Abstract
In higher plants the production of starch is orchestrated by chloroplast-localized biosynthetic enzymes, namely starch synthases, ADP-glucose pyrophosphorylase, and starch branching and debranching enzymes. Diurnal regulation of these enzymes, as well as starch-degrading enzymes, influences both the levels and composition of starch, and is dependent in some instances upon phosphorylation-linked regulation. The phosphoserine/threonine-binding 14-3-3 proteins participate in environmentally responsive phosphorylation-related regulatory functions in plants, and as such are potentially involved in starch regulation. We report here that reduction of the ɛ subgroup of Arabidopsis 14-3-3 proteins by antisense technology resulted in a 2- to 4-fold increase in leaf starch accumulation. Dark-governed starch breakdown was unaffected in these “antisense plants,” indicating an unaltered starch-degradation pathway and suggesting a role for 14-3-3 proteins in regulation of starch synthesis. Absorption spectra and gelatinization properties indicate that the starch from the antisense plants has an altered branched glucan composition. Biochemical characterization of protease-treated starch granules from both Arabidopsis leaves and maize endosperm showed that 14-3-3 proteins are internal intrinsic granule proteins. These data suggest a direct role for 14-3-3 proteins in starch accumulation. The starch synthase III family is a possible target for 14-3-3 protein regulation because, uniquely among plastid-localized starch metabolic enzymes, all members of the family contain the conserved 14-3-3 protein phosphoserine/threonine-binding consensus motif. This possibility is strengthened by immunocapture using antibodies to DU1, a maize starch synthase III family member, and direct interaction with biotinylated 14-3-3 protein, both of which demonstrated an association between 14-3-3 proteins and DU1 or DU1-like proteins.
Carbon and nitrogen apportioning in plants has a direct impact on their usefulness as agricultural commodities. Research directed toward altered regulation and reallocation of these assimilates represents a major effort in agricultural biology at both the biochemical and genetic levels. Several key enzymes in the metabolic pathways that direct carbon and nitrogen flow, process assimilates, and transfer the products into various sink tissues are of intense investigative interest. The biosynthesis of starch is one such well-regulated diurnal process (1, 2), with starch serving as a major carbon reserve as well as an energy source for plants and providing a major nutritional value of food crops. The dynamic throughput of plant carbon demands a tight, yet responsive, control of the key enzymes. To further add to the complexity, starch production occurs exclusively in membrane-bound plastids, thereby requiring import of the biosynthetic enzymes and regulators involved. However, localization within the plastid also serves to essentially distinguish enzymes directly involved in starch synthesis and therefore identify potential targets for genetic manipulation.
Starch is synthesized in leaves during the day from photosynthetically assimilated carbon derived via the reductive pentose phosphate pathway. One simple view of starch polymer production involves four types of enzymes: ADP-glucose pyrophosphorylase (AGP), starch synthases (SSs), starch-branching enzymes (SBEs), and starch-debranching enzymes (DBEs) (1). AGP forms ADP-glucose from glucose 1-phosphate. SSs add ADP-glucose to the elongating end of an α(1–4)-linked glucan chain, whereas SBEs cut α(1–4) links and rejoin them as α(1–6) branches that are subsequently trimmed by DBEs to yield short chains for further synthetic extension. However, different isoforms of SSs (soluble and granule-associated SSI, SSII, and SSIII) can participate in the production of branched glucans. For example, the granule-bound SSI, the waxy-encoded protein in maize, is directly and perhaps exclusively involved in producing amylose, an α(1–4) glucan polymer with little branching. In contrast, SSII participates in the synthesis of amylopectin, an α(1–6) branched glucan polymer that typically is found together with amylose to form starch granules. The ratio of these two glucans affects the physical characteristics of starch such as gelatinization and the absorption spectra of iodine-complexed starch. The alteration or absence of certain starch biosynthetic enzymes (3–5) has a dramatic effect on the physical characteristics of starch, as well as the level of starch accumulated by the plant. In a similar, yet opposing, manner dark-regulated starch degradation occurs by means of catabolic enzymes such as amylase, α-glucosidase, and starch phosphorylase. The resulting starch stasis is the consequence of the metabolism and catabolism orchestrated by the respective enzymes.
