Abstract
Hypersensitive response (HR), a form of programmed cell death, is frequently associated with plant disease resistance. It has been proposed that mitogen-activated protein kinase (MAPK) cascades regulate HR cell death based on pharmacological studies by using kinase inhibitors. However, direct evidence is lacking. Here, we demonstrate that NtMEK2, a MAPK kinase, is upstream of salicylic acid-induced protein kinase (SIPK) and wounding-induced protein kinase (WIPK), two tobacco MAPKs that are activated by various pathogens or pathogen-derived elicitors. Expression of a constitutively active mutant of NtMEK2 induces HR-like cell death in tobacco, which is preceded by the activation of endogenous SIPK and WIPK. In addition, NtMEK2-SIPK/WIPK cascade appears to control the expression of 3-hydroxy-3-methylglutaryl CoA reductase (HMGR) and l-phenylalanine ammonia lyase (PAL), two defense genes encoding key enzymes in the phytoalexin and salicylic acid biosynthesis pathways. These results demonstrate that a plant MAPK cascade controls multiple defense responses against pathogen invasion.
The active defense mechanisms of plants against invading pathogens often include rapid cell death, known as the hypersensitive response (HR), the activation of a complex array of defense genes, and the production of antimicrobial phytoalexins (1, 2). Several lines of evidence suggest HR cell death during plant disease resistance is a form of programmed cell death, as it requires active transcription and translation and is genetically defined (3–5). Although the details for the regulation and execution of HR remain poorly understood, production of reactive oxygen species, ion fluxes, protein phosphorylation/dephosphorylation, and gene activation have been implicated in the process (5). In addition to these local responses, the uninfected portions of the plant usually develop systemic acquired resistance, which is manifested as enhanced resistance to a subsequent challenge by the initial or even unrelated pathogens (2).
Plant-defense responses are initiated by the recognition of pathogens, which is mediated either by a gene-for-gene interaction between a plant resistance (R) gene and a pathogen avirulence (Avr) gene or by the binding of nonrace-specific elicitors such as elicitins to a putative receptor (1, 6–9). Signals generated by such interactions are transduced into cellular responses via several interlinked pathways. Recent studies suggest that plant mitogen-activated protein kinase (MAPK) cascade is one of the converging points after the perception of different pathogens and pathogen-derived elicitors (10–15).
MAPK cascades are major pathways by which extracellular stimuli are transduced into intracellular responses in yeast and mammalian cells (16, 17). The basic assembly of MAPK cascade is a three-kinase module conserved in all eukaryotes. MAPK, the last kinase in the three-kinase cascade, is activated by dual phosphorylation of Thr and Tyr residues in a TXY motif by MAPK kinase (MAPKK). MAPKK is, in turn, activated by MAPKK kinase. In mammals, three of the five subgroups of the MAPK family, the stress-activated protein kinase/Jun N-terminal kinase (SAPK/JNK), the p38 kinase, and extracellular signal-regulated protein kinase 5, are activated in response to various stress signals, including UV and ionizing radiation, hyperosmolarity, oxidative stress, and cytokines (16, 17). The outcome of SAPK/JNK and/or p38 activation depends on the magnitude and duration of their activation. A transient activation of these MAPKs induces various defense responses and allows the cells to adapt to an adverse environment. By contrast, a persistent activation leads to apoptosis (16, 17).
The induction of MAPK-like activities has been associated with HR cell death in tobacco cells treated with xylanase, a fungal elicitor from Trichoderma viride and Arabidopsis cells treated with harpin, a bacterial elicitor from Pseudomonas syringae pv. Syringae (18, 19). Through the use of kinase inhibitors, we linked the activation of salicylic acid (SA)-induced protein kinase (SIPK) and wounding-induced protein kinase (WIPK), two tobacco MAPKs, to HR cell death induced by fungal elicitins (20). SIPK was first identified as SA-induced protein kinase (21). It is also activated by various biotic and abiotic stresses, much like mammalian JNK/SAPK (12, 16). WIPK, which was first cloned as a gene whose transcript is induced by wounding (22), is activated by pathogens and pathogen-derived elicitors as well (10, 11, 20). Its ortholog in parsley, ERMK, is also implicated in plant-defense response (13). However, the exact roles of these MAPKs remain unknown. Here, we report the identification of NtMEK2, a tobacco MAPKK. as the upstream kinase for both SIPK and WIPK based on in vitro and in vivo evidence. Expression of NtMEK2DD, a constitutively active mutant of NtMEK2, activates endogenous SIPK and WIPK and leads to HR-like cell death and the activation of a subset of defense genes, demonstrating that the NtMEK2-SIPK/WIPK cascade controls multiple defense responses during plant disease resistance.
Materials and Methods
Cloning of Tobacco MAPKKs.
Two primers, 5′-YTIGARTWYATGGAYVRNGG-3′ and 5′-CKYTCNGGNSHCATRTA-3′ (where Y = T + C; I = deoxyinosine; R = A + G; W = A + T; V = A + C + G; N = A + T + C + G; K = T + G; S = C + G; and H = A + T + C), which correspond to the conserved MAPKK domains LEF/YMDG/Q/RG and YMS/APER, were used to PCR amplify the cDNA reverse transcribed from poly(A)+ RNAs prepared from tobacco (Nicotiana tabacum cv. Xanthi nc). The reverse transcription–PCR products were cloned into pGEM-T vector (Promega). Four unique clones with homology to MAPKKs were identified, which were used to screen a tobacco ZapExpress cDNA library. Positive clones containing the longest insert were sequenced.
