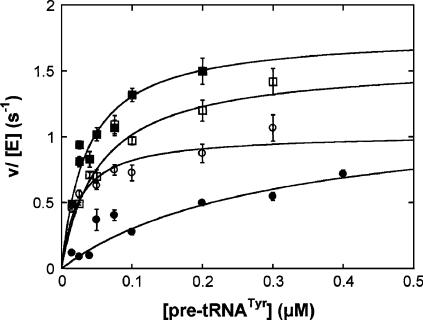
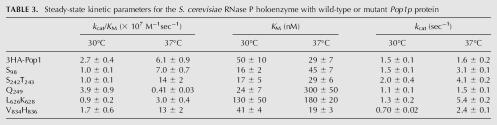
The in vitro steady-state cleavage activity catalyzed by wild-type and Q249 RNase Ps are shown in Figure 9 as examples. The dependence of the initial reaction velocity on the substrate concentration showed saturation kinetics, so the Michaelis–Menten equation is fit to the data (Fersht 1985). A summary of the steady-state kinetic parameters determined for RNase P holoenzyme with temperature-sensitive POP1 mutations (S98, S242T243, Q249, L626K628, and V384H839) affinity-purified from yeast is shown in Table 3. The rate of substrate cleavage by these isolated holoenzymes remains steady over the time course of the reactions (data not shown), indicating that these enzymes are stable under the assay conditions.
FIGURE 9.
Processing of S. cerevisiae pre-tRNATyr in vitro. The cleavage of a pre-tRNA substrate with a 12-nt leader catalyzed by RNase P holoenzyme with either the wild-type (■, at 37°C; □, at 30°C) or Q249 mutant (●, at 37°C; ○, at 30°C) Pop1p was determined at either 30°C or 37°C under steady-state conditions. The Michaelis–Menten equation is fit to the data to determine the values for k cat, K M, and k cat/K M (Table 3).
TABLE 3.
Steady-state kinetic parameters for the S. cerevisiae RNase P holoenzyme with wild-type or mutant Pop1p protein
The values of the steady-state kinetic parameters for the wild-type holoenzyme at 37°C are consistent with our previous measurement of the nontagged enzyme under the same conditions (Ziehler et al. 2000). While the k cat values are within experimental error of each other (1.3 ± 0.1 sec−1 and 1.6 ± 0.2 sec−1 for the nontagged and 3-HA tagged enzymes, respectively), the value of K M is modestly decreased for the 3-HA tagged enzyme (29 ± 7 nM) compared to the nontagged enzyme (55 ± 10 nM), and therefore the value of k cat /K M increases by ∼2.5-fold (Table 3; Ziehler et al. 2000). A similar increase in the specificity constant has been measured previously for affinity-isolated yeast nuclear RNase P holoenzyme in which RPR1 RNA is modified with an “S1” streptavidin affinity tag (Xiao et al. 2005). This similarity suggests that the increase in activity might reflect the more gentle and rapid purification methods of this unstable enzyme, rather than the presence of a specific RNA tag. The value of k cat/K M is near the diffusion-controlled limit (2.5 × 108 M−1sec−1) suggesting that pre-tRNA association is limiting under these conditions (Ziehler et al. 2000; Xiao et al. 2005). Decreasing the temperature from 37°C to 30°C has little effect on the k cat value but decreases the value of k cat/K M for the wild-type holoenzyme about twofold (Table 3).
Although at a permissive growth temperature these POP1 mutations do not exhibit significant pre-tRNA cleavage defects in vivo (Table 2), the specificity constants (k cat/K M) for the holoenzyme with the mutated S98, S242T243, and L626K628 Pop1p are decreased threefold compared to the 3HA-tagged wild-type holoenzyme, while mutations at Q249 and V834H836 in Pop1p have little effect on the value of k cat/K M. Upon switching the temperature to 37°C, the Michaelis constant for the holoenzyme with Q249 Pop1p shows a dramatic 10-fold increase. More importantly, the k cat/K M value for the holoenzyme with Q249 Pop1p decreases by 15-fold as the result of the temperature shift compared to the activity of the wild-type holoenzyme (Table 3). These results indicate that this pop1 mutation decreases the efficiency of pre-tRNA cleavage catalyzed by RNase P, which likely leads to the temperature-sensitive phenotype observed in vivo. None of the other mutations demonstrate a significantly larger catalytic defect at 37°C compared to 30°C. Interestingly, the k cat/K M values for holoenzymes with S242T243 and V834H836 Pop1p are twofold greater than the wild-type enzyme at 37°C (Table 3). Furthermore, a greater enhancement than the wild-type RNase P as a result of the temperature shift (7- to 14-fold vs. 2.2-fold) and result in twofold greater k cat/K M values than the wild-type enzyme at 37°C (Table 3). The apparent discrepancy between the increased efficiency of RNase P in vitro and pre-tRNA processing defect caused by pop1 mutations in vivo will be discussed below.