Regulation of some enzymes involved in major resource allocation is affected by allosteric effectors, substrate levels, and product levels, as well as by phosphorylation (6–8). For several key enzymes, regulation of activity is a two-step process involving phosphorylation of the enzyme, followed by formation of a complex with 14-3-3 proteins to complete the regulatory transition (9, 10). For example, the assimilation of nitrogen for production of amino acids or nucleotide bases is tightly controlled by nitrate reductase (NR). NR responds to environmental signals, such as light and metabolite levels, by phosphorylation and interacts with 14-3-3 proteins (11, 12), thereby rapidly altering nitrogen flux according to the plant's metabolic requirements. This phosphorylation-dependent interaction of NR with 14-3-3 proteins has become a paradigm for posttranslational regulation of metabolic enzymes (9). Recently, 14-3-3 proteins have been identified inside plastids (13), thereby implicating a potential role in starch regulation.
We have investigated the biological role of chloroplast-localized 14-3-3 proteins in carbon partitioning, namely starch accumulation. We found that reduced levels of starch granule-associated 14-3-3 proteins result in a dramatic increase in starch accumulation. On the basis of the presence of 14-3-3 consensus binding domains and biochemical experiments, one target of the granule 14-3-3 proteins appears to be the SSIII family.
Materials and Methods
Antisense GF14 Vector Construction and Transformation into Arabidopsis.
Clones for the Arabidopsis 14-3-3 proteins GF14 ɛ and GF14 μ, from yeast two-hybrid vectors (14), were used as templates for PCR to produce XbaI cassettes that were subsequently subcloned into the binary plant transformation vector pBI121 (CLONTECH). Gene orientation was determined by automated DNA sequencing on a Perkin–Elmer ABI 373A. Clones containing the antisense GF14 gene orientation were amplified in Escherichia coli INVαF′ and used to transform competent Agrobacterium tumefaciens strain EHA105 by the freeze–thaw method (15). The vector-harboring Agrobacterium was used to transform Arabidopsis ecotype WS seedlings by using vacuum infiltration, essentially as described by Bechtold and Pelletier (16). Transformants were screened on germination media plates using 40 μg/ml kanamycin selection as described previously (17). Seed from positive transformants were selected through three successive generations to ensure homozygous transgenic lines. A minimum of 12 antisense lines were generated for both GF14 ɛ and GF14 μ.
Plant Growth.
Arabidopsis plants were grown in constant light at 22°C on germination media plates oriented in a vertical position or in flats of Transplant mix A (Vergro, Tampa, FL). Starch degradation experiments were done by transferring the plants to dark and samples taken at three hour intervals.
Starch Analysis.
Starch was visualized by Lugol's iodine staining reagent (Sigma). Leaves from 10-day-old plants were harvested and blanched in 80% (vol/vol) ethanol. After rinsing with double-distilled water the leaves were stained with Lugol's reagent and briefly destained with water. Stained plants and leaves were photographed with an Olympus SZH10 stereo dissecting microscope and DP10 digital camera.
Enzymatic measurement of starch in leaves was performed by using a method adapted from Zeeman et al. (18). Rosettes were harvested and weighed, then boiled in 80% ethanol. After clearing, the samples were ground in a mortar and pestle in 80% ethanol and the crude starch pellet was recovered by centrifugation at 5,000 rpm for 5 min in a Beckman JA20 rotor and J2–21 centrifuge. The crude starch was resuspended in 80% ethanol and repelleted two more times. The final pellet was dried and resuspended in double-distilled water, then placed at 85°C for 10 min. The starch solution was then digested with 3 mg/ml amyloglucosidase and 20 units of amylase in 20 mM calcium acetate pH 4.5 buffer for 24 h at 37°C. The final concentration of liberated glucose was determined by using a glucose oxidase assay kit (Sigma).
Purified starch granules used for immunological and biochemical studies were extracted from plants by using the Mops-based protocol reported by Zeeman et al. (18). Essentially, whole plants minus the roots were ground in a Mops buffer system, washed with SDS-containing buffer, and finally washed extensively with deionized water. Yields were calculated on a milligrams of isolated starch per gram fresh plant weight basis.
Relative amylose/amylopectin ratios from purified starch granules were assayed by using iodine starch spectral analysis as described by Konishi et al. (19).
Immunolocalization and Blotting.
Transmission electron microscopy using GF14 isoform-specific polyclonal primary antibodies and gold secondary antibodies was used to localize 14-3-3 proteins in the starch granules of Arabidopsis leaves by the method previously described (20). Starch granules for immunoblotting were first treated with thermolysin to ensure removal of surface-associated proteins as described by Mu-Foster et al. (21), and intrinsic starch granule proteins were separated by SDS/PAGE and transferred to nitrocellulose membranes as described (13).