Preparation of Recombinant Proteins.
An NdeI site was introduced in front of the ATG start codon of MAPKs or MAPKKs by PCR, which was then ligated in-frame into the pET-28a(+) vector (Novagen). Mutations were introduced by QuickChange site-directed mutagenesis (Stratagene) and confirmed by sequencing. BL21(DE3) cells transformed with pET-28a(+) constructs were induced with 0.5 mM isopropyl β-D-thiogalactoside for 3 h. His-tagged proteins were purified by using nickel columns (Amersham Pharmacia), desalted, and concentrated by using Centricon-10 (Millipore).
Antibody Preparation and Immunoblot Analysis.
NtMEK2 antibody was raised in rabbit against purified recombinant protein corresponding to the N-terminal 156 amino acids of NtMEK2 that share the least homology with other tobacco MAPKKs (Zymed). The antibody was affinity purified before use.
For immunoblot analysis, 15 μg of total protein or 2 ng of recombinant protein per lane was separated on 10% SDS–polyacrylamide gels, and the proteins were transferred to nitrocellulose membranes (Schleicher & Schuell). The blots were then probed with either Anti-Flag (1:5,000, Sigma) or Ab-NtMEK2 antibody (0.5 μg/ml), as previously described (10).
Preparation of Protein Extracts.
Proteins were extracted from cells or leaf discs as previously described (10, 23). The concentration of protein extracts was determined by using the Bio-Rad protein assay kit with BSA as standard.
Kinase Assays.
Autophosphorylation assay was performed by incubating equal amounts (0.5 μg) of purified recombinant NtMEK2 and its mutants in reaction buffer (25 mM Tris, pH 7.5/10 mM MnCl2/1 mM EGTA/1 mM DTT) in the presence of 25 μM [γ-32P]ATP (≈3,000 dpm/pmol) at 30°C for 30 min. The reaction was stopped by the addition of SDS loading buffer, and kinase activities were detected by autoradiography after SDS/PAGE.
Phosphorylation activities of NtMEK2 and its mutants were determined by using inactive mutant MAPKs (HisMAPKR, 1 μg) as substrates under the same conditions as autophosphorylation assay, except 0.1 μg of HisNtMEK2 was used in each reaction.
MAPK activation assay was carried out by first incubating HisMAPKs (1 μg) with 0.1 μg of HisNtMEK2 or its mutant in the presence of 50 μM unlabeled ATP at 30°C for 15 min. Myelin basic protein (MBP, final concentration of 0.25 μg/μl) and [γ-32P]ATP (1 μCi per reaction) were then added. The reactions were stopped by the addition of SDS loading buffer after 30 min. The phosphorylated MBP was visualized by autoradiography after being resolved on a 15% SDS–polyacrylamide gel.
Immune complex–kinase assays of NtMEK2, SIPK, WIPK, and Flag-tagged NtMEK2 were performed by using 75 μg of protein extract with 4 μg of Ab-NtMEK2, 2 μg of Ab-p48N, 2 μg of Ab-p44N, or 2 μg of anti-Flag (Sigma), respectively, as previously described (10). The in-gel kinase activity assay was performed as described previously (21).
Agrobacterium-Mediated Transient Transformation.
Tobacco plants (N. tabacum cv. Xanthi nc [NN] and N. tabacum cv. Xanthi nc [NN]/NahG transgenic) were grown at 22°C in a growth room programmed for a 14-h light cycle. Seven to eight-week-old tobacco plants were used for experiments. Tobacco MAPKKs and their mutants with a Flag-epitope at their N termini were inserted into the XhoI/SpeI sites of pTA7002 vector (24). The 5′-untranslated region was replaced with the Ω sequence from tobacco mosaic virus. Agrobacterium tumefaciens LBA4404 carrying different constructs was grown overnight in AB medium (28) containing 100 μg/ml streptomycin, 50 μg/ml kanamycin, and 100 μM acetosyringone. Cells were collected by centrifugation (4,000 × g), resuspended to OD600 of 0.8 in MS medium (pH 5.9; Life Technologies) with 100 μM acetosyringone, and infiltrated into the fully expanded leaves. The expression of transgene was induced by infiltration of dexamethasone (DEX, 30 μM) 40–48 h later. Samples for protein and RNA preparation were collected at indicated times, quick frozen in liquid N2, and stored at −80°C until use.
Results
Identification of Upstream Kinase of SIPK and WIPK.
Attempts to manipulate the activity of SIPK or WIPK by overexpressing these two genes in sense or antisense orientation were unsuccessful (S.Z. and D. Klessig, unpublished data). As a result, we turned our attention to their upstream kinase(s). By using a yeast two-hybrid screening, a MAPKK that specifically interacts with SIPK and is likely to be SIPK kinase was identified (25). To identify WIPK kinase, we cloned six tobacco MAPKKs that belong to four distinct groups on the basis of phylogenetic analysis (data not shown). One member from each of these four groups was studied in detail. Quite unexpectedly, our data demonstrated that a single MAPKK, NtMEK2, is the upstream kinase for both SIPK and WIPK. The deduced amino acid sequence of NtMEK2 is shown in Fig. 1. It is most closely related to Arabidopsis AtMEK4 of unknown function.
Figure 1.