DISCUSSION
Alignment of the protein sequences of Pop1p and its homologs reveals four conserved regions. Randomization mutagenesis of conserved amino acids in these regions of yeast Pop1p showed that most positions are tolerant of at least limited substitutions and that conditionally defective alleles could be identified in each of the conserved regions, with COR1 being least tolerant to changes. It is somewhat unexpected that the highly conserved amino acids in Pop1p are not absolutely required for viability, since Pop1p is essential. Characterization of the ts mutant strains has uncovered that the mutations weaken various aspects of pre-tRNA processing, 5.8S rRNA processing, maturation of the RPR1 RNA, or stability of the NME1 RNA at the restrictive temperature (37°C). It is difficult to assign precise molecular defects conferred by the various mutations since, as with all large macromolecular complexes, effects on subunit and substrate interactions could be either direct or indirect. However, several interesting observations emerged.
As expected, many Pop1p mutations in this study affect both pre-tRNA and 5.8S rRNA processing, suggesting that the mutated positions play similar roles in RNase P and RNase MRP functions. In addition, mutations that preferentially affect either the pre-tRNA or 5.8S rRNA biogenesis pathway have been obtained. Thus, different subsets of conserved amino acids in Pop1p contribute differentially to RNase P and MRP functions, although it is not yet possible to determine whether these differential effects are at the level of interaction with substrates or different subunits of the holoenzymes. It seems unlikely that any entire COR is exclusively required for the function of RNase P or MRP, since some mutations in all of the CORs affect both pre-tRNA processing and 5.8S rRNA maturation.
Functional defects of RNase P and MRP in several Pop1p mutant strains are coincident with stability defects of the RNA subunit of the corresponding enzymes, indicating that the mutations might affect enzyme function by damaging the assembly or stability of RNP complexes. Previous studies have shown that the yeast Pop1p directly binds to the RPR1 RNA and is involved in several protein–protein interactions within RNase P holoenzyme (Houser-Scott et al. 2002). Pop1p also binds to the NME1 RNA subunits of RNase MRP, which has the conserved RNA binding site that is the primary target of Pop1 binding in RPR1 RNA (Ziehler et al. 2001), but the interactions with that site in the two RNAs could be subtly different or Pop1p could have secondary contact points in the two RNAs that are different. In addition, Pop1p contacts with other protein subunits might be differentially altered in RNase P versus RNase MRP. The Pop1p mutations described here that seem to block assembly of RNase P or MRP will provide a guide for future assessment of contacts between Pop1p and other subunits.
We also found that some mutations slow the processing of pre-tRNA or precursor to 5.8S rRNA in vivo without affecting the stability of the corresponding enzymes. This raises the possibility that Pop1p might be involved directly in substrate recognition or catalysis. We therefore measured the in vitro RNase P activity at both 30°C and 37°C to test this hypothesis. Most of the RNase Ps with temperature-sensitive pop1 mutations retained reasonably stable steady-state turnover activity even at the nonpermissive temperature. Only the Q249 and L626K628 mutations significantly lower the value of the specificity constant (k cat/K M) at 37°C compared to wild-type RNase P, indicating that these two mutations affect the pre-RNA cleavage activity of the holoenzyme. Interestingly, S242T243 and V834H836 mutations increase the specificity constant of RNase P. Similar effects have been previously observed for the RNAse P holoenzyme with a mutation in the RPR1 subunit, which also shows an in vivo tRNA processing defect (Xiao et al. 2005). Although these mutations enhance catalytic efficiency in vitro for this pre-tRNA substrate they also allow binding and cleavage of a wider variety of RNA substrates, thus leading to a decrease in the apparent specificity constant for cleavage of a specific pre-tRNA substrate in the cellular milieu containing a wide variety of other RNA species. These mutations thus could lead to an in vivo tRNA processing defect by becoming less specific for binding and cleaving pre-tRNA substrates.
A direct contribution of the protein subunit to substrate recognition has been demonstrated by biochemical characterization of Bacillus subtilis RNase P, where the protein specifically enhances the binding affinity of the pre-tRNA substrate by directly contacting the leader (Kurz et al. 1998; Niranjanakumari et al. 1998). Whether the same mode of substrate recognition by the protein subunit is conserved in eukaryotes remains unknown, but the yeast nuclear RNase P has strong binding sites for single-stranded RNAs, which could possibly contribute to recognition of the 5′ leader or 3′ trailer sequences of pre-tRNAs (Ziehler et al. 2000). Consistent with this, a protein component of the human RNase P, rather than the RNA subunit, is reported to form a cross-link to a photoreactive substrate (True and Celander 1998). Although the yeast nuclear RNase P is still an RNA-based enzyme, it is entirely possible that the increased protein content has been accompanied by transfer of some of the roles of the catalytic bacterial RNA subunit or catalytic metal ions to the protein subunits.
