Abstract
The chemical structures and accumulation kinetics of several major soluble as well as wall-bound, alkali-hydrolyzable compounds induced upon infection of Arabidopsis thaliana leaves with Pseudomonas syringae pathovar tomato were established. All identified accumulating products were structurally related to tryptophan. Most prominent among the soluble substances were tryptophan, β-d-glucopyranosyl indole-3-carboxylic acid, 6-hydroxyindole-3-carboxylic acid 6-O-β-d-glucopyranoside, and the indolic phytoalexin camalexin. The single major accumulating wall component detectable under these conditions was indole-3-carboxylic acid. All of these compounds increased more rapidly, and camalexin as well as indole-3-carboxylic acid reached much higher levels, in the incompatible than in the compatible P. syringae/A. thaliana interaction. The only three prominent phenylpropanoid derivatives present in the soluble extract behaved differently. Two kaempferol glycosides remained largely unaffected, and sinapoyl malate decreased strongly upon bacterial infection with a time course inversely correlated with that of the accumulating tryptophan-related products. The accumulation patterns of both soluble and wall-bound compounds, as well as the disease resistance phenotypes, were essentially the same for infected wild-type and tt4 (no kaempferol glycosides) or fah1 (no sinapoyl malate) mutant plants. Largely different product combinations accumulated in wounded or senescing A. thaliana leaves. It seems unlikely that any one of the infection-induced compounds identified so far has a decisive role in the resistance response to P. syringae.
Arabidopsis thaliana has a unique role as a study object not only for various aspects of plant molecular genetics and development, but also for the analysis of disease resistance mechanisms (1, 2). In the latter context, emphasis has so far been placed largely on pathogen-derived signal perception (3–5), the nature and role of resistance genes and their products (6, 7), intracellular and intercellular signal transduction (8–13), and the mechanisms of defense-related gene activation (14, 15). In contrast, little is known about the intermediates and end products of primary and secondary metabolism, although their modulations are likely to be the major purpose of all of the numerous induced changes in transcriptional activity occurring in infected and the surrounding plant cells.
Phenylpropanoid biosynthetic pathways are among the most frequently observed metabolic activities that are transcriptionally induced upon infection of plants with pathogens or treatment of plant tissue or cultured plant cells with pathogen-derived elicitors. However, with the possible exceptions of an apparent role of salicylic acid in defense-related signal transduction (10, 16, 17) and an as yet unexplained close relationship between the A. thaliana RPM1 disease resistance locus and the induction of an aromatic alcohol dehydrogenase (18, 19), no direct functional link between phenylpropanoid metabolism and pathogen defense in A. thaliana has been demonstrated. In particular, not a single phenylpropanoid compound has been identified in this species that would have a decisive role in disease resistance, despite the invariably observed strong induction of phenylpropanoid biosynthetic mRNAs and enzymes upon infection or elicitor treatment of A. thaliana (20, 21). This apparent discrepancy is in contrast to the UV-light response in A. thaliana, which has been shown to be closely related to the accumulation of flavonoid glycosides and sinapoyl esters as well as the transcriptional activation of the responsible genes (22–24).
A. thaliana is a member of the Brassicaceae family, which is known for the frequent occurrence of various indolic constituents, including several indolyl glucosinolates and closely related indolyl thiazoles, including camalexin and 6-methoxycamalexin (25, 26). Both camalexin and 6-methoxycamalexin have antibiotic activity and are induced upon infection and hence are considered to act as phytoalexins (27). In A. thaliana, camalexin, but not 6-methoxycamalexin, has been shown to accumulate in response to infections and to fulfill all of the criteria of a bona fide phytoalexin. However, an active role in disease resistance could be demonstrated only in some of the plant/pathogen interactions tested (28–30).
In extension of previous studies on the regulation and functional significance of secondary product accumulation during plant/pathogen interactions (31–34), we now report on the chemical nature and timing of changes in the levels of both phenolic and indolic compounds at pathogen infection sites in A. thaliana leaves.
Materials and Methods
Plant Material.
A. thaliana seeds, ecotype Col-0 or Ler-1, were germinated overnight at 4°C, and seedlings were grown for 14 days on Murashige–Skoog plates (Sigma) without sucrose (Sigma M5524) and then transplanted to soil. The total period of plant growth was 5–6 weeks under controlled short-day conditions (8 h light/16 h dark) or an additional 10 days in the dark to obtain senescing leaves. Wounding was performed by squeezing with a pair of forceps without injuring the midrib. After the appropriate treatment, leaves or leaf discs (see below) were harvested, frozen in liquid N2, and stored at −80°C.
Bacterial Infections.
Pseudomonas syringae pathovar tomato, strains DC3000 (Pstc) or DC3000 containing the avirulence gene avrRpm1 (Psti), were grown overnight at 28–30°C in 50 ml medium as described (35), centrifuged for 5 min at 4,000 rpm, washed twice with 50 ml 10 mM MgCl2, and adjusted to an OD600 of 0.004, or 2 × 106 colony-forming units/ml, in 10 mM MgCl2. About 10–20 μl of the bacterial suspension was injected with a syringe via the stomata into the lower surface of half of a leaf. Control leaves were treated similarly with a sterile solution of 10 mM MgCl2. At the appropriate times after inoculation, 4-mm discs were removed from the leaves with a cork borer.
Extraction Procedure and HPLC Analysis.