14-3-3 Protein-Binding Motif Analysis of Starch Granule-Associated Proteins.
A blast search (22) for the 14-3-3 phosphoserine/threonine-binding consensus motif (RXXS/TXP) was conducted on the available plant starch-associated protein sequences by using the National Institutes of Health blast web server.
Immunocapture Experiments.
Commercial corn starch (Argo, Englewood Cliffs, NJ) was used as a source of protein complexes for the immunocapture experiments. The starch was first digested with thermolysin to remove surface-associated proteins (21), then washed and digested at 25°C with α-amylase and amyloglucosidase in 100 mM Tris–acetate buffer, pH 7.5, containing 100 mM KCl, 2.5 mM DTT, 10% (vol/vol) glycerol, 25 mM NaF, 3 mM CaCl2, and 0.1% BSA for 3 h by using a protocol adapted from MacDonald and Preiss (23). Undigested material was removed by ultracentrifugation in a Beckman SW55 Ti rotor at 4°C at 50,000 rpm for 30 min. Supernatant was transferred to a plastic conical tube and BSA was added to a final concentration of 0.1%. The supernatant was passed over anti-14-3-3 ɛ- and μ-conjugated Sepharose made from CNBr-activated Sepharose (Amersham Pharmacia Biotech) and the 14-3-3 protein antisera IgG fractions (13). A control column containing antibodies raised against the transcriptional cofactor GIP1 (unpublished data) was used as a negative control. The columns were loaded with the starch-derived protein extract, then washed three times with phosphate-buffered saline (PBS), pH 7.6, containing 25 mM NaF. The processed beads were boiled for 1 min in 2× SDS/PAGE sample buffer. The beads were removed by centrifugation and supernatant was loaded onto 10% polyacrylamide gels before SDS/PAGE. The proteins were transferred to nitrocellulose and blocked overnight with Blotto Tween (24). The membranes were probed with antiserum to the Zea mays (Zm)SSIII DU1 (25). The membrane was washed and incubated with horseradish peroxidase-conjugated antibodies to rabbit IgG. Labeled bands were identified by the process of chemiluminescence, using SuperSignal West Pico Chemiluminescent Substrate according to the supplier's instructions (Pierce).
Biotinylated 14-3-3 Protein Overlay Experiments.
To identify corn starch proteins that are potential targets for 14-3-3 protein binding, proteins from corn starch were separated by electrophoresis and assayed by using a blot overlay procedure with biotinylated recombinant 14-3-3 Zm GF14–12. Zm GF14–12 was expressed in E. coli and purified by nickel-Sepharose chromatography as described previously (26). The protein was dialyzed against 100 mM sodium borate, pH 8.8, overnight before addition of biotinamidocaproate N-hydroxysuccinimide ester in DMSO at a ratio of 50 μg of ester per mg of protein. After 4 h at room temperature, the reaction was terminated by the addition of 1 M ammonium chloride, pH 8.0. The biotinylated 14-3-3 protein was dialyzed exhaustively against PBS over the course of 2 days at 4°C. Proteins from 10 mg of corn starch boiled in SDS/PAGE sample buffer were separated by PAGE and transferred to nitrocellulose, then incubated overnight at 4°C with biotinylated 14-3-3 protein in PBS containing 1% BSA. The blot was washed three times with PBS/1% BSA and incubated for 30 min with streptavidin-conjugated horseradish peroxidase diluted in PBS/1% BSA. The blot was washed three additional times and the 14-3-3-bound protein was identified by using chemiluminescence as described above.