One anomaly arising from these data is that mutation S827S829 does not have any obvious defects in either pre-tRNA or 5.8S rRNA processing at the restrictive temperature. The cause of cell death of this strain at 37°C is not known at this point, but it is possible that RNase P, RNase MRP, or Pop1p in some unknown complex has additional biological functions. Recent studies have indicated that yeast RNase MRP not only processes precursors to 5.8S rRNA, but also cleaves selected messenger RNAs to promote cell cycle progression (Gill et al. 2004). Eukaryotic RNase P might well have other substrates also, since the natural substrates for bacterial RNase P include precursors to 4.5S RNA, tm RNA, and mRNA in addition to pre-tRNAs. A genome-wide screen for additional substrates of RNase P is in progress.
MATERIALS AND METHODS
Plasmids and strains
A copy of the wild-type POP1 gene from the yeast S. cerevisiae was cloned into plasmid p413TEF with three haemagglutinin (3HA) epitopes fused to the 5′ end of the gene. A NotI restriction site was inserted immediately before and after the sequence of the 3HA tag due to the cloning strategy. The TEF1 promoter in the resulting plasmid was replaced with the endogenous POP1 promoter (233695–234411 on chromosome XIV) amplified from the genomic DNA of S. cerevisiae. The resulting clone p413–3HA-POP1, which contains a HIS3 selectable marker, was used as the template for PCR mutagenesis of the POP1 gene and served as a control for subsequent characterization of the Pop1p mutations.
Screen for Pop1p mutant strains with conditionally defective growth was performed in the S. cerevisiae haploid strain SXY2 (MAT a ade2-1 his3-11, 15 leu2-3, 112 trp1-1 ura3-1 can1-100 pop1Δ NAT1 RPR1:: kan r). The chromosomal copies of POP1 and RPR1 were disrupted by drug-resistant markers nat1 (Goldstein and McCusker 1999) and kanr (Wach et al. 1994), respectively, via PCR-based homologous recombination (Wach et al. 1994). To support viability of the strain, a wild-type POP1 allele was expressed on plasmid p416ADH-POP1 and a copy of the S1-affinity tagged RPR1 gene was expressed on pRS315-S1-RPR1 (Srisawat and Engelke 2001). Previous studies had shown that presence of the S1-tag on the RPR1 RNA did not affect growth at 30°C, 37°C, or 16°C (Srisawat and Engelke 2001; Xiao et al. 2005). The selectable markers on plasmids p416ADH-POP1 and pRS315-S1-RPR1 are URA3 and LEU2, respectively.
Effects of the Pop1p mutations on the functions and assembly of RNases P and MRP were examined in haploid strain SXY1 (MAT a ade2-1 his3-11, 15 leu2-3, 112 trp1–1 ura3-1 can1-100 pop1ΔNAT1), in which the POP1 gene was deleted.
Random mutagenesis of the conserved amino acids of Pop1p
Two conserved amino acids of Pop1p were randomized at a time. DNA fragments containing POP1 mutations were amplified by overlap-extension PCR (Ling and Robinson 1997) with primers containing random nucleotides at the targeted positions. The mutated DNA fragments were ligated into p413–3HA-POP1 plasmid at unique restriction sites. Plasmids carrying POP1 mutations have a selectable marker HIS3. The complexity of each library of p413–3HA-pop1 with two randomized amino acids is 256. The library complexity is increased to 4096 molecules when three amino acids are randomized (library R97R98R99; Table 1).
Yeast transformation and screen for conditionally defective mutants for growth
Mutated pop1 libraries in p413–3HA-pop1 (1.5 μg) were transformed into strain SXY2 containing p416ADH-POP1 and pRS315-S1-RPR1 plasmids using lithium acetate (Gietz et al. 1995). After transformation, cells were allowed to recover at 30°C for 1 h in 30 mL standard synthetic medium containing dextrose and lacking uracil, leucine, and histidine (SDC-ULH). Transformation efficiency was determined by plating on SDC-ULH medium. More than 2000 transformants were tested per mutated pop1 library, which covered at least seven times the complexity of most libraries. Since the theoretical complexity of library R97R98R99 (Table 1) was 4096, ∼105 transformants were screened.