Frozen leaf material (≈100–150 mg) was homogenized with a small pistil in an Eppendorf vessel, shaken at room temperature for 15 min with 50% aqueous methanol (vol/vol; ≈20 μl/10 mg fresh weight), and then centrifuged for 15 min at 15,000 × g. This extraction procedure was repeated once, and supernatants were combined where appropriate. The solvent was removed at 40°C with a Speed-Vac (Eppendorf), and the residue was redissolved in 50% aqueous methanol (20 μl/10 mg initial fresh weight) and subjected to HPLC on a Nucleosil C-18 column (ET 250/4, 120–5; Macherey & Nagel), with 0.1% trifluoroacetic acid as solvent A and 98% acetonitrile/0.1% trifluoroacetic acid as solvent B, at a flow rate of 1.3 ml/min at 30°C (gradient solvent A: 100% at 0, 100% at 1, 66% at 25, 46% at 30, 20% at 35, 20%/100% at 37, and 100% at 50 min), and a Photodiode Array Detector 540 at 254 nm as part of the Biotech System (Solvent Delivery System 522, Autosampler 565, Jet-Stream plus, Degasy DG 1210, software chroma 2000; Biotech, Neufahrn, Germany). For preparative HPLC, a SS 250/0.5"/10 Nucleosil 7 column and the respective part of the gradient were used under otherwise identical conditions.
NMR Spectroscopy.
General procedure.
NMR analyses were carried out on a Bruker DRX 500 spectrometer at 500.13 MHz (1H) and 125.76 MHz (13C) in MeOH-d4. 1H NMR, homonuclear correlation spectra [1H-(1)H correlated spectroscopy], heteronuclear multiple bond correlation, and heteronuclear multiple quantum coherence experiments were recorded in a 2.5-mm inverse detection microprobe head with the use of standard Bruker pulse sequences. Broadband decoupled 13C and distortionless enhancement by polarization transfer (DEPT) spectra were run with the use of a 2.5-mm broadband microprobe head. Tetramethylsilane was used as an internal reference. Metabolites were identified as follows.
Tryptophan (C).
1H NMR: δ 7.70, dd, J = 8.0, 1.0 Hz, H-4′; 7.36, dd, J = 8.3, 1.0 Hz, H-7′; 7.20, s, H-2′; 7.13, ddd, J = 8.3, 7.0, 1.0 Hz, H-6′; 7.04, ddd, J = 8.0, 7.0, 1.0 Hz, H-5′; 3.85, dd, J = 9.5, 3.9 Hz, H-2; 3.52, dd, J = 15.3, 3.9 Hz, H-3a; 3.14, dd, J = 15.3, 9.5 Hz, H-3b. 13C NMR chemical shifts were obtained from heteronuclear multiple bond correlation and heteronuclear multiple quantum coherence spectra: δ 138.4, C-8′; 128.4, C-9′; 124.9, C-2′; 123.5, C-6′, 120.0, C-5′; 119.0, C-4′, 112.7, C-7′; 109.6, C-3′; 56.6, C-2; 28.3, C-3; C-1 not detected.
Indole-3-carboxylic acid (L).
1H NMR: δ 8.06, dd, J = 7.7, 1.3 Hz, H-4; 7.94, s, H-2; 7.43, dd, J = 7.7, 1.3 Hz, H-7; 7.19, ddd, J = 7.7, 7.7, 1.3 Hz, H-6; 7.16, ddd, J = 7.7, 7.7, 1.3 Hz, H-5. 13C NMR: δ 138.2, C-8; 133.4, C-2; 127.6, C-9; 123.6, C-6; 122.4, C-5; 122.0, C-4; 112.9, C-7; 108.7, C-3; COOH not detected. The 13C NMR shift values of C-3 and C-9 were obtained from the heteronuclear multiple bond correlation spectrum.
6-Hydroxyindole-3-carboxylic acid 6-O-β-d-glucopyranoside (D).
1H NMR: δ 7.94, d, J = 8.8 Hz, H-4; 7.87, s, H-2; 7.21, d, J = 2.1 Hz, H-7; 7.02, dd, J = 8.8, 2.1 Hz, H-5; 4.91, d, J = 7.6 Hz, H-1′; 3.92, dd, J = 12.1, 2.1 Hz, H-6′a; 3.72, dd, J = 12.1, 5.3 Hz, H-6′b; 3.47, m, H-2′ and H-3′; 3.44, m, H-5′; 3.41, m, H-4′. 13C NMR: δ 156.0, C-6; 138.7, C-8; 133.5, C-2; 123.5, C-9; 122.5, C-4; 114.1, C-5; 109.5, C-3; 103.5, C-1′; 100.7, C-7; 78.2 and 78.1, C-3′ and C-5′; 75.1, C-2′; 71.5, C-4′; 62.6, C-6′; COOH not detected. The 13C NMR shift values of C-3 and C-9 were obtained from the heteronuclear multiple bond correlation spectrum.
β-d-Glucopyranosyl indole-3-carboxylic acid (E).
