Abstract
Unlinked autosomal microsatellites in six Jewish and two non-Jewish populations were genotyped, and the relationships among these populations were explored. Based on considerations of clustering, pairwise population differentiation, and genetic distance, we found that the Libyan Jewish group retains genetic signatures distinguishable from those of the other populations, in agreement with some historical records on the relative isolation of this community. Our methods also identified evidence of some similarity between Ethiopian and Yemenite Jews, reflecting possible migration in the Red Sea region. We suggest that high-resolution statistical methods that use individual multilocus genotypes may make it practical to distinguish related populations of extremely recent common ancestry.
The first reliable evidence of Jewish immigration into the area now contained in Libya records the settlement of Jews from Egypt around 312 BCE (1, 2), and the ancient historian Josephus reports the size of the Libyan Jewish population as 500,000 in the 1st century CE (3). This community, however, was decimated by Roman rulers during revolts ending in 118 CE (2). After a lengthy, severe, and poorly documented bottleneck in population size, Libyan Jews engaged in cultural interactions with Berber tribes that lasted through the 6th century CE (4). During this time, the population absorbed Berber converts, although the proportion of Berber genetic contribution to the Libyan Jews is not known (1, 5). A small number of additional Jewish immigrants may have entered the region from Spain in the 6th century (1), and others may have arrived from Arabia and Syria with the Moslem conquest of Libya in the 7th century (6). The Jewish population seems to have been significant by the 11th century (7), but after persecution and emigration under the Spanish and the Knights of Malta, from 1510 to 1551, it may have been small and mostly rural by the time of the Ottoman conquest in 1551 (1, 4). Unlike other parts of North Africa, Libya did not serve as a major destination of Iberian Jews seeking refuge after their 1492 expulsion from Spain (2). Although occasional Jewish immigrants arrived in Libya from Jewish communities in Italy and elsewhere, over the last 400 years the Libyan Jews were mostly isolated from all other Jewish populations (1, 4). The Libyan Jews eventually numbered more than 30,000 before the emigration of 1949–1951, when most members of the group moved to Israel (4).
Precise population size estimates of the Libyan Jews do not exist before this century (4). One traveler reported the Jewish population of Tripoli to be about 3,000 in 1783 (1); a 1906 study estimated 12,000 Jews in Tripoli and about 20,000 in Libya as a whole (4). Because Tripoli was the largest Jewish city and because little migration appears to have taken place into the Libyan Jewish population over the last 400 years, it seems reasonable to suggest that the group reached its largest population size of modern times at the time of its evacuation.
The few early records of the Libyan Jewish community indicate diverse origins and a series of population size fluctuations. Because of a dearth of information and the potential for preservation bias among sources documenting influential Jewish immigrants from Italy and Spain, it is difficult to quantify the contributions of different genetic groups to the population that eventually became the modern Libyan Jews—the ancient Jews, the Berber converts, and the possible sources of immigrants between the 5th and 15th centuries CE. A few facts appear to be clear from available records. First, this population was relatively secluded over the past 400 or more years, with its greatest demographic changes involving in situ population growth and urbanization of rural communities (1, 2, 6). Thus, for at least 400 years and possibly longer, the group can be regarded as a small population isolate. Second, the demographic influences that acted on the Jews of Libya were different from those of other Jewish communities of North Africa. The Jewish populations of Algeria, Morocco, and Tunisia likely had later origins than the Libyan Jews had; they were subject to different conquests from those that brought immigrants to Libya; their hypothesized Berber admixture would have involved tribes different from those who lived in Libya; and importantly, they received a sizeable influx of Jews fleeing Spain and Portugal during the 14th to 16th centuries (2, 3).