Transformants were washed with SDC-ULH medium and plated on SDC-LH plates at 30°C for 3 d to select for plasmid uptake. Transformants were replica plated onto SDC-HL medium containing 5-fluoroorotic acid (5-FOA) and grown at 30°C for 2–3 d to select for cells that had lost the p416ADH-POP1 plasmid. One hundred colonies showing 5-FOA resistence from each library were streaked again onto SDC-HL plates containing 5-FOA to confirm their growth. The viable colonies were grown on SDC-HL plates at 30°C, 37°C, and 16°C to screen for ts and cs alleles. Incubation at 30°C and 37°C took 2–3 d, while the 16°C incubation was for 10 d.
The p413–3HA-pop1 plasmids in the ts or cs mutants were extracted from yeast, amplified in Escherichia coli, and retransformed into yeast strains SXY1/p416ADH-POP1 and SXY2/(p416ADH-POP1, pRS315-S1-RPR1) to confirm phenotypes. The pop1 mutations were determined by DNA sequencing.
Growth of the control and ts mutant strains in liquid synthetic minimum medium was monitored after a shift to 37°C. The cultures were diluted with medium prewarmed to 37°C to keep in exponential growth phases.
Northern blotting of RNAs
Wild-type and ts pop1 mutant strains with SXY1 strain background were cultured in SDC-H medium at 30°C to OD600 of 0.6–1.0. A fraction of each culture was harvested for total cellular RNA extraction. Another fraction of the culture was diluted with SDC-histidine medium prewarmed to 37°C, followed by incubation at 37°C for up to 30 h. A portion of each culture was collected after 6, 18, and 30 h of incubation at 37°C, diluting as necessary with prewarmed (37°C) SDC-H medium. Total yeast RNAs were extracted from cells harvested at 30°C and at 37°C with hot acidic phenol (Kohrer and Domdey 1991) and concentrations determined by UV absorbance.
Total yeast RNAs (10 μg) from each strain were electrophoresed on a denaturing 8% polyacrylamide gel and were electrotransferred to a Nytran SuperCharge membrane (Schleicher & Schuell Bioscience). The processing profiles of tRNALeu, RPR1 RNA, 5.8S rRNA, and NME1 RNA were probed with 32P-radiolabeled oligodeoxynucleotides complementary to the corresponding RNAs. The probe to tRNALeu was 5′-TGCTAAGAGATTCGAACTCTTGCA-3′, the RPR1 RNA probe was 5′-GCTGGAACAGCAGCAGTAATCGGTA-3′, the 5.8S rRNA probe was 5′-CGCATTTCGCTGCGTTCTTCATCG-3′, and the NME1 probe was 5′-GCAATAGAGGTACCAGGTCAAGAAG-3′. A small nucleolar RNA, SNR190 RNA, was also probed on the same Northern blot as an internal loading control (probe: 5′-ATGGTCGAATCGGACGAG-3′). Signals on the Northern blots were detected with a PhosphorImager (Molecular Dynamics 445 SI) and were quantified with IPlab Gel software (Signal Analytics). Averages are provided for three or more independent experiments.
HA immunoprecipitation of RNP complexes
SXY1 strains containing wild-type Pop1p-HA or various pop1 mutations were cultured in SDC-H medium at 30°C to OD600 of 0.6–1. The cultures were diluted with prewarmed SDC-H medium and were incubated at 30°C or 37°C for 6 h. Yeast extracts were prepared with glass beads as described (Xiao et al. 2005). The HA-tagged Pop1 proteins were the only copies of Pop1p in these strains. Yeast extract equivalent to 10 mg of protein was incubated with 20 μg of anti-HA antibody (12CA5; Roche) and 1% bovine serum albumin at 4°C for 1 h. To increase the immunoprecipitation efficiency, 20 μg of rabbit anti-mouse antibody (AffiniPure Rabbit Anti-Mouse IgG (H+L), Jackson ImmunoResearch) was subsequently added to the yeast extract and was incubated at 4°C for another hour. The mixture was bound at 4°C for 2 h to protein A-agarose beads (100 μL; BioRad Affi-Gel), preequilibrated with Buffer A (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 5 mM EDTA, 0.1% Triton-X 100, 10% glycerol, 1 mM DTT, Complete protease inhibitors-EDTA free [Roche]). The beads were washed five times with ice-cold Buffer A and were pelleted at 1500 rpm for 2 min.
To examine the association of the mutated Pop1p with the RPR1 and NME1 RNAs, complexes containing 3HA-Pop1p were extracted from the beads in 100 μL sample buffer (60 mM Tris-HCl at pH 6.8, 2% SDS, 15% glycerol, 0.025% bromophenol blue, 5% β-mercaptoethanol, 6 M urea) at 55°C for 10 min. The beads were pelleted at 1500 rpm for 2 min. RNAs in 50 μL of the eluate were directly precipitated with sodium acetate and ethanol with glycogen as carriers. The RNA samples were separated on a denaturing 6% polyacrylamide gel followed by Northern blotting as above except that only RPR1 RNA and NME1 RNA were probed.