Results and Discussion
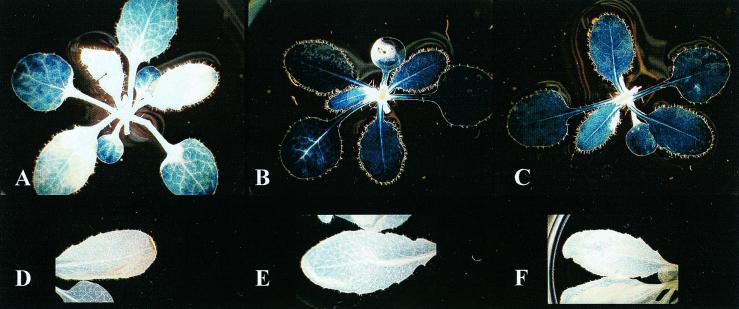
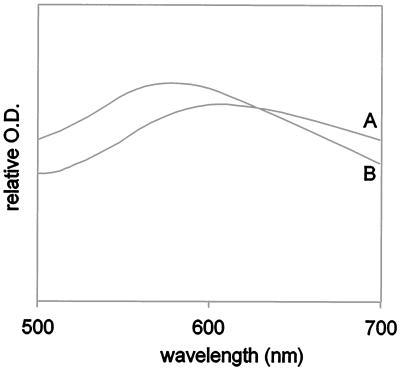
Transgenic Arabidopsis plants expressing antisense cDNA of At 14-3-3s GF14 ɛ and μ, two members of the ɛ subgroup of 14-3-3 proteins, displayed normal growth behavior but demonstrated phenotypic changes relative to wild-type plants with regard to starch accumulation in leaves. Although the absolute level of starch present in the leaves of Arabidopsis depended upon culture conditions and the lines examined, the leaves of plants from all 12 GF14 ɛ and GF14 μ antisense lines consistently accumulated increased starch levels relative to leaves of wild-type plants. Iodine staining indicated that the increased starch accumulation was equally distributed throughout the leaves of the antisense plants (Fig. 1 A–C). Quantitative measurements of the starch present in the leaves of plants grown in constant light revealed an approximately 2-fold increase in total starch content in antisense plants over wild-type plants (28 ± 7 mg of starch per g fresh weight in transgenic plants vs. 15 ± 3 mg of starch per g fresh weight in wild-type plants). The extractable starch from antisense plants was approximately 4-fold higher than that from wild-type plants (43 ± 5 mg of starch per g fresh weight vs. 9 ± 2 mg of starch per g fresh weight, respectively). Isolated starch granules from antisense plants were used to evaluate the absorption spectra of the iodine/starch complex, as an indicator of unbranched and branched glucan ratios. The absorption spectrum of the iodine/starch complex from antisense plants (Fig. 2, spectrum B) was blue-shifted relative to the absorption spectrum of the iodine/starch complex from wild-type plants (Fig. 2, spectrum A), suggesting that the starch from antisense plants has an increase in branched glucan content. This premise is further supported by the observation that the percentage of gelatinizable starch from antisense plants was reduced relative to that found in wild-type plants (data not shown).
Figure 1.
Starch accumulation in transgenic GF14 antisense plants. Starch levels in wild-type (A) and GF14 ɛ (B) and μ (C) antisense plants grown under constant light were assayed by iodine staining. The density of staining clearly indicates increased starch levels in the leaves of antisense plants. Identical photographic lighting and exposure conditions were used for A–C so that the intensities of the staining are directly comparable. Similar plants were subjected to an 18-h dark period to allow for starch degradation before staining (D, E, and F, respectively), and the results indicate that starch degradation is uninhibited in the 14-3-3 antisense plants.
Figure 2.
Altered starch composition of antisense starch granules. The absorption spectra of iodine/starch complexes of wild-type (A) and 14-3-3 ɛ antisense (B) Arabidopsis starch granules indicate that the 14-3-3 antisense plants contain an increase in the relative content of branched glucans.
To determine whether altered degradation rates might be responsible for the elevated starch accumulation in 14-3-3 antisense plants, plants were grown in constant light and harvested after a dark period of 18 h. Iodine staining of leaves at the end of the dark period was indistinguishable between wild-type (Fig. 1D) and antisense plants (Fig. 1 E and F). To measure the rate of starch breakdown, leaf samples were taken every 3 h after the plants were placed in the dark. Wild-type plants degraded starch at a rate of approximately 1 mg of starch per g fresh weight per h, whereas the antisense plants cleared starch from their leaves at rates of approximately 1.3 to 1.5 mg of starch per g fresh weight per h. This result indicates that the starch degradation pathway is fully functional in the antisense plants and suggests that reduced negative regulation of starch biosynthesis is responsible for increased starch in the 14-3-3 antisense plants. The 14-3-3 proteins would therefore appear to function as inhibitory proteins in starch metabolism by normally shutting down starch biosynthesis, thereby playing a key regulatory role in carbon allocation that is similar to their role in nitrogen fixation.