Despite historical information suggesting the possibility that Libyan Jews were relatively secluded from other Jewish groups, genetic evidence that Libyan Jews have a distinctive ancestry among Jewish groups has previously been inconclusive. Some studies using classical (8, 9), mitochondrial (10, 11), and Y-chromosomal (12–17) markers have considered North African Jews in their treatments of genetic relationships among Jewish populations, but these studies did not assess the relationship of the Libyan Jews to other groups. Studies that treated Libyan Jewish and other North African Jewish populations separately identified hereditary disorders in Libyan Jews that are rare in other populations (6, 18), but these studies did not find a consistent pattern of relationships among Libyan and other North African Jewish populations (6, 19–25). Genetic studies of Jewish populations based on classical markers (6, 19, 20) initially found substantial allele frequency differences and high genetic distances between the Libyan and Moroccan Jews, and suggested that these geographically related Jewish populations may have maintained a considerable degree of isolation from each other. Although more recent analyses have not contradicted these reports, they have been unable to make strong conclusions about the Libyan Jews, placing them in different positions with respect to other Jewish and non-Jewish populations (21–24). These studies have generally found that most Jewish populations show genetic relationships closer to each other than to most non-Jewish populations. Most recently, this evidence of shared ancestry among Jewish populations has been strengthened by the discovery of an otherwise uncommon Y-chromosomal haplotype frequent in the widely geographically dispersed Cohanim (26) and by analysis of Y chromosomes in a variety of Jewish and non-Jewish populations from Africa, Asia, and Europe (16).
In this article, we consider the genetic relationships among six Jewish and two non-Jewish populations using a statistical technique (27) that allows finer resolution of population structure than has heretofore been possible. Although all populations of our study are very closely related, we report that the Libyan Jews have been genetically isolated from other Jewish groups, including the Moroccan Jews. The same methods suggest some affinity between the Jewish populations of Ethiopia and Yemen.
Materials and Methods
Subjects.
We analyzed 159 males from eight populations. These individuals were Ashkenazi Jews from Poland (20), Druze (20), Ethiopian Jews (19), Iraqi Jews (20), Libyan Jews (20), Moroccan Jews (20), Palestinian Arabs (20), and Yemenite Jews (20). DNA samples were obtained from The National Laboratory for the Genetics of Israeli Populations (www.tau.ac.il/medicine/NLGIP/nlgip.htm). Jewish DNA samples were contributed by second-generation immigrants to Israel from the various source populations. To avoid sampling from small communities whose genotypes were not representative of the whole population, the Druze in this study were identified from a large settlement in the Galilee region of northern Israel, and Palestinians were sampled in large cities.
Genotyping.
We genotyped individuals for 20 unlinked microsatellites spread across 14 autosomes. These included seven dinucleotide polymorphisms (D1S235, D3S1311, D4S403, D6S305, D14S53, D20S103, D20S851), one trinucleotide (D4S2361), and 12 tetranucleotides (D1S1679, D2S410, D2S1400, D3S2387, D5S1456, D7S2846, D8S1128, D9S934, D10S677, D10S1426, D11S446, D17S1298). A Stratagene thermal cycler was used for the PCR reactions, which were performed in a 20-μl final reaction volume containing 50–60 ng of genomic DNA, 10 mM dNTP, 200 μM of each forward primer and of each reverse primer, 25 mM MgCl, 2 μl of 10× PCR reaction buffer, and 0.5 unit of Taq polymerase. Primers were purchased from Research Genetics (Huntsville, AL). Forward primers were fluorescently labeled. PCR cycling was performed as recommended by Research Genetics, with one alteration: a predenaturation step of 3 min and an extension time of 15 min. PCR products were run in an ABI 377 sequencer. Allele sizes were calibrated using a Genescan-350 TAMRA size standard run along with each sample. genescan 672 and genotyper 1.1 software packages were used to determine allele sizes. Alleles were designated by fragment size, which was measured in base pairs.
Statistical Analysis.
For each pair of populations, we tested the null hypothesis that the two populations had identical allele frequencies. A contingency table was constructed with absolute allele frequencies for each pair of populations and each locus. Rare alleles were pooled as in ref. 28. The chi-squared test statistic of association was computed for each locus (29), and these test statistics were summed across loci. Significance levels for this overall test statistic were obtained from the χ2 distribution whose number of degrees of freedom equaled the sum of the numbers of degrees of freedom for the single-locus tests.