The amount of 3HA-Pop1p in the HA immunoprecipitates was determined by Western blotting. Proteins in 30 μL of each eluate were electrophoresed on a 7.5% polyacrylamide-Tris-HCl gel (BioRad) and were electrotransferred onto a PVDF membrane. The blot was incubated with anti-HA antibody (12CA5, Roche), goat anti-mouse HRP antibody (Chemicon, AP308), and chemiluminescent substrate (ECL Plus Western Blotting Detection System, Amersham Biosciences). The Western blot was exposed to an X-ray film (Kodak BIOMAX MR), followed by quantification of the signals on a densitometer (IS-1000 Digital Imaging System). Although signal intensity in film is nonlinear, the signal was examined as a ratio to the RNA signal in Northern blots and was reproducible within 30% in two or more independent experiments.
RNase P assays
RNase P enzymes carrying Pop1p mutations were immunoprecipitated with HA antibody as described above. To gently elute RNases P and MRP from the immunoprecipitates, beads were incubated with 300 μL of 5 mg/mL HA peptide (in Buffer A without EDTA, synthesized by the University of Michigan Protein Structure Facility) for 4 h at 4°C. Eluate was collected by centrifugation at 1500 rpm for 2 min. Concentration of the RNase P in the eluate was determined by Northern blot analysis, compared with known amounts of in vitro transcribed RPR1 RNA.
Wild-type yeast pre-tRNATyr with the intervening sequence, 5′ 12-nt leader, and 3′ UUUUU trailer was prepared as described (Beebe and Fierke 1994) by in vitro transcription from linearized plasmid by T7 RNA polymerase (plasmid kindly provided by Prof. W.T. McAllister [He et al. 1997]). Pre-tRNA substrate labeled at the 5′ end with 32P was prepared after transcription by first treating pre-tRNA with calf intestinal alkaline phosphatase (New England BioLabs) to hydrolyze the 5′ triphosphate group, followed by incubating with [γ-32P] ATP and T4 polynucleotide kinase (New England BioLabs) to add a labeled phosphate group at the 5′ terminus of the transcript.
The steady-state kinetic turnover of the yeast nuclear RNase P holoenzyme was characterized under conditions where the substrate concentration was in large excess compared to the enzyme concentration ([S] >> [E]) in Buffer B (10 mM HEPES at pH 8.0, 100 mM KCl, and 10 mM MgCl2) as previously described (Ziehler et al. 2000; Xiao et al. 2005). Unlabeled pre-tRNA substrate with trace amount of the 5′ 32P-labeled substrate was refolded by incubating at 95°C for 2 min in 10 mM Tris (pH 8.0), and 0.1 mM EDTA followed by incubating at 37°C for 30 min prior to the addition of an equal volume of 2× Buffer B. The enzyme sample was diluted directly into Buffer B. Both the enzyme and the substrate were incubated at the reaction temperature for 10 min before initiating the experiment. The assay was started by the addition of RNase P (0.1–2 pM, final concentration) to various concentrations of the substrate (25–300 nM). At various times, an aliquot of the reaction mixtures was removed and mixed with an equal volume of the stop solution (200 mM Na3·EDTA, 20 mM Tris at pH 8.0, 0.05% bromophenol blue, 0.05% xylene cyanol, and 8 M urea). Uncleaved substrate and products were separated by PAGE (12% polyacrylamide-7 M urea), and were visualized and quantified by using a PhosphorImager (Molecular Dynamics). The initial cleavage rate constants were determined from the time region when <10% of total substrate was cleaved (the initial velocity condition). Steady-state kinetic parameters k cat, K M, and k cat/K M were determined by fitting the Michaelis–Menten equation (Fersht 1985) to the initial velocities using KaleidaGraph software (Synergy Software). The reported errors are the asymptotic standard errors.
ACKNOWLEDGMENTS
We thank F. Houser-Scott, P. Good, S. Walker, C. Srisawat, S. Chen, J.J. Day, and members of the Engelke and Fierke laboratories for helpful discussions and technical assistance. We are grateful to M. Tsoi for technical assistance. This work was supported by NIH grants GM34869 (D.R.E.) and GM55387 (C.A.F.), and a fellowship from the Horace H. Rackham Graduate School at the University of Michigan (S.X.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.23206.