Alignment of tobacco NtMEK2 with Arabidopsis AtMEK4 (BAA28830) and human MEK1 (Q02750). Roman numerals indicate the 11 conserved subdomains of the kinase catalytic domain. Numbers in parentheses indicate the percentage of amino acid sequence identity to the NtMEK2. The conserved Ser/Thr residues (Thr-227, Ser-233, and Thr-237) between subdomains VII and VIII for MAPKKs are marked with asterisks underneath. The conserved Lys-111 that is important for the ATP binding is marked with a dot.
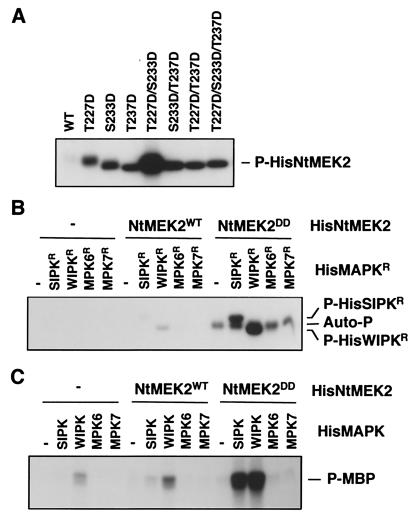
In yeast and animals, MAPKKs are activated through the phosphorylation of two Ser/Thr residues in a conserved S/TxxxS/T motif by MAPKK kinases (17). When these two Ser/Thr residues are replaced with Glu (E) or Asp (D), the mutant MAPKK becomes constitutively active (26, 27). To generate such a mutant of NtMEK2 for gain-of-function studies, we mutated several Ser/Thr residues between the kinase subdomains VII and VIII to D as single, double, or triple mutants. The activities of the His-tagged recombinant proteins were determined by an autophosphorylation assay. Fig. 2 shows that HisNtMEK2DD, where the conserved Thr-227 and Ser-233 were replaced by Asp, has much higher kinase activity than the wild-type protein. Mutagenesis of other tobacco MAPKKs at corresponding positions also leads to increases in kinase activity (data not shown). These results suggest that plant MAPKKs have an activation motif with two Ser/Thr residues separated by five amino acids instead of the three in MAPKKs from yeast and animals (Fig. 1).
Figure 2.
Constitutively active NtMEK2 mutant phosphorylates and activates SIPK and WIPK. (A) NtMEK2DD, the T227D/S233D double mutant of NtMEK2, has elevated kinase activity. Autophosphorylation activities of 0.5 μg of His-tagged wild-type (WT) NtMEK2 and its mutants were determined as described in Materials and Methods. (B) NtMEK2DD preferentially phosphorylates SIPK and WIPK. Phosphorylation activities of HisNtMEK2WT and HisNtMEK2DD (0.1 μg) were determined by using inactive mutant MAPKs (HisMAPKR, 1 μg) as substrates. Reactions in the absence (−) of either MAPK or MAPKK were used as controls. (C) Phosphorylation by NtMEK2DD activates SIPK and WIPK. Wild-type MAPKs (HisMAPK, 1 μg) were incubated in the absence (−) or with 0.1 μg of HisNtMEK2WT or HisNtMEK2DD in the presence of 50 μM unlabeled ATP. MBP (final concentration of 0.25 μg/μl) and [γ-32P]ATP (1 μCi per reaction) were then added. Phosphorylated MBP (P-MBP) that reflects the activity of MAPK was visualized by autoradiography after SDS/PAGE.
For a MAPKK to function as the upstream kinase of either SIPK or WIPK, this MAPKK should be able to phosphorylate and activate SIPK or WIPK. In addition, this MAPKK should be activated before or concurrently with the activation of SIPK or WIPK in vivo. To test whether NtMEK2 is able to phosphorylate SIPK or WIPK, inactive HisMAPKR proteins with the catalytic essential Lys (K) residue substituted with an Arg (R) were used as substrates. HisMAPKR proteins lack autophosphorylation activity (Fig. 2B, first five lanes) and are, therefore, ideal substrates for assaying MAPKK activity. As shown in Fig. 2B, HisNtMEK2DD phosphorylated HisSIPKR and HisWIPKR, but not HisMPK6R or HisMPK7R, two other tobacco MAPKs, whereas HisNtMEK2WT showed little activity. Phosphorylation of HisSIPK and HisWIPK by the active HisNtMEK2DD enhanced their activities toward MBP (Fig. 2C). There is a 50- to 100-fold increase in the activity of NtMEK2DD over that of NtMEK2WT, which correlates with its ability to activate SIPK and WIPK (Fig. 2). That NtMEK2 might be the upstream kinase for both SIPK and WIPK is not too surprising, as SIPK and WIPK share almost 75% identity and belong to the two most closely related subfamilies of plant MAPKs (21).
NtMEK2 Is Activated Concurrently with SIPK and WIPK in Elicitin-Treated Cells.