For each population, we also considered the normalized average differentiation test statistic across the seven pairwise comparisons with other populations. This overall statistic equaled the average of the ratios of the seven chi-squared test statistics to their numbers of degrees of freedom. Using the fact that the mean of a χ2 distribution with k degrees of freedom is k (30), and the fact that this statistic is an average of seven normalized chi-squared random variables Xi/ki (i = 1 to 7) each with mean 1, the expectation of the statistic is 1.
To test the correspondence of genetic clusters with culturally labeled groups, we used the computer program structure (27), which identifies clusters of genetically similar diploid individuals from multilocus genotypes without prior knowledge of their population affinities. For this analysis, we assumed that each individual had ancestry in all clusters, so that fractions of ancestry in the various clusters were estimated. Using a version of structure that ignored population affiliation when clustering individuals, we ran the program for 1,000,000 iterations with a burn-in period of 30,000, with the number of specified clusters equaling one up to eight. The posterior probability that the proper number of clusters was 3 was essentially 1. An individual was assigned to a cluster if the fraction of his genome assigned to that cluster was greater than 75%. Although the value of this criterion was somewhat arbitrary, it did not greatly affect assignments of individuals to clusters. Repeated runs of structure produced nearly identical results to those shown.
We used the neighbor-joining algorithm (31) with the proportion-of-shared-alleles distance measure between individuals (32) to construct an unrooted tree of individuals. This genetic distance was computed using microsat (33). In the implementation of the neighbor-joining algorithm with the neighbor program (34), we randomized the input order of the individuals using the “jumble” option. The population-level analog of this distance (35, 36) was also calculated. For each locus, this allele-sharing distance summed the lower of corresponding allele frequencies in two populations across all alleles. These sums were then averaged across all loci genotyped, yielding an overall proportion of shared alleles (33). For population-level distance, we used the negative logarithm of the proportion of shared alleles, and for individual-level distance, we used one minus the proportion of shared alleles.
We also computed Fst distances according to ref. 37 (equation 5.3).
Results
Using the differentiation test, all population pairs were distinguishable at P < 0.0005, indicating that no two populations had the same set of allele frequencies (Table 1). Five of the six highest pairwise differences involved the Libyan Jews. When corrected for the number of degrees of freedom, the average chi-squared test statistic across comparisons was substantially higher for the Libyan Jews than for other populations.
Table 1.
Pairwise population differentiation tests and normalized average differentiation test statistics
| Ashkenazi Jews | Druze | Ethiopian Jews | Iraqi Jews | Libyan Jews | Moroccan Jews | Palestinians | Yemenite Jews | Average of X2/df | |
|---|---|---|---|---|---|---|---|---|---|
| Ashkenazi Jews | 151.9 | 229.4 | 162.3 | 279.2 | 193.9 | 169.0 | 168.3 | 2.436 | |
| Druze | 5 | 247.4 | 155.0 | 267.0 | 175.9 | 131.8 | 207.9 | 2.412 | |
| Ethiopian Jews | 5 | 8 | 220.9 | 283.6 | 195.4 | 293.4 | 160.1 | 2.863 | |
| Iraqi Jews | 2 | 4 | 5 | 236.8 | 130.6 | 170.0 | 162.3 | 2.268 | |
| Libyan Jews | 7 | 7 | 6 | 4 | 215.0 | 263.8 | 294.9 | 3.187 | |
| Moroccan Jews | 5 | 6 | 5 | 2 | 6 | 195.6 | 175.0 | 2.284 | |
| Palestinians | 3 | 2 | 8 | 4 | 8 | 5 | 251.2 | 2.574 | |
| Yemenite Jews | 4 | 6 | 2 | 2 | 7 | 5 | 8 | 2.501 |
All pairwise tests were significant below the 0.0005 level. Above the diagonal are values of the chi-squared test statistic; numbers of degrees of freedom for the tests ranged from 75 to 85. Below the diagonal lie the number of loci that produced pairwise population differentiation test statistics significant at the 0.01 level. For each population, the last column reports the average of the ratios of its chi-squared statistics to their numbers of degrees of freedom (df).