1H NMR: δ 8.08, dd, J = 7.6, 1.5 Hz, H-4; 8.06, s, H-2; 7.45, dd, J = 7.6, 1.5 Hz, H-7; 7.21, ddd, J = 7.6, 7.6, 1.5 Hz, H-6; 7.20, ddd, J = 7.6, 7.6, 1.5 Hz, H-5; 5.73, d, J = 7.9 Hz, H-1′; 3.87, dd, J = 12.0, 2.1 Hz, H-6′a; 3.73, dd, J = 12.0, 5.0 Hz, H-6′b; 3.54, dd, J = 7.9, 9.5 Hz, H-2′; 3.50, dd, J = 9.5, 8.6 Hz, H-3′; 3.46, ddd, 9.0, 2.1, 5.0 Hz, H-5′; 3.43, dd, 8.6, 9.0 Hz, H-4′. 13C NMR: δ 138.3, C-8; 134.3, C-2; 127.5, C-9; 124.0, C-6; 122.8, C-5; 122.0, C-4; 113.2, C-7; 107.5, C-3; 95.4, C-1′; 78.9, C-5′; 78.3, C-3′; 74.3, C-2′; 71.3, C-4′; 62.5, C-6′; COOH not detected.
Mass Spectrometry.
MS analysis was performed on a Quattro II (Micromass, Manchester, UK) tandem quadrupole mass spectrometer (geometry quadrupole-hexapole-quadrupole) equipped with either an atmospheric pressure chemical ionization or an electrospray ionization source. For atmospheric pressure chemical ionization measurements of indole-3-carboxylic acid (L), the corona pin was operated at 3.5 kV and the sample cone at 17 V. A mixture of acetonitrile and water (1:1) was used as the solvent, at a flow rate of 0.5 ml/min. Vaporization was achieved with a nitrogen sheath gas (300 liters/h) and drying gas (150 liters/h) at 400°C. For electrospray ionization measurements of D and E, the capillary voltage was 3.5 kV and the cone voltage was 14 V. Water (containing 0.1% trifluoroacetic acid) was used as a spraying solvent, at a flow rate of 0.1 ml/min, and nitrogen was used as both the electrospray ionization nebulization (20 liters/h) and drying gases (150 liters/h) at 150°C. Standard mass spectra were measured by using the first quadrupole analyzer only. Product ion mass spectra were recorded by setting the first quadrupole to transmit the parent ion of interest and scanning the second quadrupole from m/z 50 to m/z 350. Argon was used as a collision gas at 2 × 10−3 mbar. A collision energy of 8 eV was used to achieve fragmentation of the glucose ester and glucoside, and a collision energy of 18 eV was used for the free aglycone. PseudoMS (3) analysis of D and E was achieved by setting the cone voltage to 27 eV and selecting source-generated fragment ions (m/z 178 and 163, respectively) with the use of the first quadrupole. Their product ions, generated in the collision cell at 18 eV, were analyzed by scanning the second quadrupole from m/z 50 to m/z 185. Purified samples were injected directly into the atmospheric pressure chemical ionization or electrospray ionization solvent flow with a loop injection valve (Rheodyne, Cotati, CA).
Results
Time Courses of Bacterial Infections.
Most of the following experiments were carried out on 6-week-old A. thaliana ecotype Col-0 plants that showed a fully compatible interaction with Pstc and a strongly incompatible interaction with Psti. Inoculation of leaves with Psti resulted within 12 h in callose deposition and tissue disintegration at the bacterial infiltration site, whereas neither response occurred at this time point with Pstc under otherwise identical conditions. Not until 36–48 h after inoculation were the plant responses similar between the two types of interaction with respect to the formation of both callose and water-soaked lesions.
Secondary Product Identification.
Pstc- or Psti-infected leaves were harvested, and discs of infected tissue were prepared 48 h after inoculation, along with similar leaf discs from either mock-treated or, for further comparison, wounded or senescing plants. Multiple samples were combined for each of the respective treatments and extracted with 50% methanol, and the residual tissue was extensively washed, hydrolyzed with alkali, acidified, and extracted with ethyl acetate. The two extracts represented soluble and wall-bound substances, respectively, and were subjected separately to reversed-phase HPLC.
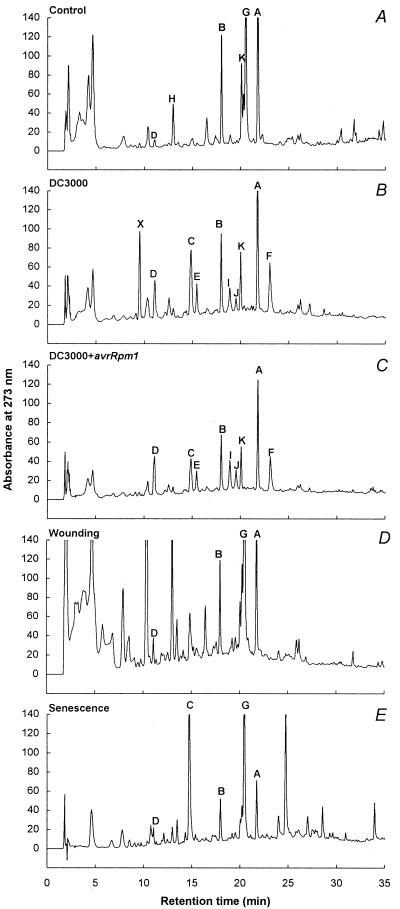
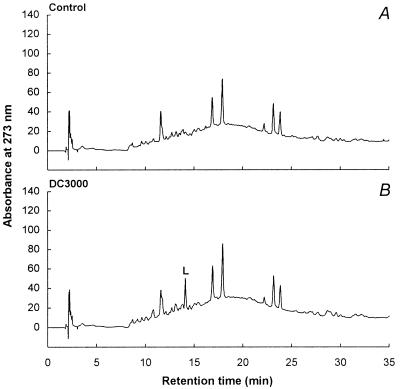
The HPLC chromatograms obtained with soluble products from infected leaves 48 h after inoculation with Pstc or Psti are shown in Fig. 1 A–C. While compounds A and B remained largely unchanged in several independent experiments, compounds C–H and X were strongly affected by both types of infection. Comparatively simple patterns were observed with wall-bound substances (Fig. 2), among which only one strongly Pstc- or Psti-induced product (compound L) was detected. Of all clearly discernible induced peaks in Figs. 1 and 2, those representing compounds I and J occurred with low reproducibility, and a seeming slight decrease in compound K was statistically insignificant. Therefore, compounds I–K were not further analyzed.