REFERENCES
- Aravind L., Koonin E.V. G-patch: A new conserved domain in eukaryotic RNA-processing proteins and type D retroviral polyproteins. Trends Biochem. Sci. 1999;24:342–344. doi: 10.1016/s0968-0004(99)01437-1. [DOI] [PubMed] [Google Scholar]
- Beebe J.A., Fierke C.A. A kinetic mechanism for cleavage of precursor tRNA(Asp) catalyzed by the RNA component of Bacillus subtilis ribonuclease P. Biochemistry. 1994;33:10294–10304. doi: 10.1021/bi00200a009. [DOI] [PubMed] [Google Scholar]
- Boomershine W.P., McElroy C.A., Tsai H.Y., Wilson R.C., Gopalan V., Foster M.P. Structure of Mth11/Mth Rpp29, an essential protein subunit of archaeal and eukaryotic RNase P. Proc. Natl. Acad. Sci. 2003;100:15398–15403. doi: 10.1073/pnas.2535887100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusca E.M., True H.L., Celander D.W. Novel RNA-binding properties of Pop3p support a role for eukaryotic RNase P protein subunits in substrate recognition. J. Biol. Chem. 2001;276:42543–42548. doi: 10.1074/jbc.M107293200. [DOI] [PubMed] [Google Scholar]
- Chamberlain J.R., Lee Y., Lane W.S., Engelke D.R. Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes & Dev. 1998;12:1678–1690. doi: 10.1101/gad.12.11.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S., Archer R.H., Zengel J.M., Lindahl L. The RNA of RNase MRP is required for normal processing of ribosomal RNA. Proc. Natl. Acad. Sci. 1994;91:659–663. doi: 10.1073/pnas.91.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falquet L., Pagni M., Bucher P., Hulo N., Sigrist C.J., Hofmann K., Bairoch A. The PROSITE database, its status in 2002. Nucleic Acids Res. 2002;30:235–238. doi: 10.1093/nar/30.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht A. Enzyme structure and mechanism. W.H. Freeman; New York: 1985. [Google Scholar]
- Forster A.C., Altman S. Similar cage-shaped structures for the RNA components of all ribonuclease P and ribonuclease MRP enzymes. Cell. 1990;62:407–409. doi: 10.1016/0092-8674(90)90003-w. [DOI] [PubMed] [Google Scholar]
- Frank D.N., Pace N.R. Ribonuclease P: Unity and diversity in a tRNA processing ribozyme. Annu. Rev. Biochem. 1998;67:153–180. doi: 10.1146/annurev.biochem.67.1.153. [DOI] [PubMed] [Google Scholar]
- Frank D.N., Adamidi C., Ehringer M.A., Pitulle C., Pace N.R. Phylogenetic-comparative analysis of the eukaryal ribonuclease P RNA. RNA. 2000;6:1895–1904. doi: 10.1017/s1355838200001461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R.D., Schiestl R.H., Willems A.R., Woods R.A. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- Gill T., Cai T., Aulds J., Wierzbicki S., Schmitt M.E. RNase MRP cleaves the CLB2 mRNA to promote cell cycle progression: Novel method of mRNA degradation. Mol. Cell. Biol. 2004;24:945–953. doi: 10.1128/MCB.24.3.945-953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein A.L., McCusker J.H. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae . Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- Hall T.A., Brown J.W. Interactions between RNase P protein subunits in archaea. Archaea. 2004;1:247–254. doi: 10.1155/2004/743956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B., Rong M., Lyakhov D., Gartenstein H., Diaz G., Castagna R., McAllister W.T., Durbin R.K. Rapid mutagenesis and purification of phage RNA polymerases. Protein Expr. Purif. 1997;9:142–151. doi: 10.1006/prep.1996.0663. [DOI] [PubMed] [Google Scholar]
- Houser-Scott F., Xiao S., Millikin C.E., Zengel J.M., Lindahl L., Engelke D.R. Interactions among the protein and RNA subunits of Saccharomyces cerevisiae nuclear RNase P. Proc. Natl. Acad. Sci. 2002;99:2684–2689. doi: 10.1073/pnas.052586299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrous N. Human ribonuclease P: Subunits, function, and intranuclear localization. RNA. 2002;8:1–7. doi: 10.1017/s1355838202011184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T., Altman S. Protein–protein interactions with subunits of human nuclear RNase P. Proc. Natl. Acad. Sci. 2001;98:920–925. doi: 10.1073/pnas.021561498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakuta Y., Ishimatsu I., Numata T., Kimura K., Yao M., Tanaka I., Kimura M. Crystal structure of a ribonuclease P protein Ph1601p from Pyrococcus horikoshii OT3: An archaeal homologue of human nuclear ribonuclease P protein Rpp21. Biochemistry. 2005;44:12086–12093. doi: 10.1021/bi050738z. [DOI] [PubMed] [Google Scholar]