To ensure that high population differentiation statistics did not derive from a single locus that was unusually variable in one group but not in other populations, we considered individual values of the differentiation test statistic, computed for each locus. For each pairwise comparison of populations, at least two loci produced significant test statistics at P < 0.01 (Table 1). The mean number of loci significant at P < 0.01 in the seven pairwise comparisons between the Libyan Jews and the other populations was 6.43.
Genetic distances were low in all pairwise comparisons: allele-sharing distances ranged from 0.299 to 0.552, and Fst ranged from 0.0091 to 0.0656 (Table 2). Distances that involved the Libyan Jews were generally larger than for other populations.
Table 2.
Genetic distances between populations
| Ashkenazi Jews | Druze | Ethiopian Jews | Iraqi Jews | Libyan Jews | Moroccan Jews | Palestinians | Yemenite Jews | |
|---|---|---|---|---|---|---|---|---|
| Ashkenazi Jews | 0.0267 | 0.0386 | 0.0208 | 0.0562 | 0.0236 | 0.0265 | 0.0343 | |
| Druze | 0.355 | 0.0522 | 0.0250 | 0.0491 | 0.0237 | 0.0178 | 0.0445 | |
| Ethiopian Jews | 0.420 | 0.479 | 0.0421 | 0.0570 | 0.0402 | 0.0656 | 0.0253 | |
| Iraqi Jews | 0.334 | 0.367 | 0.415 | 0.0419 | 0.0091 | 0.0221 | 0.0255 | |
| Libyan Jews | 0.470 | 0.470 | 0.506 | 0.417 | 0.0379 | 0.0532 | 0.0582 | |
| Moroccan Jews | 0.358 | 0.364 | 0.411 | 0.299 | 0.393 | 0.0262 | 0.0256 | |
| Palestinians | 0.362 | 0.325 | 0.552 | 0.342 | 0.496 | 0.384 | 0.0517 | |
| Yemenite Jews | 0.388 | 0.442 | 0.364 | 0.371 | 0.507 | 0.358 | 0.460 |
Above the diagonal: Fst. Below the diagonal: allele-sharing distance. The Mantel correlation coefficient (29) for the two distance measures was 0.984 (P < 0.0001, 10,000 permutations).
Because significant allele frequency differences were found across the populations, it seemed reasonable to expect that analyses based on the clustering of individuals would identify some genetic clusters that corresponded well to populations. In fact, structure revealed a cluster that almost coincided with our sample of Libyan Jews (Fig. 1). Of 20 Libyan Jewish individuals in our sample, 19 fell into Cluster 3 (Table 3), while only five other individuals also fell into this cluster. This grouping of the Libyan Jews into Cluster 3 was highly statistically significant (X2 = 107.2, P < 10−9, 2 df), indicating that the Libyan Jewish appellation labeled not only a cultural group, but also a genetic cluster. Cluster 2 contained only Ethiopian and Yemenite Jews, whereas most sampled individuals fell into Cluster 1. Nine individuals did not belong to any cluster because no cluster was inferred to contain more than 75% of their ancestry. These unassigned individuals included one Ashkenazi Jew, two Druze, three Ethiopian Jews, two Iraqi Jews, and one Moroccan Jew.
Figure 1.
Diagram of three inferred clusters of individuals. For each individual and each inferred cluster, the fraction of the individual's ancestry estimated by structure to derive from that cluster is represented by its distance to the opposite side of the triangle. The sum of the distances from any point to all three sides is 1. Although pairwise genetic distances between clusters are not equal, clusters are graphically represented as equidistant from each other.
Table 3.