Figure 1.
HPLC analysis of soluble extracts from uninfected (A), Pstc- (B) or Psti- (C) infected, wounded (D), and senescing (E) A. thaliana ecotype Col-0 leaves. Plants were grown for 5–6 weeks (A–D) or for an additional 10 days (E), and appropriate leaf areas were harvested 48 h after the indicated treatment. Each analytical run contained 40 μl methanolic extract (50% aqueous methanol). See text and Fig. 3 for an explanation of peak labeling.
Figure 2.
HPLC analysis of extracts derived from cell wall-associated, alkali-hydrolyzed uninfected (A) and Pstc- (B) or Psti- (C) infected A. thaliana leaves. See the legend of Fig. 1 for further explanation. Compound L, indole-3-carboxylic acid.
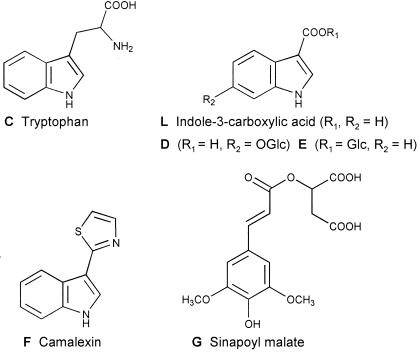
All substances reproducibly occurring in amounts sufficient for purification and chemical identification (A–H, X, and L) were isolated by preparative HPLC and subjected to structural analysis by MS and NMR. Compounds A and B proved to be kaempferol derivatives with unidentified glycosidic residues, compound H could not be unequivocally identified, and compound X is probably a bacterial product, the chemical nature and modes of induction of which will be described elsewhere. The chemical structures of all major plant substances that were strongly affected by Pstc or Psti infections are depicted in Fig. 3: Compounds C–F and L were identified as tryptophan (C), indole-3-carboxylic acid (L), and three biosynthetically related substances (D–F), including camalexin. The only major phenylpropanoid derivative was sinapoyl malate (G).
Figure 3.
Chemical structures of all major compounds accumulating (C, D, E, F, and L) or declining (G) in infected A. thaliana leaf tissue, except for the putative bacterial metabolite X (see text for further explanation). Structural identification was achieved by a combination of UV, MS, and NMR analysis.
At 48 h after inoculation, the patterns of induced changes (Fig. 1 B and C) were very similar for the two infection types. In contrast, wounding (Fig. 1D) and senescence (Fig. 1E) caused partially overlapping but, in each case, distinctly different changes in the profiles of accumulating secondary products. The same applies to wall-bound substances, all of which occurred in relatively small amounts and therefore remained unidentified, except for indole-3-carboxylic acid, which accumulated during senescence but not upon wounding (data not shown).
Timing of Product Accumulation.
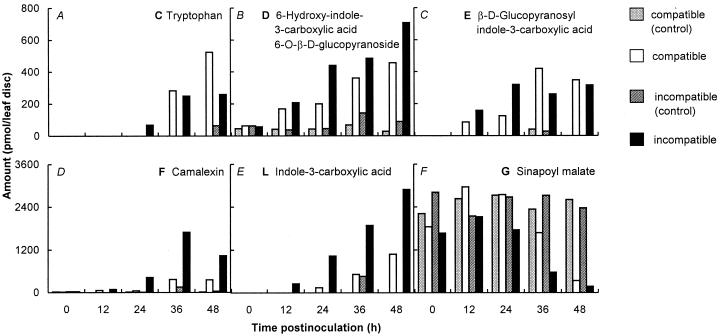
The time courses of induction or repression during the first 48 h after inoculation with Pstc or Psti were determined for all compounds listed in Fig. 3. The five induced substances increased from low or undetectable levels to about 400–600 pmol (C–E) or even 1.5–3 nmol (F and L) per 4-mm infected leaf disk (Fig. 4 A–E). In all cases, the induction was more rapid, and camalexin and indole-3-carboxylic acid reached much higher levels, in Psti than in Pstc infections. The inverse behavior was observed for sinapoyl malate (Fig. 4F), which decreased between 24 and 48 h after inoculation more rapidly and more drastically in the incompatible than in the compatible interaction.
Figure 4.
Timing of product accumulation (A–E) or disappearance (F) in infected A. thaliana leaves. HPLC profiles analogous to those shown in Figs. 1 and 2 were established for all time points given and used for quantification of the indicated compounds. Bars represent data from Pstc control (white) or infected (light gray) and Psti control (dark gray) or infected (black) leaf discs. All data are mean values from two independent experiments.
Effects of tt4 and fah-1 Mutations.