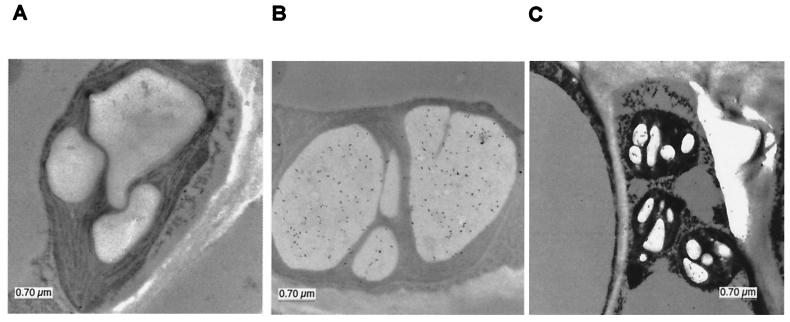
Antibodies to 14-3-3 proteins were used in an immunolocalization electron microscopy experiment looking at starch granules in the leaves of wild-type Arabidopsis. The inside of chloroplast starch granules was densely decorated by antibodies that recognize eight non-ɛ subgroup members (Fig. 3B). Antibodies specific to GF14 ɛ also decorated the inside of starch granules, but more sparsely (Fig. 3C). This limited amount of ɛ in the starch granules of wild-type plants may explain why the antisense plants displayed reduced levels of starch-associated GF14 ɛ, whereas the cytoplasmic levels of ɛ remained reasonably normal (data not shown). These data also indicate that non-ɛ 14-3-3 proteins may be involved in starch biosynthesis, although no phenotypic data yet exist to support this conclusion. The relationship among the 14-3-3 isoforms present in starch grains, as well as the question of whether active forms of 14-3-3 proteins exist as homodimers or heterodimers, is not well established and therefore will need to be addressed in future studies.
Figure 3.
Immunolocalization of 14-3-3 proteins in starch granules. Arabidopsis leaves were processed for electron microscopy (20) and immunolabeled with GF14 antibodies. Control antibodies to Dictyostelium spores (A) did not immunodecorate the granules; however, antibodies that recognize both ɛ (C) and non-ɛ (B) 14-3-3 proteins were concentrated inside the starch granules.
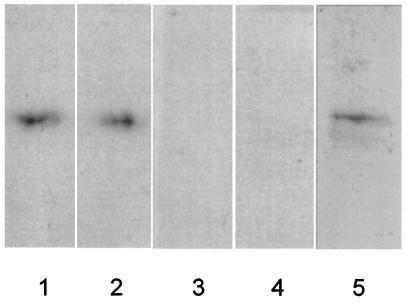
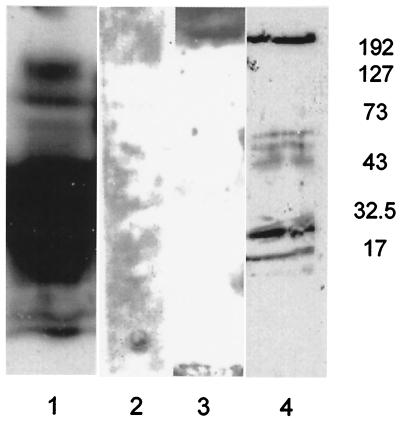
To confirm that 14-3-3 proteins are present within chloroplast starch granules and that increased starch production is a result of decreased 14-3-3 proteins, starch granules from wild-type, GF14 ɛ, and GF14 μ antisense plant leaves were biochemically analyzed for the presence of 14-3-3 proteins. Purified starch granules were incubated with the protease thermolysin to remove external proteins, washed, boiled in SDS/PAGE sample buffer, and analyzed on SDS/PAGE by Western analysis with antibodies specific to 14-3-3 proteins GF14 ɛ or μ (13). Wild-type starch contained both GF14 ɛ and μ (Fig. 4 lanes 1 and 2), whereas antisense starch did not contain detectable amounts of either (Fig. 4 lanes 3 and 4). This coregulated suppression is not surprising, as the identity between cDNAs is ≈70% and therefore both mRNAs are presumably reduced by antisense regulation in planta. Western analysis of whole-leaf extracts did not demonstrate a pronounced decrease in GF14 ɛ and μ proteins (data not shown). Starch granule-specific reduction of GF14 ɛ and μ may be reflective of a selection process for chloroplastid 14-3-3 proteins, perhaps pressured by an as-yet-uncharacterized import mechanism (13). The presence of ɛ and μ 14-3-3 proteins in starch granules is significant in that they appear essential for proper regulation of leaf starch biosynthesis in Arabidopsis. In addition, commercial starch from maize also possesses 14-3-3 proteins (Fig. 4 lane 5), suggesting that 14-3-3 protein regulation of starch synthesis is used by crops and occurs in other plastids, such as amyloplasts, and is not limited to photosynthetically active plastids.