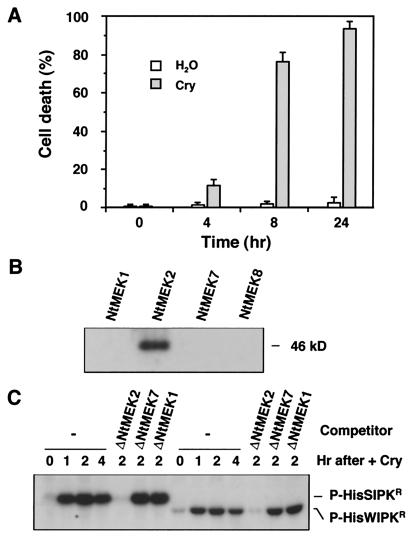
Our previous results demonstrated that the long-lasting activation of SIPK and delayed activation of WIPK are associated with the cell death induced by cryptogein, a fungal elicitin from phytopathogenic Phytophthora spp. (20, 23). As shown in Fig. 3A, treatment of tobacco cells with 25 nM cryptogein led to ≈80% cell death within 8 h, as determined by the fluorescein diacetate method (20). Within 24 h, the shrinkage of cytoplasm, a hallmark of HR cell death, was clearly visible (data not shown). To study the activation of endogenous MAPKK(s) in these cells, antibody was raised against recombinant protein corresponding to the N-terminal 156-aa unique region of NtMEK2. The specificity of Ab-NtMEK2 antibody was tested by immunoblot analysis against different full-length recombinant tobacco MAPKKs. As shown in Fig. 3B, Ab-NtMEK2 recognized NtMEK2 but not NtMEK1, NtMEK7, or NtMEK8. After the specificity of the Ab-NtMEK2 was established, it was used in an immune complex–kinase assay to determine whether NtMEK2 is activated during elicitin-induced HR cell death. As shown in Fig. 3C, NtMEK2 was activated before HR cell death and concurrently with the activation of SIPK and WIPK in vivo (20), indicating that NtMEK2 is the upstream kinase of SIPK and WIPK in the same pathway. The truncated NtMEK2, against which the antibody was raised, but not the corresponding regions of NtMEK1 and NtMEK7, could compete for the binding of the Ab-NtMEK2, demonstrating the specificity of the immune complex–kinase assay. The N-terminal portion of NtMEK8, ΔNtMEK8, is insoluble. As a result, it cannot be used as a competitor in the immune complex–kinase assay. Nonetheless, immunoblot analysis indicated that Ab-NtMEK2 does not crossreact with NtMEK8 (Fig. 3B).
Figure 3.
Activation of NtMEK2 during HR-like cell death initiated by fungal elicitin. (A) Cryptogein (Cry), a fungal elicitin from Phytophthora cryptogea, induces HR-like cell death in tobacco suspension cells. Cells were treated with cryptogein at a final concentration of 25 nM, and cell viability was monitored by the fluorescein diacetate method (20) at the indicated times. Data represent the mean of three replicates ±SE. (B) Ab-NtMEK2 specifically recognized NtMEK2 protein. Two nanograms each of the purified recombinant NtMEK1, NtMEK2, NtMEK7, and NtMEK8 were subjected to immunoblot analysis with Ab-NtMEK2. (C) Activation of NtMEK2 in cells treated with cryptogein. Protein extracts (75 μg) from cryptogein-treated cells were immunoprecipitated with purified Ab-NtMEK2 (4 μg) in the absence (−) or the presence of the N-terminal portion of NtMEK2 (ΔNtMEK2), NtMEK7 (ΔNtMEK7), or NtMEK1 (ΔNtMEK1) as competitor. Kinase activity of the immune complex was subsequently assayed with HisSIPKR or HisWIPKR as a substrate.
NtMEK2 Activates SIPK and WIPK in Vivo.
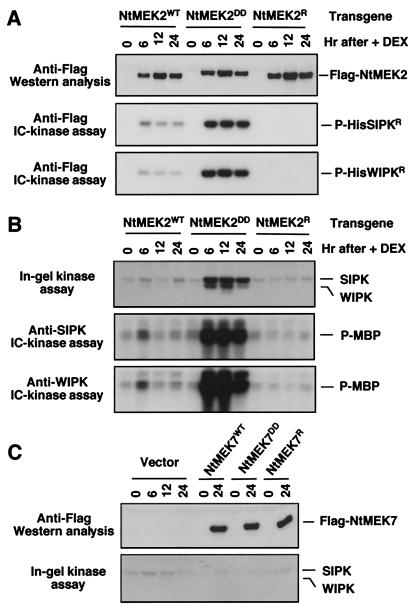
To study the function of NtMEK2 in plant disease resistance signaling, we used a steroid-inducible transient transformation system (24, 28). Agrobacterium cells carrying Flag-tagged NtMEK2WT or active NtMEK2DD constructs were infiltrated into tobacco leaves. An inactive mutant of NtMEK2, NtMEK2R, was used as a negative control. Expression of transgene was induced by the application of DEX 40–48 h later. As shown in Fig. 4A Top, expression of all three proteins was detectable 6 h after the application of DEX, whereas no Flag-tagged protein was present before induction (0 h) or in leaves infiltrated with vector control (Fig. 4C Upper). Expression of neither NtMEK2WT nor NtMEK2R activated any MAPKs as determined by an in-gel kinase assay (Fig. 4B Top); however, cells expressing NtMEK2DD showed high and long-lasting activation of a 48- and a 44-kDa kinase (Fig. 4B Top), which is similar to the activation of SIPK and WIPK in tobacco challenged by tobacco mosaic virus or suspension cells treated with fungal elicitins (10, 20, 23). Immune complex–kinase assay by using SIPK- and WIPK-specific antibodies confirmed that they were SIPK and WIPK (Fig. 4B Middle and Bottom). Activation of SIPK and WIPK in vivo correlated with high MAPKK activity of Flag-NtMEK2DD as demonstrated by immune complex–kinase assay by using HisSIPKR and HisWIPKR as substrates (Fig. 4A Middle and Bottom). The very low basal activity of NtMEK2WT was not sufficient to activate either SIPK or WIPK in vivo (Fig. 4B).