The surprising lack of phenylpropanoid derivatives among all major pathogen-induced compounds prompted us to analyze two A. thaliana mutants [tt4 (= chs) and fah1–7] blocked in the flavonoid and ferulate/sinapate pathways, respectively, for possible effects on the resistance phenotype and the accumulation patterns of tryptophan-related products. In this case A. thaliana ecotype Ler-1, which harbored the tt4 mutation, served as the control. As expected, the tt4 mutant lacked the two kaempferol glycosides (A and B), and the fah1 mutant was devoid of sinapoyl malate (G) and possibly a few minor related compounds, but otherwise the patterns were essentially similar in the respective controls as well as after infection.
Likewise, similar results for wild-type and mutants were obtained for wall-bound substances, where indole-3-carboxylic acid (L) again was the only major induced compound. Moreover, the accumulation profiles of soluble as well as wall-bound products 48 h after inoculation with Psti were also indistinguishable within experimental error between the Ler-1 plants used in these experiments (except for the differences caused by mutations) and the Col-0 plant described above. Finally, neither the compatible nor the incompatible interaction with P. syringae was significantly affected by the tt4 or the fah-1–7 mutation.
Discussion
These results on induced changes in secondary product metabolism in infected A. thaliana leaves are surprising for two reasons. Not only the absence of phenylpropanoid derivatives, but also the abundance of indolic compounds among the major detectable infection-induced compounds was unexpected. Although the extraction procedure may have missed certain compounds with extreme properties, its broad applicability and efficacy had been amply demonstrated before and were confirmed in this study by the efficient and, in most cases, highly reproducible isolation of a wide spectrum of indolic as well as phenolic glycosides and aglycones.
All five major induced compounds isolated by this procedure from infected A. thaliana leaves could be structurally identified and turned out to be indolic substances. One of them, indole-3-carboxylic-acid, accumulated as a cell-wall constituent, probably in the form of an alkali-labile ester, and was not detectable in the soluble extract. Conversely, all other accumulating compounds were found in the soluble extract, but not in association with the cell wall. Of all five indolic substances, only camalexin has been reported previously to be induced at infection sites in A. thaliana. Its antibiotic activity, classification as a phytoalexin, and accumulation mode upon infection have been studied extensively (36, 37). However, a significant role in the defense response of A. thaliana to P. syringae could not be demonstrated; even camalexin-less mutants showed the same disease resistance phenotype as observed for the corresponding wild-type plants (28). Much less is known about possible antibiotic properties of the other induced indolic metabolites, except that some closely related compounds, including indole-3-propionic and -3-acrylic acids, were shown to possess antibacterial activity (38).
Of the five induced compounds, tryptophan is the classical type of primary metabolite, whereas camalexin is a typical product of secondary metabolism. Less unequivocal is the classification of indole-3-carboxylic acid and its glucosidic derivatives, particularly the role of indole-3-carboxylic acid as a cell-wall constituent at infection sites. Plausible assumptions are either a function as an antibiotic agent, a contribution to the formation of a rigid, impenetrable cell-wall barrier within and around infection sites, or a combination of both. In contrast, the two glucosides D and E probably serve as intermediates in indolic secondary metabolism rather than being either primary metabolites or metabolic end products. Compound E in particular has a high probability of serving as an energy-rich intermediate for transfer reactions of the carboxylic acid moiety, because of the high free energy of hydrolysis contained in the glucose ester bond of aromatic or quasiaromatic carboxylic acids (39). Such a role as secondary metabolic intermediate would imply that compound E might be considered to be a precursor of camalexin, either in addition to, or instead of, the previously proposed indole-3-carboxaldehyde (40). In addition, or alternatively, compound E could serve as an energy-rich transport intermediate for the incorporation of indole-3-carboxylic acid into the cell wall.
Probably somewhat lower, but still rather high, is the free energy of hydrolysis contained in aromatic O-glucosides, such as compound D. Thus this compound could likewise have a role as an energy-rich biosynthetic intermediate. 6-O-Substituted indolic compounds have not been reported previously for A. thaliana. However, the occurrence of 6-methoxycamalexin along with camalexin in other Brassicaceae species (25), together with the now established presence of compound D in A. thaliana, suggests that so far undetected amounts of 6-methoxycamalexin and/or other 6-O-substituted indolic end products may occur in this plant as well.
Particularly wide open to interpretation is the accumulation of tryptophan at infection sites. On the one hand, tryptophan-biosynthetic mRNAs and enzymes are strongly induced by infection (41–43). On the other hand, one might intuitively have expected the tryptophan level to decrease rather than increase, considering the strong and rapid accumulation of various indolic secondary metabolites. However, indolic metabolism in Brassicaceae is very complex and includes numerous functionally distinct classes of compounds, including the phytohormone auxin (indole-3-acetic acid; ref. 44) and the herbivore, and possibly pathogen, defense-related indolic glucosinolates (26, 27). None of the respective biosynthetic pathways, nor their interdependence, have been fully elucidated. Even for the extensively investigated biosynthesis of auxin, the picture is rather unclear. Several alternatives have been proposed, including the existence of both tryptophan-dependent and tryptophan-independent pathways in A. thaliana (44–47). Furthermore, although indolic glucosinolates were not among the major substances identified under the present conditions, they are known to be derived from tryptophan and believed to act as phytoanticipins in pathogen defense (26, 27, 48, 49). Thus they may constitute a relatively low-abundance or otherwise undetected group of compounds. A striking example of the large biochemical and physiological complexity of tryptophan-related secondary metabolism, particularly in A. thaliana and other Brassicaceae, is the existence not only of two rather common alternative routes for auxin biosynthesis, but also of a third one involving glucosinolate catabolites (50), indicating a widely ramified metabolic grid for indolic intermediates and end products.