- Kifusa M., Fukuhara H., Hayashi T., Kimura M. Protein–protein interactions in the subunits of ribonuclease P in the hyperthermophilic archaeon Pyrococcus horikoshii OT3. Biosci. Biotechnol. Biochem. 2005;69:1209–1212. doi: 10.1271/bbb.69.1209. [DOI] [PubMed] [Google Scholar]
- Kohrer K., Domdey H. Preparation of high molecular weight RNA. Methods Enzymol. 1991;194:398–405. doi: 10.1016/0076-6879(91)94030-g. [DOI] [PubMed] [Google Scholar]
- Kurz J.C., Niranjanakumari S., Fierke C.A. Protein component of Bacillus subtilis RNase P specifically enhances the affinity for precursor-tRNAAsp. Biochemistry. 1998;37:2393–2400. doi: 10.1021/bi972530m. [DOI] [PubMed] [Google Scholar]
- Lee J.Y., Evans C.F., Engelke D.R. Expression of RNase P RNA in Saccharomyces cerevisiae is controlled by an unusual RNA polymerase III promoter. Proc. Natl. Acad. Sci. 1991a;88:6986–6990. doi: 10.1073/pnas.88.16.6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.Y., Rohlman C.E., Molony L.A., Engelke D.R. Characterization of RPR1, an essential gene encoding the RNA component of Saccharomyces cerevisiae nuclear RNase P. Mol. Cell. Biol. 1991b;11:721–730. doi: 10.1128/mcb.11.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Frank D.N., Pace N., Zengel J.M., Lindahl L. Phylogenetic analysis of the structure of RNase MRP RNA in yeasts. RNA. 2002;8:740–751. doi: 10.1017/s1355838202022082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zaman S., Langdon Y., Zengel J.M., Lindahl L. Identification of a functional core in the RNA component of RNase MRP of budding yeasts. Nucleic Acids Res. 2004;32:3703–3711. doi: 10.1093/nar/gkh689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl L., Zengel J.M. RNase MRP and rRNA processing. Mol. Biol. Rep. 1995;22:69–73. doi: 10.1007/BF00988708. [DOI] [PubMed] [Google Scholar]
- Lindahl L., Archer R.H., Zengel J.M. A new rRNA processing mutant of Saccharomyces cerevisiae . Nucleic Acids Res. 1992;20:295–301. doi: 10.1093/nar/20.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling M.M., Robinson B.H. Approaches to DNA mutagenesis: An overview. Anal. Biochem. 1997;254:157–178. doi: 10.1006/abio.1997.2428. [DOI] [PubMed] [Google Scholar]
- Lygerou Z., Mitchell P., Petfalski E., Seraphin B., Tollervey D. The POP1 gene encodes a protein component common to the RNase MRP and RNase P ribonucleoproteins. Genes & Dev. 1994;8:1423–1433. doi: 10.1101/gad.8.12.1423. [DOI] [PubMed] [Google Scholar]
- Lygerou Z., Allmang C., Tollervery D., Seraphin B. Accurate processing of a eukaryotic precursor ribomsomal RNA by ribonuclease MRP in vitro. Science. 1996a;272:268–270. doi: 10.1126/science.272.5259.268. [DOI] [PubMed] [Google Scholar]
- Lygerou Z., Pluk H., van Venrooij W.J., Seraphin B. hPop1: An autoantigenic protein subunit shared by the human RNase P and RNase MRP ribonucleoproteins. EMBO J. 1996b;15:5936–5948. [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., Bryant S.H. CD-Search: Protein domain annotations on the fly. Nucleic Acids Res. 2004;32:W327–W331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niranjanakumari S., Stams T., Crary S.M., Christianson D.W., Fierke C.A. Protein component of ribozyme ribonuclease P alters substrate recognition by directly contacting pre-tRNA. Proc. Natl. Acad. Sci. 1998;95:15212–15217. doi: 10.1073/pnas.95.26.15212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numata T., Ishimatsu I., Kakuta Y., Tanaka I., Kimura M. Crystal structure of archaeal ribonuclease P protein Ph1771p from Pyrococcus horikoshii OT3: An archaeal homolog of eukaryotic ribonuclease P protein Rpp29. RNA. 2004;10:1423–1432. doi: 10.1261/rna.7560904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly T.H., Schmitt M.E. The yeast, Saccharomyces cerevisiae, RNase P/MRP ribonucleoprotein endoribonuclease family. Mol. Biol. Rep. 1995;22:87–93. doi: 10.1007/BF00988711. [DOI] [PubMed] [Google Scholar]