Figure 4.
Expression of NtMEK2DD activates SIPK and WIPK in tobacco. (A) Induction of NtMEK2 and its mutants by steroid in Agrobacterium-mediated transient transformation. Tobacco leaves were infiltrated with Agrobacterium carrying pTA7002 constructs. DEX (30 μM) was infiltrated 40 h later, and samples were taken at indicated times. The expression of transgenes was monitored by immunoblot analysis by using anti-Flag antibody (Top). The kinase activities of Flag-tagged NtMEK2 and its mutants were determined by immune complex (IC)-kinase assay with HisSIPKR (Middle) or HisWIPKR (Bottom) as a substrate. (B) Induction of NtMEK2DD expression activates SIPK and WIPK in vivo. The MAPK activities in cells after DEX treatment were determined by an in-gel kinase assay with MBP substrate (Top). The identities of the two kinases were confirmed by immune complex–kinase assays by using SIPK-specific (Ab-p48N; Middle) or WIPK-specific (Ab-p44N; Bottom) antibody. (C) Expression of NtMEK7DD does not activate SIPK or WIPK. The levels of NtMEK7 transgene expression were determined by immunoblot analysis (Upper). Endogenous SIPK and WIPK activities were detected by an in-gel kinase assay with MBP as a substrate (Lower).
Activation of SIPK and WIPK by NtMEK2DD is selective, as the expression of active mutants of other tobacco MAPKKs, including NtMEK1DD, NtMEK7DD, and NtMEK8DD, did not activate SIPK or WIPK, even though these proteins were expressed at a similar level (Fig. 4C and data not shown).
Induction of NtMEK2DD Expression Activates Defense Responses.
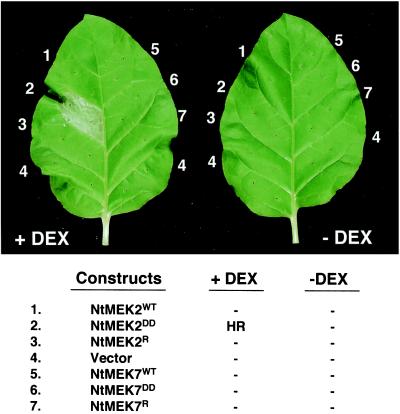
The activation of SIPK and WIPK by ectopically expressed NtMEK2DD leads to the activation of several defense responses. Within 8–12 h after the application of DEX, cell death in small areas could be seen. By 24–36 h, the whole area infiltrated with Agrobacterium carrying NtMEK2DD collapsed, a phenotype identical to the HR elicited by various pathogens or pathogen-derived elicitors (Fig. 5). The timing for the appearance of cell death depends on the age of the leaves. In older leaves, cell death happens much later because of delayed and lower level expression of transgene after DEX application. Besides the similarity at the macroscopic level, cell death induced by NtMEk2DD is associated with membrane leakage, as determined by Evens–Blue staining and shrinkage of cytoplasm (data not shown), which are diagnostic for HR cell death. These results support our hypothesis based on inhibitor studies that SIPK and WIPK are involved in regulating HR cell death (20). Active mutants of other tobacco MAPKKs, including NtMEK7DD, do not cause HR-like cell death under the same condition (Fig. 5 and data not shown), suggesting that this phenotype is caused by a specific MAPK pathway.
Figure 5.
Induction of NtMEK2DD expression leads to HR-like cell death. Different sections of a tobacco leaf were infiltrated with Agrobacterium carrying indicated constructs or vector control, and DEX (30 μM) was applied 40 h later. Solvent (0.1% of ethanol) was used in the control leaf (−DEX). Photo was taken 30 h after application of DEX. In the table, HR denotes the development of an HR-like necrosis in the leaf section; — indicates no visible phenotype.
SA plays important roles in the activation of several defense responses (2, 5). To test whether SA is required for NtMEK2DD-induced cell death, transgenic Xanthi nc plants expressing the NahG gene were used. NahG encodes the SA-metabolizing enzyme salicylate hydroxylase (29). As a result, the SA-dependent responses in such transgenic plants are compromised. Comparison of the phenotype of NahG and wild-type plants indicated that SA is not required for NtMEK2DD-induced cell death. Interestingly, we noticed consistently that the leaves collapse a few hours earlier in the NahG plants (data not shown).