The classification of the two indolic glucosides D and E as metabolic intermediates, indole-3-carboxylic acid and camalexin as more or less stable end products, and tryptophan as a comparatively versatile central metabolite would be in accord with the relative timing of induction (Fig. 4). The levels of compounds D and E in infected tissue started increasing strongly around 12 h after inoculation and did not exceed values of a few hundred picomoles per 4-mm leaf disk during the period of experimentation. In contrast, the accumulation of substantial amounts of camalexin and indole-3-carboxylic acid did not commence until about 24 h after inoculation and reached considerably higher values. Tryptophan increased with similar slow timing and was the only compound analyzed that reached significantly higher levels in the compatible than in the incompatible interaction. It may be interesting to note in this connection that this pattern correlates with the inverse behavior, that is, the accumulation to much lower levels in the compatible interaction, of the two strongly accumulating, putative defense-related compounds, camalexin and indole-3-carboxylic acid, suggesting either lower rates of synthesis or higher rates of degradation, or both, in the compatible interaction for these presumed metabolic end products.
Usage of the term “defense-related” in this context requires further elaboration. Whereas resistance genes encoding functionally decisive proteins with essential roles in defense-related signal transduction have been identified in several plant pathosystems, including the RPM1 gene in A. thaliana (51, 52), single equally decisive secondary metabolites have not been reported in A. thaliana and are still rare in other plant species. Attempts to verify a role in disease resistance for camalexin by using camalexin-less mutants gave negative results in the case of P. syringae (28), but were positive, e.g., for the interaction with the fungus Alternaria brassicicola (30). Comparison of the results shown here in Figs. 1 and 4 indicates that major, most probably decisive, differences between the plant's responses in compatible and incompatible interactions occur well before the accumulation of camalexin and other secondary products, which apparently represents an intermediate stage in the overall defense response. This overall response can be grossly divided into three successive stages: first, pathogen recognition, intracellular and intercellular signaling, oxidative burst, and highly localized barrier formation, often associated with hypersensitive cell death (stage 1); second, defense-related metabolic reprogramming within a confined area of surrounding tissue, including transcriptional induction of phytoalexins and other soluble or wall-bound secondary products (stage 2); and third, the “systemic acquired resistance” response throughout the whole organ or even the entire plant (stage 3). In the present case, the decisive difference between compatible and incompatible interactions obviously lies within stage 1, whereas secondary product accumulation is part of stage 2. Thus the probably rather universal, multicomponent defense response of A. thaliana leaves to various types of pathogens appears to include the local accumulation of secondary compounds as a stereotypical part that may be important or essential for defense in certain cases but not in others, such as the present one.
Of the three nonindolic compounds A, B, and G, the last, sinapoyl malate, has a proven role in UV resistance (22) but not in pathogen defense. Its decline upon infection is in line with both this functional assignment and the previous observation in parsley that the response to UV irradiation is strongly overridden by treatment with a pathogen-derived elicitor (53). However, as this latter effect includes transcriptional repression of previously UV light-induced chalcone synthase, that is, of all branches of flavonoid biosynthesis, it is surprising that the two kaempferol glycosides A and B remained unaltered under the present conditions. This lack of change could be explained in two ways. Either their turnover rates are very low or A. thaliana and parsley differ principally in this respect.
The tt4 and fah1–7 mutants lacking flavonoids and sinapoyl malate, respectively, did not differ significantly from the corresponding wild type in their responses to P. syringae infections. They were included in this study to test their possible effects on indolic pathways and on the numerous interconnections within cellular metabolism. This question seemed particularly relevant in view of both the previously established flavonoid and ferulate/sinapate pathway interactions in A. thaliana (22) and more recent results, again obtained with parsley, indicating that induction of flavonoid biosynthesis has major effects on various supply pathways from primary metabolism (54) and thus may indirectly affect other biosynthetic capacities as well. Clearly, for the three classes of compounds investigated, flavonoid glycosides, sinapoyl esters, and indole derivatives, evidence for mutual interference with product accumulation rates was not obtained. Whether this lack of evidence indicates true metabolic independence or elaborate compensatory mechanisms has yet to be determined.
Another open question concerns the significance of tryptophan accumulation in wounded and senescing leaves, which contrasts with the lack of accumulation of all other infection-induced indolic compounds, except for indole-3-carboxylic acid induction during senescence. This low degree of metabolic overlap was unexpected, considering the frequently observed similarities at the gene expression level (12). Here again, more extensive analyses including less abundant as well as less easily extractable products from various pathways are required to obtain a more detailed answer.