The number of individuals in the three clusters inferred by using structure
| Cluster 1 | Cluster 2 | Cluster 3 | |
|---|---|---|---|
| Ashkenazi Jews | 19 | 0 | 0 |
| Druze | 18 | 0 | 0 |
| Ethiopian Jews | 3 | 11 | 2 |
| Iraqi Jews | 17 | 0 | 1 |
| Libyan Jews | 1 | 0 | 19 |
| Moroccan Jews | 19 | 0 | 0 |
| Palestinians | 18 | 0 | 2 |
| Yemenite Jews | 16 | 4 | 0 |
Nine individuals who were not assigned to any cluster are not shown. When this table was treated as a contingency table, the 8 × 3 test of association (27) yielded a highly significant clustering (X2 = 187.0, P < 10−9, 14 df).
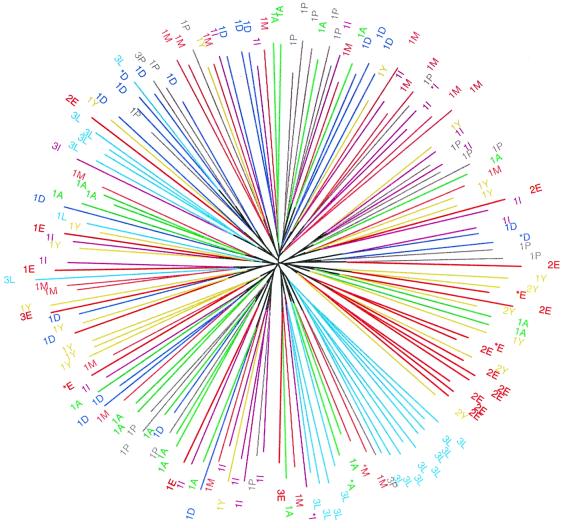
A neighbor-joining tree of individuals based on the proportion of alleles shared between individuals clustered Libyan Jews in a few regions of the tree, including nine in one clade (Fig. 2). For all pairs of populations, neighbor-joining trees that used only the individuals from those two groups could not be partitioned into exactly two clades, each of which contained the individuals from only one population (not shown).
Figure 2.
Neighbor-joining tree of individual genotypes. Each individual is labeled by his inferred cluster (based on Table 3) and his population affiliation. A, Ashkenazi Jews; D, Druze; E, Ethiopian Jews; I, Iraqi Jews; L, Libyan Jews; M, Moroccan Jews; P, Palestinians; Y, Yemenite Jews. Clusters are denoted 1, 2, and 3 as in Fig. 1. Nine individuals unassigned to any cluster are denoted by asterisks.
Discussion
It is consistent with historical sources that the Libyan Jews should separate from and show strong differentiation from the other populations of our study. This population has a unique history among North African Jewish communities, including an early founding, a harsh bottleneck, possible admixture with local Berbers, limited contact with other Jewish communities, and small size in the recent past (1, 2, 4, 38). Although the few Iberian and European Jews who settled in Libya probably had cultural influence disproportionate to their numbers, it may be that they and other Jewish immigrants left no sizable genetic signature. The characteristic genotypes detected here are therefore most likely attributable to the relative isolation of and possible genetic drift in the Libyan Jewish group. It is also notable that although cluster analysis separated the Libyan and Moroccan Jews, the lowest differentiation test statistic involving Libyan Jews was with Moroccan Jews, perhaps reflecting shared ancestral Jewish, Iberian Jewish, or Berber contributions to these populations, or gene flow among them.
In addition to highlighting the Libyan Jews, structure also grouped 11 Ethiopian Jews and 4 Yemenite Jews in Cluster 2 (Fig. 1 and Table 3), and the neighbor-joining tree identified clades with Ethiopian and Yemenite Jews intermixed (Fig. 2). The differentiation statistic and genetic distances for the Ethiopian and Yemenite Jews were quite low, among the smallest of comparisons that involved either of these populations. Many hypotheses for the origin of the Ethiopian Jews have been proposed, including Jewish migrations from ancient Israel, Egypt, or Arabia, or conversions of native Ethiopians by North African or Arabian Jews (39, 40). Both linguistic and historical evidence demonstrate that Ethiopian and Arabian peoples have communicated with each other since at least the 6th century BCE (41). In addition, Yemen was ruled by governors from Ethiopia during 525–573 CE (42). Although some authors have argued that Ethiopian Jews derive mostly from Africans (15, 43), or that the Ethiopian Jews are distant from all other populations that they studied (24), others have claimed that this group, as well as Ethiopian non-Jewish populations in general, may contain some African and some Middle Eastern ancestry (11, 44–47). Ethiopian Jewish Y-chromosomal haplotypes are often present in Yemenite and other Jewish populations (see table 1 in ref. 16), but analysis of Y-chromosomal haplotype frequencies does not indicate a close relationship between Ethiopian and other Jewish groups (see figure 2 in ref. 16).