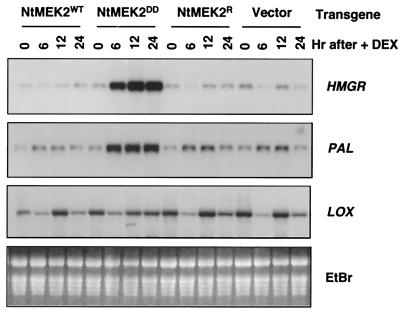
In addition to the HR-like cell death, expression of NtMEK2DD also activated a subset of defense genes. As shown in Fig. 6, the steady-state levels of 3-hydroxy-3-methylglutaryl CoA reductase (HMGR) and L-phenylalanine ammonia lyase (PAL) mRNAs were greatly elevated in leaves expressing NtMEK2DD. Within 6 h after DEX application, the levels of HMGR and PAL transcripts nearly peaked, which was significantly earlier than cell death at 12 h in this particular experiment. This result strongly argues against the possibility that the defense gene activation is a secondary effect caused by cell death. HMGR catalyzes the rate-limiting step in the biosynthesis of all isoprenoid compounds including capsidiol, the major tobacco phytoalexin accumulated during a defense response (30), whereas PAL catalyzes the first committed step in the biosynthesis of phenylpropanoid by converting L-phenylalanine to trans-cinnamic acid, the precursor for lignins and SA biosynthesis (31). Activation of these two genes is prolonged and correlates with SIPK/WIPK activity in the cells. Again, expression of the active mutants of NtMEK1, NtMEK7, and NtMEK8 under the same conditions failed to give this phenotype (data not shown). Previously, we have linked fungal elicitor-induced PAL expression with SIPK activation by the use of kinase inhibitor (23). In contrast, the mRNA levels of lipoxygenase (LOX) (32), another pathogen-activated defense gene, did not correlate with the SIPK/WIPK activities (Fig. 6), suggesting that LOX expression is not controlled by this MAPK pathway.
Figure 6.
Induction of NtMEK2DD expression leads to the activation of a subset of defense genes. Total RNA was isolated from samples collected at the same time as those for protein analyses (Fig. 4). Equal amounts of RNA (10 μg) were electrophoresed on a 1.2% formaldehyde-agarose gel and transferred to Zeta-Probe membrane (Bio-Rad). The transcript levels of HMGR, PAL, and LOX were determined by sequential probing with [α-32P]CTP random primer-labeled cDNA inserts as previously described (23). An ethidium bromide-stained gel was used to show equal loading of samples.
Discussion
Similar defense responses are elicited in plants by viral, bacterial, and fungal pathogens, suggesting shared signaling pathway(s) (1, 8). Increasing evidence demonstrated that SIPK/WIPK or their orthologs in other plant species are converging points after the perception of different pathogens (11–13). In this paper, we report that NtMEK2 is an upstream kinase for both SIPK and WIPK based on in vitro and in vivo evidence. Expression of a constitutively active mutant of NtMEK2 induces HR-like cell death in tobacco, which is preceded by the activation of endogenous SIPK and WIPK. In addition, NtMEK2-SIPK/WIPK cascade appears to control the expression of HMGR and PAL, two defense genes encoding key enzymes in the phytoalexin and SA biosynthesis pathways. These results demonstrate that the NtMEK2-SIPK/WIPK cascade controls multiple defense responses during plant disease resistance. By the use of a conditional system, the correlation between SIPK/WIPK activation and the induction of defense responses is strengthened in this gain-of-function study. Furthermore, parallel studies of three other tobacco MAPKKs, which failed to activate SIPK/WIPK and give similar phenotypes, demonstrated the specificity of the NtMEK2-SIPK/WIPK cascade. Because MAPKs are the sole substrates of MAPKKs, the phenotypes generated by expressing NtMEK2DD are likely the results of endogenous SIPK and WIPK activation. This greatly reduced the possibility of nonspecific effects, as the magnitude of SIPK/WIPK activation induced by expression of NtMEK2DD is similar to that induced by pathogens or pathogen-derived elicitors (10, 20, 23).
The finding that SIPK and WIPK share a common upstream MAPKK could explain why the activation of SIPK and WIPK during stress responses is frequently associated (10, 11, 20). In tobacco suspension cells treated with fungal elicitins (20, 23) or plants infected by tobacco mosaic virus (10), there is a prolonged activation of SIPK, followed by a delayed activation of WIPK. This delay is caused by the requirement of gene activation and de novo synthesis of WIPK protein (20). In contrast, SIPK is transiently activated alone in wounded tobacco leaves or suspension cells treated with fungal cell wall elicitor (23, 33). This is likely a result of the combination of a low basal level of WIPK protein in untreated tobacco plants/cells and a short duration of NtMEK2 activation. In cells treated with fungal cell wall elicitor, WIPK protein eventually accumulates to a high level (20). However, it is not accompanied by the activation of WIPK, possibly because of the absence of NtMEK2 activity. As a result, it appears that the presence or absence of WIPK protein and the duration of NtMEK2 activation could influence whether only SIPK or both SIPK and WIPK are activated in response to a stress signal.
An alternative explanation for differential activation of SIPK and WIPK is the presence of other unidentified MAPKKs that can act on SIPK or WIPK alone. By using a yeast two-hybrid screening, a tobacco MAPKK was identified as a SIPK-interacting protein (25). This MAPKK was named SIPKK, although its phosphorylation activity toward SIPK is yet to be demonstrated. NtMEK1 is identical to SIPKK except for three amino acid residues, which may represent variation of the same gene in different tobacco cultivars. NtMEK1 shares 30.7% amino acid identity with NtMEK2. Overexpression of NtMEK1DD did not activate SIPK or WIPK in tobacco (data not shown), suggesting that NtMEK1 is not the upstream kinase.