Figure 4.
Reduction in GF14 ɛ and μ protein levels in the starch granules of antisense plants and presence of 14-3-3 proteins in commercial corn starch. Isolated starch granules from wild-type and antisense Arabidopsis were treated with thermolysin to remove externally attached proteins and subjected to SDS/PAGE Western analysis with 14-3-3 protein antibodies (29). Protein extracts from 3 mg of starch from wild-type (lanes 1 and 2), GF14 ɛ antisense (lane 3), and GF14 μ antisense (lane 4) plants were probed with antibodies recognizing GF14 ɛ (lanes 1 and 3) and μ (lanes 2 and 4). A clear reduction of these 14-3-3 isoforms is observed in the starch-granule proteins of antisense plants. A 3-mg sample of commercial corn starch was processed as described above and the blot was probed with antibodies that recognize maize 14-3-3 proteins (lane 5), indicating the presence of 14-3-3 proteins in starch grains from maize.
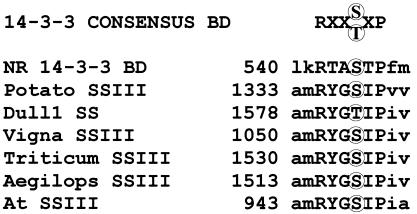
Although a chloroplast-localized 14-3-3 protein partner in starch synthesis has not been reported, a search of all available starch-related enzyme sequences for the consensus 14-3-3-binding motif revealed the SSIII family as an obvious potential target within the plastid (Fig. 5). SSIII members from potato, Arabidopsis, Vigna unguiculata, Aegilops tauschii, Triticum aestivum, and maize all contain a conserved hexapeptide motif very similar to the 14-3-3 protein binding site of NR. This is the only example of an entire family sharing such a highly conserved potential binding site among the plastid enzyme sequences currently available. It is interesting to note that SSIII is directly involved in the production of amylopectin and has significant control over other SS isoforms (4), perhaps explaining both starch accumulation and the qualitative shift in branched glucan content observed in 14-3-3 antisense plants.
Figure 5.
Consensus 14-3-3-binding sites in SSIII coding sequences. The phosphoserine/threonine-containing binding sequence for 14-3-3 proteins is present in all known members of the SSIII family listed in GenBank: SSIII from Vigna unguiculata (Vigna SSIII, AJ225088), SSIII from Solanum tuberosum (Potato SSIII, X94400 and X95759), SSIII DU1 from Zea mays (Dull1 SS, AF023159), SSIII from Triticum aestivum (Triticum SSIII, AF258608), SSIII from Aegilops tauschii (Aegilops SSIII, AF258609), and a predicted SSIII from Arabidopsis thaliana (At SSIII, AL021713). The 14-3-3 protein consensus binding domain (BD) and the NR 14-3-3 binding domain are shown for comparison.
Immunocapture experiments with anti-GF14 column and proteins isolated from processed corn starch were used to experimentally determine whether starch granule 14-3-3 proteins associate directly with SSIIIs. Commercial corn starch was chosen as a source of proteins because of its bulk availability and antibodies to the maize SSIII enzyme were available (25). SDS/PAGE and Western analysis of immunocaptured proteins identified ZmSSIII DU1 as a starch 14-3-3 partner protein (Fig. 6). The molecular masses of the captured protein bands were lower than the mass of intact ZmSSIII DU1 (see below); however, this can be attributed to breakdown of SSIII DU1 during the starch degradation process (25). Although ZmSSIII DU1 was reported as primarily located in the soluble fractions of kernel extracts, low levels of ZmSSIII DU1 in starch were observed in starch granules (25). To confirm that SSIII DU1 is present inside the corn starch grains, and to avoid the degradation observed in the immunocapture experiment, protease-treated commercial starch was boiled in SDS/PAGE sample buffer, separated by electrophoresis, and transferred to nitrocellulose. The blot was then probed with biotinylated recombinant 14-3-3 protein, and bound bands were detected by chemiluminescence (Fig. 6, lane 3). Biotinylated 14-3-3 protein bound to a protein of approximately 200 kDa, whose migration corresponds to a main band recognized by ZmSSIII DU1 antibodies (Fig. 6, lane 4). These data provide correlative support for an interaction between 14-3-3 proteins and DU1 or DU1-like proteins within starch grains, but confirmation of the interaction awaits detailed characterization of the protein complex.