Because structure easily separates African and Asian individuals into different clusters (27), it is possible that the 11 Ethiopian Jews that it placed into Cluster 2 are individuals who have significant native African ancestry. If this is the case, then the four Yemenite Jews who also fell into this cluster may be descendants of reverse migrants of African origin, who crossed from Ethiopia to Yemen. Gene flow and distant common ancestry are confounded here. However, the evidence of an African contribution to the ancestry of Ethiopian Jews and the evidence of communication across the Red Sea suggest that gene flow between these populations would be a more plausible explanation for our clustering of some Yemenite Jews with some Ethiopian Jews. Recent studies suggest that the Lemba of southern Africa derive partly from Yemenite Jews or other Semitic peoples of this region (17), and that Ethiopians share a combination of African and Middle Eastern genotypes and languages (41, 46). Together with these results, our data lend support to the theory that the Red Sea straits between Ethiopia and Arabia have been an important crossing point for migrating peoples.
Although gene flow between the Ethiopian and Yemenite Jewish populations is one explanation of our results, it is also possible that gene flow did not occur directly between these two populations, but rather took place between non-Jewish populations of Ethiopia and Arabia, between Ethiopian Jews and Ethiopian non-Jews, and also between Yemenite Jews and Yemenite non-Jews. This interpretation of indirect gene flow is not only consistent with the historical record of communication and the linguistic similarities of Ethiopia and Arabia, but also with the absence of evidence regarding communication between the two Jewish communities and the general ambivalence of previous studies about a direct Ethiopian Jewish and Yemenite Jewish genetic link. Thus, Cluster 2 may reflect the general distinctiveness of some peoples of this region with respect to the other groups in our study. A more satisfactory resolution of this issue will require assessment of Jewish and non-Jewish populations of the region.
The differentiation test statistics were smaller for the other populations in our study than for comparisons that involved the Ethiopian and Libyan Jews. Additionally, subclustering analysis of the individuals placed by structure into Cluster 1 showed that this largest of the three clusters could not be further divided. Following the method of ref. 27 for estimating the number of clusters (K) in a data set, we did not find that K > 1 for the 111 individuals in Cluster 1.
In addition, when only those individuals from any two of the six populations represented mainly by Cluster 1 were included in structure analysis, the individuals did not split into distinct clusters corresponding to their population affinities (not shown). The inability of structure or the neighbor-joining tree to subdivide the populations that fell mostly into Cluster 1 may be attributable to a combination of several factors: gene flow among the groups, recent common ancestry, and population sizes too large to experience rapid genetic drift. Another complication may be that our sample sizes and number of loci may have been insufficient for structure or the neighbor-joining tree to identify genetic subgroups. Perhaps we can only comment regarding these six populations that, because genetic distances were fairly small (generally lower than or similar to within-continent allele-sharing distances for 11 geographically diverse human populations from ref. 48, which ranged from 0.429 to 0.659), the populations of this study are indeed closely related genetically.
Between-cluster distances were also similar to previous comparisons of populations from the same continent: allele-sharing distances equaled 0.556 (Clusters 2 and 3), 0.359 (Clusters 3 and 1), and 0.439 (Clusters 1 and 2). The allele-sharing distance between the Libyan Jewish cluster and the main cluster (Clusters 3 and 1) was less than all within-continent genetic distances for the worldwide data set of ref. 48. Although the Libyan Jews are closely related to the other groups in our study, with typical allele-sharing and Fst distances for intracontinental comparisons, the Libyan Jews separate into a unique cluster and they are distinguishable when our methods are used.