In conclusion, the relative predominance of indole derivates among the compounds accumulating at Psti and Pstc infection sites in A. thaliana leaves is taken as an indication of their functional relevance in pathogen defense in general, even though they might be irrelevant in the particular incompatible interaction analyzed and insufficient in the corresponding compatible interaction. However, relative abundance is not a measure of importance. With the use of additional biochemically defined mutants as well as modified or more refined methods for extraction and analysis of less abundant substances, we expect it to be possible to considerably expand the spectrum of metabolite classes accumulating upon infection in A. thaliana. Phenylpropanoid derivatives will continue to be of prime interest, considering both the strong infection-related induction of many of the responsible mRNAs and enzymes and the apparent importance of at least one such compound, salicylic acid, in defense-related signaling. Furthermore, it should be emphasized that the present investigations were confined to A. thaliana leaves and do not necessarily apply to other organs. To mention but one example of major organ-specific differences, sesquiterpenoid phytoalexins are completely absent in infected potato leaves but accumulate strongly in tubers from the same plant upon infection with the same pathogen (55). A similar degree of system complexity may exist in A. thaliana.
Acknowledgments
We thank Dr. G. Glombitza for help with the identification of camalexin; Dr. J. Dangl for a gift of P. syringae strains; Drs. A. Gierl, I. Somssich, and E. Weiler for helpful comments on the manuscript; and the Fonds der Chemischen Industrie for financial support.
Abbreviations
- Pstc
P. syringae pathovar tomato (strain causing compatible interaction)
- Psti
P. syringae pathovar tomato (strain causing incompatible interactions)
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.021551098.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.021551098
References
- 1.Innes R W. Curr Opin Plant Biol. 1998;1:299–304. doi: 10.1016/1369-5266(88)80050-5. [DOI] [PubMed] [Google Scholar]
- 2.Parker J E, Feys B J, van der Biezen E, Noel L, Aarts N, Austin M J, Botella M A, Frost L N, Daniels M J, Jones J D G. Mol Plant Pathol. 2000;1:17–24. doi: 10.1046/j.1364-3703.2000.00003.x. [DOI] [PubMed] [Google Scholar]
- 3.Ritter C, Dangl J F. Plant Cell. 1996;8:251–257. doi: 10.1105/tpc.8.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Wit P J G M. Trends Plant Sci. 1997;2:452–458. [Google Scholar]
- 5.Grant J J, Mansfield J. Curr Opin Plant Biol. 1999;2:312–319. doi: 10.1016/S1369-5266(99)80055-7. [DOI] [PubMed] [Google Scholar]
- 6.Bent A F. Plant Cell. 1996;8:1757–1771. doi: 10.1105/tpc.8.10.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hammond-Kosack K E, Jones J D G. Annu Rev Plant Physiol Mol Biol. 1997;48:575–607. doi: 10.1146/annurev.arplant.48.1.575. [DOI] [PubMed] [Google Scholar]
- 8.Hammond-Kosack K E, Jones J D G. Plant Cell. 1996;8:1773–1791. doi: 10.1105/tpc.8.10.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baker B, Zambrynski P, Staskawicz B, Dinesh-Kumar S P. Science. 1997;276:726–733. doi: 10.1126/science.276.5313.726. [DOI] [PubMed] [Google Scholar]
- 10.Aarts N, Metz M, Holub E, Staskawicz B J, Daniels M J, Parker J E. Proc Natl Acad Sci USA. 1998;95:10306–10311. doi: 10.1073/pnas.95.17.10306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong X. Curr Opin Plant Biol. 1998;1:316–323. doi: 10.1016/1369-5266(88)80053-0. [DOI] [PubMed] [Google Scholar]
- 12.Maleck K, Dietrich R A. Trends Plant Sci. 1999;6:215–219. doi: 10.1016/s1360-1385(99)01415-6. [DOI] [PubMed] [Google Scholar]
- 13.Grant J J, Loake G J. Plant Physiol. 2000;124:21–29. doi: 10.1104/pp.124.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rushton P J, Somssich I E. In: Plant Microbe Interactions. Stacey G, Keen N T, editors. St. Paul, MN: APS Press; 1999. pp. 251–274. [Google Scholar]
- 15.Eulgem T, Rushton P J, Robatzek S, Somssich I E. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/s1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- 16.Delaney T P. Plant Physiol. 1997;113:5–12. doi: 10.1104/pp.113.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reymond P, Farmer E E. Curr Opin Plant Biol. 1998;1:404–411. doi: 10.1016/s1369-5266(98)80264-1. [DOI] [PubMed] [Google Scholar]
- 18.Kiedrowski S, Kawalleck P, Hahlbrock K, Somssich I E, Dangl J L. EMBO J. 1992;11:4677–4684. doi: 10.1002/j.1460-2075.1992.tb05572.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Somssich I E, Wernert P, Kiedrowski S, Hahlbrock K. Proc Natl Acad Sci USA. 1996;93:14199–14203. doi: 10.1073/pnas.93.24.14199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wanner L A, Li D, Ware I, Somssich I E, Davis K R. Plant Mol Biol. 1995;27:327–338. doi: 10.1007/BF00020187. [DOI] [PubMed] [Google Scholar]