HR cell death, as well as a number of other defense responses such as the generation of reactive oxygen species and activation of defense genes, can be blocked by protein kinase inhibitors (18, 20, 23, 34, 35). Potential targets for the kinase inhibitor could be at the recognition step, as several R genes, including Pto and Xa21, encode protein kinases (8) or kinases immediately downstream of R protein, such as Pto-interacting kinase, Pti (36). By adding kinase inhibitor at various times after the cells were challenged with fungal elicitin, we demonstrated that a prolonged activation of a kinase and/or a kinase activated late in the signal pathway is required for plant cells to commit to the death program (20). The kinetics of SIPK and WIPK activation matches this profile. In this study, we provide more direct evidence linking SIPK and WIPK with HR cell death and defense gene activation. Activation of SIPK and WIPK by NtMEK2DD is not accompanied by increase in H2O2 production (data not shown). This is in accord with a previous finding by using kinase inhibitor that these two MAPKs are not required for H2O2 production during plant-defense responses (11). Interestingly, we found that H2O2 can activate SIPK in tobacco cells (S.Z. and D. Klessig, unpublished data). Very recently, it was reported that AtMPK6 and AtMPK3, the SIPK and WIPK orthologs in Arabidopsis, can also be activated by H2O2 (37). It is likely that the signaling pathway leading to H2O2 burst separates from the pathway leading to MAPK activation after the perception of pathogens or elicitors, and the H2O2 generated can then feed into the MAPK pathway, forming a positive feedback loop. The potential role(s) of such a positive feedback in plant disease resistance remains to be evaluated.
In mammalian cells, activation of the SAPK/JNK and p38 kinase plays important roles in stress-induced apoptosis (16, 17). Recently, it was shown that SAPK/JNK regulates cytochrome c release in the mitochondrial death signaling pathway (38). How plant HR cell death, a form of programmed cell death, is regulated and how NtMEK2-SIPK/WIPK cascade is integrated in the signaling pathway(s) remain to be discovered. Nevertheless, the finding that a plant MAPK pathway may play a role similar to JNK/SAPK is very intriguing. The role(s) that SIPK and WIPK each plays in the NtMEK2DD-induced HR-like cell death and defense gene activation is currently unknown. On the basis of kinase inhibitor experiments, we hypothesized that SIPK is involved in the activation of defense gene expression (23), whereas the prolonged activation of SIPK, perhaps in combination with the activation of WIPK, turns on the HR cell death pathway (20, 23). Future analyses of transgenic plants or mutants in which the expression/activation of either SIPK or WIPK is altered should clarify the role of each MAPK in the signaling pathway(s).
Acknowledgments
We thank Dr. N.-H. Chua (Rockefeller Univ., New York) for pTA7002 vector; Drs. D. Klessig and A. Guo (Rutgers Univ., Piscataway, NJ) for tobacco cDNA library and HMGR and LOX cDNA clones; Drs. W. Folk and A. Kenzior (Univ. of Missouri, Columbia) for plasmid-carrying Ω-leader sequence and Flag epitope; and Drs. T. Guilfoyle, D. Randall, and J. Walker for critical reading of the manuscript. S.Z. is supported by National Science Foundation and University of Missouri–Monsanto grants. K.Y. was partially supported by a Korea Science and Engineering Foundation fellowship.
Abbreviations
- MAPK
mitogen-activated protein kinase
- MAPKK
MAPK kinase
- SA
salicylic acid
- SIPK
salicylic acid-induced protein kinase
- WIPK
wounding-induced protein kinase
- HR
hypersensitive response
- MBP
myelin basic protein
- SAPK
stress-activated protein kinase
- JNK
Jun N-terminal kinase
- DEX
dexamethasone
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF325168).
References
- 1.Scheel D. Curr Opin Plant Biol. 1998;1:305–310. doi: 10.1016/1369-5266(88)80051-7. [DOI] [PubMed] [Google Scholar]
- 2.Yang Y, Shah J, Klessig D F. Genes Dev. 1997;11:1621–1639. doi: 10.1101/gad.11.13.1621. [DOI] [PubMed] [Google Scholar]
- 3.Pennell R I, Lamb C. Plant Cell. 1997;9:1157–1168. doi: 10.1105/tpc.9.7.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greenberg J T, Guo A, Klessig D F, Ausubel F M. Cell. 1994;77:551–563. doi: 10.1016/0092-8674(94)90217-8. [DOI] [PubMed] [Google Scholar]
- 5.Richberg M H, Aviv D H, Dangl J L. Curr Opin Plant Biol. 1998;1:480–485. doi: 10.1016/s1369-5266(98)80039-3. [DOI] [PubMed] [Google Scholar]
- 6.Keen N T. Annu Rev Genet. 1990;24:447–463. doi: 10.1146/annurev.ge.24.120190.002311. [DOI] [PubMed] [Google Scholar]