Figure 6.
14-3-3 proteins bind to DU1 or DU1-like SS. Proteins isolated from digested starch were passed over an anti-14-3-3 column and a control column. Bound proteins were eluted, separated by electrophoresis, transferred to nitrocellulose, and probed with antiserum to ZmSSIII DU1. The anti-GF14 column retained the DU1 cross-reactive protein (largely degraded from multiple processing steps) (lane 1), whereas the negative control column did not (lane 2). Proteins extracted directly from gelled starch were separated by electrophoresis and transferred to nitrocellulose. Probing with biotinylated Zm GF14–12 identified a 14-3-3-binding protein of approximately 200 kDa (lane 3). Probing with antiserum to ZmSSIII DU1 labeled proteins of a similar size (lane 4).
The biological significance of choroplastid 14-3-3 proteins, specifically the ɛ subgroup, in starch metabolism is clearly demonstrated through the use of 14-3-3 antisense plants. While we cannot rule out a contributory role in starch accumulation by cytosolic and/or other upstream enzymatic perturbations caused by reduction of 14-3-3 proteins (27, 28), a direct role of in plastid 14-3-3 proteins in regulation of starch accumulation is clearly indicated. Additionally, the increase in branched glucans vs. nonbranched glucans in the antisense plants would seem contrary to simply increasing the cytosolic flux of starch precursors, as would be the effect of altered upstream regulation of starch metabolism. Further experiments are necessary to confirm the interaction between 14-3-3 proteins and SSIIIs or other enzymes regulated in this pathway, and the possibility of other plastid enzymes being regulated by 14-3-3 proteins is not excluded. However, the specific localization of the 14-3-3 proteins in the starch granules should, in this instance, serve to limit the range of possible 14-3-3 protein targets to those enzymes located within starch-producing plastids.
These results suggest a model in which starch composition and accumulation are directly regulated by plastid 14-3-3 proteins. The data presented herein are consistent with a mechanism whereby starch production in continuously illuminated plants is limited through inactivation of SSs by phosphorylation and 14-3-3 protein binding. Without 14-3-3 proteins to complete the inactivation step, starch continues to accumulate beyond normal levels. Further understanding of the regulatory events limiting starch accumulation could result in improved starch productivity in crop plants.
Acknowledgments
We thank Curt Hannah for advice and guidance in experiments involving starch, Alan Myers (Iowa State University) for generously providing the DU1-specific antisera, the Interdisciplinary Center for Biotechnology Research Electron Microscopy Core (University of Florida) for immunomicroscopic analysis, and the Interdisciplinary Center for Biotechnology Research Sequencing Core (University of Florida) for DNA sequence analysis. This research was supported by the U.S. Department of Agriculture Grants 00-35304-9601 and 97-35304-4942 (to R.J.F. and P.C.S.). This article is Florida Agricultural Experiment Station journal series number R-07869.