- Salinas K., Wierzbicki S., Zhou L., Schmitt M.E. Characterization and purification of Saccharomyces cerevisiae RNase MRP reveals a new unique protein component. J. Biol. Chem. 2005;280:11352–11360. doi: 10.1074/jbc.M409568200. [DOI] [PubMed] [Google Scholar]
- Schmitt M.E., Clayton D.A. Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae . Mol. Cell. Biol. 1993;13:7935–7941. doi: 10.1128/mcb.13.12.7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt M.E., Clayton D.A. Characterization of a unique protein component of yeast RNase MRP: An RNA-binding protein with a zinc-cluster domain. Genes & Dev. 1994;8:2617–2628. doi: 10.1101/gad.8.21.2617. [DOI] [PubMed] [Google Scholar]
- Shuai K., Warner J.R. A temperature sensitive mutant of Saccharomyces cerevisiae defective in pre-rRNA processing. Nucleic Acids Res. 1991;19:5059–5064. doi: 10.1093/nar/19.18.5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidote D.J., Hoffman D.W. NMR structure of an archaeal homologue of ribonuclease P protein Rpp29. Biochemistry. 2003;42:13541–13550. doi: 10.1021/bi030170z. [DOI] [PubMed] [Google Scholar]
- Sidote D.J., Heideker J., Hoffman D.W. Crystal structure of archaeal ribonuclease P protein aRpp29 from Archaeoglobus fulgidus . Biochemistry. 2004;43:14128–14138. doi: 10.1021/bi048578z. [DOI] [PubMed] [Google Scholar]
- Srisawat C., Engelke D.R. Streptavidin aptamers: Affinity tags for the study of RNAs and ribonucleoproteins. RNA. 2001;7:632–641. doi: 10.1017/s135583820100245x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srisawat C., Houser-Scott F., Bertrand E., Xiao S., Singer R.H., Engelke D.R. An active precursor in assembly of yeast nuclear ribonuclease P. RNA. 2002;8:1348–1360. doi: 10.1017/s1355838202027048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub E., Fiziev P., Rosenthal A., Hinzmann B. Insights into the evolution of the nucleolus by an analysis of its protein domain repertoire. Bioessays. 2004;26:567–581. doi: 10.1002/bies.20032. [DOI] [PubMed] [Google Scholar]
- Takagi H., Watanabe M., Kakuta Y., Kamachi R., Numata T., Tanaka I., Kimura M. Crystal structure of the ribonuclease P protein Ph1877p from hyperthermophilic archaeon Pyrococcus horikoshii OT3. Biochem. Biophys. Res. Commun. 2004;319:787–794. doi: 10.1016/j.bbrc.2004.05.055. [DOI] [PubMed] [Google Scholar]
- Tollervey D. Genetic and biochemical analyses of yeast RNase MRP. Mol. Biol. Rep. 1995;22:75–79. doi: 10.1007/BF00988709. [DOI] [PubMed] [Google Scholar]
- True H.L., Celander D.W. Protein components contribute to active site architecture for eukaryotic ribonuclease P. J. Biol. Chem. 1998;273:7193–7196. doi: 10.1074/jbc.273.13.7193. [DOI] [PubMed] [Google Scholar]
- Wach A., Brachat A., Pohlmann R., Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae . Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Welting T.J., van Venrooij W.J., Pruijn G.J. Mutual interactions between subunits of the human RNase MRP ribonucleoprotein complex. Nucleic Acids Res. 2004;32:2138–2146. doi: 10.1093/nar/gkh539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S., Scott F., Fierke C.A., Engelke D.R. Eukaryotic ribonuclease P: A plurality of ribonucleoprotein enzymes. Annu. Rev. Biochem. 2002;71:165–189. doi: 10.1146/annurev.biochem.71.110601.135352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S., Day-Storms J.J., Srisawat C., Fierke C.A., Engelke D.R. Characterization of conserved sequence elements in eukaryotic RNase P RNA reveals roles in holoenzyme assembly and tRNA processing. RNA. 2005;11:885–896. doi: 10.1261/rna.7282205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziehler W.A., Day J.J., Fierke C.A., Engelke D.R. Effects of 5′ leader and 3′ trailer structures on pre-tRNA processing by nuclear RNase P. Biochemistry. 2000;39:9909–9916. doi: 10.1021/bi000603n. [DOI] [PubMed] [Google Scholar]
- Ziehler W.A., Morris J., Scott F.H., Millikin C., Engelke D.R. An essential protein-binding domain of nuclear RNase P RNA. RNA. 2001;7:565–575. doi: 10.1017/s1355838201001996. [DOI] [PMC free article] [PubMed] [Google Scholar]