- 7.Staskawicz B J, Ausubel F M, Baker B J, Ellis J G, Jones J D G. Science. 1995;268:661–667. doi: 10.1126/science.7732374. [DOI] [PubMed] [Google Scholar]
- 8.Martin G B. Curr Opin Plant Biol. 1999;2:273–279. doi: 10.1016/S1369-5266(99)80049-1. [DOI] [PubMed] [Google Scholar]
- 9.Ricci P. In: Plant-Microbe Interaction. Stacey G, Keen N T, editors. Vol. 3. New York: Chapman & Hall; 1997. pp. 53–75. [Google Scholar]
- 10.Zhang S, Klessig D F. Proc Natl Acad Sci USA. 1998;95:7433–7438. doi: 10.1073/pnas.95.13.7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Romeis T, Piedras P, Zhang S, Klessig D F, Hirt H, Jones J D G. Plant Cell. 1999;11:273–287. doi: 10.1105/tpc.11.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang S, Klessig D F. Results Probl Cell Differ. 2000;27:65–84. doi: 10.1007/978-3-540-49166-8_6. [DOI] [PubMed] [Google Scholar]
- 13.Ligterink W, Kroj T, zur Nieden U, Hirt H, Scheel D. Science. 1997;276:2054–2057. doi: 10.1126/science.276.5321.2054. [DOI] [PubMed] [Google Scholar]
- 14.Lebrun-Garcia A, Ouaked F, Chiltz A, Pugin A. Plant J. 1998;15:773–781. doi: 10.1046/j.1365-313x.1998.00269.x. [DOI] [PubMed] [Google Scholar]
- 15.Stratmann J W, Ryan C A. Proc Natl Acad Sci USA. 1997;94:11085–11089. doi: 10.1073/pnas.94.20.11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kyriakis J M, Avruch J. BioEssays. 1996;18:567–577. doi: 10.1002/bies.950180708. [DOI] [PubMed] [Google Scholar]
- 17.Widmann C, Gibson S, Jarpe M B, Johnson G L. Physiol Rev. 1999;79:143–180. doi: 10.1152/physrev.1999.79.1.143. [DOI] [PubMed] [Google Scholar]
- 18.Suzuki K, Yano A, Shinshi H. Plant Physiol. 1999;119:1465–1472. doi: 10.1104/pp.119.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Desikan R, Clarke A, Atherfold P, Hancock J T, Neill S J. Planta. 1999;210:97–103. doi: 10.1007/s004250050658. [DOI] [PubMed] [Google Scholar]
- 20.Zhang S, Liu Y, Klessig D F. Plant J. 2000;23:339–347. doi: 10.1046/j.1365-313x.2000.00780.x. [DOI] [PubMed] [Google Scholar]
- 21.Zhang S, Klessig D F. Plant Cell. 1997;9:809–824. doi: 10.1105/tpc.9.5.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seo S, Okamoto M, Seto H, Ishizuka K, Sano H, Ohashi Y. Science. 1995;270:1988–1992. doi: 10.1126/science.270.5244.1988. [DOI] [PubMed] [Google Scholar]
- 23.Zhang S, Du H, Klessig D F. Plant Cell. 1998;10:435–449. doi: 10.1105/tpc.10.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aoyama T, Chua N-H. Plant J. 1997;11:605–612. doi: 10.1046/j.1365-313x.1997.11030605.x. [DOI] [PubMed] [Google Scholar]
- 25.Liu Y, Zhang S, Klessig D F. Mol Plant–Microbe Interact. 2000;13:118–124. doi: 10.1094/MPMI.2000.13.1.118. [DOI] [PubMed] [Google Scholar]
- 26.Mansour S J, Matten W T, Hermann A S, Candia J M, Rong S, Fukasawa K, VandeWounde G F, Ahn N G. Science. 1994;265:966–970. doi: 10.1126/science.8052857. [DOI] [PubMed] [Google Scholar]
- 27.Wurgler-Murphy S M, Maeda T, Witten E A, Saito H. Mol Cell Biol. 1997;17:1289–1297. doi: 10.1128/mcb.17.3.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang X, Frederick R D, Zhou J, Halterman D A, Jia Y, Martin G B. Science. 1996;274:2060–2063. doi: 10.1126/science.274.5295.2060. [DOI] [PubMed] [Google Scholar]
- 29.Delaney T P, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gut-Rella M, Kessmann H, Ward E, Ryals J. Science. 1994;266:1247–1250. doi: 10.1126/science.266.5188.1247. [DOI] [PubMed] [Google Scholar]
- 30.Hammerschmidt R. Annu Rev Phytopathol. 1999;37:285–306. doi: 10.1146/annurev.phyto.37.1.285. [DOI] [PubMed] [Google Scholar]
- 31.Dixon R A, Paiva N L. Plant Cell. 1995;7:1085–1097. doi: 10.1105/tpc.7.7.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veronesi C, Rickauer M, Fournier J, Pouenat M L, Esquerre-Tugaye M T. Plant Physiol. 1996;112:997–1004. doi: 10.1104/pp.112.3.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang S, Klessig D F. Proc Natl Acad Sci USA. 1998;95:7225–7230. doi: 10.1073/pnas.95.12.7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levine A, Tenhaken R, Dixon R, Lamb C. Cell. 1994;79:583–595. doi: 10.1016/0092-8674(94)90544-4. [DOI] [PubMed] [Google Scholar]
- 35.He S Y, Bauer D W, Collmer A, Beer S V. Mol Plant-Microbe Interact. 1994;7:289–292. doi: 10.1094/mpmi-7-0573. [DOI] [PubMed] [Google Scholar]
- 36.Zhou J, Loh Y-T, Bressan R A, Martin G B. Cell. 1995;83:925–935. doi: 10.1016/0092-8674(95)90208-2. [DOI] [PubMed] [Google Scholar]
- 37.Kovtun Y, Chiu W-L, Tena G, Sheen J. Proc Natl Acad Sci USA. 2000;97:2940–2945. doi: 10.1073/pnas.97.6.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tournier C, Hess P, Yang D D, Xu J, Turner T K, Nimnual A, Bar-Sagi D, Jones S N, Flavell R A, Davis R J. Science. 2000;288:870–874. doi: 10.1126/science.288.5467.870. [DOI] [PubMed] [Google Scholar]