Abbreviations
- SS
starch synthase
- NR
nitrate reductase
- Zm-
Zea mays
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.021304198.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.021304198
References
- 1.Smith A M. Curr Opin Plant Biol. 1999;2:223–229. doi: 10.1016/S1369-5266(99)80039-9. [DOI] [PubMed] [Google Scholar]
- 2.Preiss J, Sivak M N. Genet Eng. 1998;20:177–223. doi: 10.1007/978-1-4899-1739-3_10. [DOI] [PubMed] [Google Scholar]
- 3.Craig J, Lloyd J R, Tomlinson K, Barber L, Edwards A, Wang T L, Martin C, Hedley C L, Smith A M. Plant Cell. 1998;10:413–426. doi: 10.1105/tpc.10.3.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edwards A, Fulton D C, Hylton C M, Jobling S A, Gidley M, Rossner U, Martin C, Smith A M. Plant J. 1999;17:251–261. [Google Scholar]
- 5.Lloyd J R, Landschutze V, Kossmann J. Biochem J. 1999;338:515–521. [PMC free article] [PubMed] [Google Scholar]
- 6.Sokolov L N, Dejardin A, Kleczkowski L A. Biochem J. 1998;336:681–687. doi: 10.1042/bj3360681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun J, Okita T W, Edwards G E. Plant Physiol. 1999;119:267–276. doi: 10.1104/pp.119.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Imparl-Radosevich J M, Nichols D J, Li P, McKean A L, Keeling P L, Guan H. Arch Biochem Biophys. 1999;362:131–138. doi: 10.1006/abbi.1998.1028. [DOI] [PubMed] [Google Scholar]
- 9.Chung H J, Sehnke P C, Ferl R J. Trends Plant Sci. 1999;4:367–371. doi: 10.1016/s1360-1385(99)01462-4. [DOI] [PubMed] [Google Scholar]
- 10.Sehnke P C, Ferl R J. Curr Biol. 1996;6:1403–1405. doi: 10.1016/s0960-9822(96)00742-7. [DOI] [PubMed] [Google Scholar]
- 11.Bachmann M, Huber J L, Liao P C, Gage D A, Huber S C. FEBS Lett. 1996;387:127–131. doi: 10.1016/0014-5793(96)00478-4. [DOI] [PubMed] [Google Scholar]
- 12.Moorhead G, Douglas P, Morrice N, Scarabel M, Aitken A, MacKintosh C. Curr Biol. 1996;6:1104–1113. doi: 10.1016/s0960-9822(02)70677-5. [DOI] [PubMed] [Google Scholar]
- 13.Sehnke P C, Henry R, Cline K, Ferl R J. Plant Physiol. 2000;122:235–242. doi: 10.1104/pp.122.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu K, Rooney M F, Ferl R J. Plant Physiol. 1997;114:1421–1431. doi: 10.1104/pp.114.4.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holsters M, de Waele D, Depicker A, Messens E, van Montagu M, Schell J. Mol Gen Genet. 1978;163:181–187. doi: 10.1007/BF00267408. [DOI] [PubMed] [Google Scholar]
- 16.Bechtold N, Pelletier G. Methods Mol Biol. 1998;82:259–266. doi: 10.1385/0-89603-391-0:259. [DOI] [PubMed] [Google Scholar]
- 17.Daugherty C J, Rooney M F, Miller P W, Ferl R J. Plant Cell. 1996;8:1239–1248. doi: 10.1105/tpc.8.8.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeeman S C, Northrop F, Smith A M, Rees T. Plant J. 1998;15:357–365. doi: 10.1046/j.1365-313x.1998.00213.x. [DOI] [PubMed] [Google Scholar]
- 19.Konishi Y, Mojima H, Okuno K, Asaoka M, Fuwa H. Agric Biol Chem. 1985;49:1965–1971. [Google Scholar]
- 20.Bihn E A, Paul A L, Wang S W, Erdos G W, Ferl R J. Plant J. 1997;12:1439–1445. doi: 10.1046/j.1365-313x.1997.12061439.x. [DOI] [PubMed] [Google Scholar]
- 21.Mu-Forster C, Huang R, Powers J R, Harriman R W, Knight M, Singletary G W, Keeling P L, Wasserman B P. Plant Physiol. 1996;111:821–829. doi: 10.1104/pp.111.3.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacDonald F D, Preiss J. Plant Physiol. 1983;73:175–178. doi: 10.1104/pp.73.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 25.Cao H, Imparl-Radosevich J, Guan H, Keeling P L, James M G, Myers A M. Plant Physiol. 1999;120:205–215. doi: 10.1104/pp.120.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Vetten N C, Ferl R J. Plant Physiol. 1994;106:1593–1604. doi: 10.1104/pp.106.4.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moorhead G, Douglas P, Cotelle V, Harthill J, Morrice N, Meek S, Deiting U, Stitt M, Scarabel M, Aitken A, MacKintosh C. Plant J. 1999;18:1–12. doi: 10.1046/j.1365-313x.1999.00417.x. [DOI] [PubMed] [Google Scholar]
- 28.Toroser D, Athwal G S, Huber S C. FEBS Lett. 1998;435:110–114. doi: 10.1016/s0014-5793(98)01048-5. [DOI] [PubMed] [Google Scholar]
- 29.Mu-Forster C, Wasserman B P. Plant Physiol. 1998;116:1563–1571. doi: 10.1104/pp.116.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]