- 21.Ehlting J, Büttner D, Li Q, Douglas C J, Somssich I E, Kombrink E. Plant J. 1999;19:1–12. doi: 10.1046/j.1365-313x.1999.00491.x. [DOI] [PubMed] [Google Scholar]
- 22.Li J, Ou-Lee T-M, Raba R, Amundson R G, Last R L. Plant Cell. 1993;5:171–179. doi: 10.1105/tpc.5.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Landry L G, Chapple C C S, Last R. Plant Physiol. 1995;109:1159–1166. doi: 10.1104/pp.109.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bharti A K, Khurana J P. Photochem Photobiol. 1997;65:765–776. doi: 10.1111/j.1751-1097.1997.tb01923.x. [DOI] [PubMed] [Google Scholar]
- 25.Browne L M, Conn K L, Ayer W A, Tewari J P. Tetrahedron. 1991;47:3909–3914. [Google Scholar]
- 26.Chapple C C S, Shirley B W, Zook M, Hammerschmidt R, Somerville C. In: Arabidopsis: Secondary Metabolism in Arabidopsis. Meyerowitz E M, Somerville C R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. pp. 989–1030. [Google Scholar]
- 27.VanEtten H D, Mansfield J W, Balley J A, Farmer E E. Plant Cell. 1994;6:1191–1192. doi: 10.1105/tpc.6.9.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glazebrook J, Ausubel F M. Proc Natl Acad Sci USA. 1994;91:8955–8959. doi: 10.1073/pnas.91.19.8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rogers E E, Glazebrook J, Ausubel F M. Mol Plant–Microbe Interact. 1996;9:748–757. doi: 10.1094/mpmi-9-0748. [DOI] [PubMed] [Google Scholar]
- 30.Thomma B P H J, Nelissen I, Eggermont K, Broekaert W F. Plant J. 1999;19:163–171. doi: 10.1046/j.1365-313x.1999.00513.x. [DOI] [PubMed] [Google Scholar]
- 31.Hahlbrock K, Scheel D, Logemann E, Nürnberger T, Parniske M, Reinold S, Sacks W R, Schmelzer E. Proc Natl Acad Sci USA. 1995;92:4150–4157. doi: 10.1073/pnas.92.10.4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Batz O, Logemann E, Reinold S, Hahlbrock K. Biol Chem. 1998;379:1127–1135. doi: 10.1515/bchm.1998.379.8-9.1127. [DOI] [PubMed] [Google Scholar]
- 33.Somssich I E, Hahlbrock K. Trends Plant Sci. 1998;3:86–90. [Google Scholar]
- 34.Hagemeier J, Batz O, Schmidt J, Wray V, Hahlbrock K, Strack D. Phytochemistry. 1999;51:629–635. [Google Scholar]
- 35.Debener T, Lehnacker H, Arnold M, Dangl J L. Plant J. 1991;1:289–302. doi: 10.1046/j.1365-313X.1991.t01-7-00999.x. [DOI] [PubMed] [Google Scholar]
- 36.Tsuji J, Jackson E P, Gage D A, Hammerschmidt R, Somerville S C. Plant Physiol. 1992;98:1304–1309. doi: 10.1104/pp.98.4.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsuda K, Toyoda H, Yokoyama K, Wakita K, Nishio H, Nishida T, Dogo M, Kakutani K, Hamda M, Ouchi S. Biosci Biotechnol Biochem. 1993;57:1766–1767. [Google Scholar]
- 39.Mock H-P, Strack D. Phytochemistry. 1993;32:575–579. [Google Scholar]
- 40.Zook M, Hammerschmidt R. Plant Physiol. 1997;113:463–468. doi: 10.1104/pp.113.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keith B, Dong X, Ausubel F M, Fink G R. Proc Natl Acad Sci USA. 1991;88:8821–8825. doi: 10.1073/pnas.88.19.8821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao J, Last R L. Plant Cell. 1996;8:2235–2244. doi: 10.1105/tpc.8.12.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou N, Tootle T L, Glazebrook J. Plant Cell. 1999;11:2419–2428. doi: 10.1105/tpc.11.12.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Normanly J, Bartel B. Curr Opin Plant Biol. 1999;2:207–213. doi: 10.1016/s1369-5266(99)80037-5. [DOI] [PubMed] [Google Scholar]
- 45.Radwanski E R, Last R L. Plant Cell. 1995;7:921–934. doi: 10.1105/tpc.7.7.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Normanly J. Physiol Plant. 1997;100:422–431. [Google Scholar]
- 47.Müller A M, Hillebrand H, Weiler E W. Planta. 1998;206:362–369. doi: 10.1007/s004250050411. [DOI] [PubMed] [Google Scholar]
- 48.Halkier B A, Du L. Trends Biochem Sci. 1997;2:425–431. [Google Scholar]
- 49.Osbourn A E. Plant Cell. 1996;8:1821–1831. doi: 10.1105/tpc.8.10.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bartling D, Seedorf M, Mithöfer A, Weiler E W. Eur J Biochem. 1992;205:417–424. doi: 10.1111/j.1432-1033.1992.tb16795.x. [DOI] [PubMed] [Google Scholar]
- 51.Grant M R, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes R W, Dangl J F. Science. 1995;269:843–846. doi: 10.1126/science.7638602. [DOI] [PubMed] [Google Scholar]
- 52.Boyes D C, Nam J, Dangl J L. Proc Natl Acad Sci USA. 1998;95:15849–15854. doi: 10.1073/pnas.95.26.15849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lozoya E, Block A, Lois R, Hahlbrock K, Scheel D. Plant J. 1991;1:227–234. [Google Scholar]
- 54.Logemann E, Tavernaro A, Schulz W, Somssich I E, Hahlbrock K. Proc Natl Acad Sci USA. 2000;97:1903–1907. doi: 10.1073/pnas.97.4.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rohwer F, Fritzemeier K H, Scheel D, Hahlbrock K. Planta. 1987;170:556–561. doi: 10.1007/BF00402991. [DOI] [PubMed] [Google Scholar]