Although this separation of the Libyan Jews is consistent with their historical context, probable high levels of isolation of other populations in our study make it surprising that this group was the easiest to differentiate. Of the populations that did not form their own clusters, the failure of the Druze to separate is particularly puzzling. The Druze, an endogamous sect of Arab origin, derive from a founder population that numbered in the thousands and that was immediately closed to converts in the 11th century CE (49). The group has been isolated long enough to develop private Y-chromosomal haplotypes of high frequency (50), but the details of its genetic history remain unknown. As diverse followers of a particular religious leader, Druze founders may have had genetically heterogeneous origins. A larger study will be required to determine whether we did not separate the Druze and other population isolates in our study because of insufficient information or because of the genetic heterogeneity among founders and the action of other population genetic forces.
From a statistical perspective, it is of interest that structure classified populations more finely than the neighbor-joining algorithm, as was found for a considerably more diverged data set of Asians and Europeans (27). This result was due to the fact that neighbor-joining trees of individuals compress the information in two individual genotypes to a single distance and therefore use individual multilocus genotypes less efficiently than structure does (27). Tree representations of individual relationships, which have generally not recognized distinct populations from the same continent (32, 35), even when individuals were taken from only two populations at a time (32), did not completely separate the groups of our study, although they did find the same trend as structure in isolating Libyan Jewish clades.
The discrepancy between the distinctive Libyan Jewish cluster identified by structure and the suggestive trends of Libyan Jewish isolation revealed by genetic distance and genetic differentiation calculations might provide a reason to be cautious about overinterpreting our results. However, we suggest alternatively that the additional information of full multilocus genotypes may give the structure program considerable extra power to resolve population relationships beyond that of analysis based solely on allele frequencies. In particular, the use of individual multilocus genotypes for clustering allows the incorporation of linkage and higher-order disequilibria among the loci into the determination of population relationships. As an extreme example, consider two populations that have identical allele frequencies at many loci. Suppose one of these populations has complete linkage equilibrium, while the other population is a recent mixture of two distinct genetic populations and has considerable linkage disequilibrium. Most genetic distance measures would find zero distance between these two populations, and differentiation tests would not reject the null hypothesis of identical allele frequencies. However, structure could use the different frequencies of multiocus genotypes to potentially identify two distinct genetic clusters. A simulation-based comparison of the resolving power of structure and other methods at varying levels of linkage and higher-order disequilibria will be required to determine the extent of this effect. Disequilibrium, when present, needs large samples for its detection, and using the exact test implemented in genepop (51), we did not find unusual deviation from linkage equilibrium in any of the populations in our study (not shown). Future studies may quantify sample sizes and linkage disequilibrium levels that allow structure to identify separate clusters from a collection of individuals in situations where other methods are ineffective. An advantage of structure is that clustering seems to improve with larger sample sizes in each population (27), while the consistency of neighbor-joining trees with population affiliation (32) need not be systematically increased with more individuals.
As a secondary consideration, individual analysis allows for the consideration of Hardy–Weinberg disequilibrium as well as linkage disequilibrium. However, because Hardy–Weinberg disequilibrium breaks down over time faster than linkage disequilibrium, the information contained in linkage disequilibrium seems more important to the clustering of individuals from real populations than the information from Hardy–Weinberg disequilibrium.
Perhaps because most divergences among Jewish groups have taken place during the last 3,000 years, previous genetic studies based on allele frequencies and genetic distance have had insufficient power to resolve relationships among Jewish populations, including the Libyan Jews. Although these methods have achieved considerable success in understanding human genetic history at the level of continental differentiations, they have consistently been unable to distinguish populations within continents with great accuracy (32, 35, 48, 52, 53). Future studies of human evolution that use autosomal loci to unravel relationships among closely related populations should make use of methods that go beyond treating loci independently, and which tap into the potential information lying in the associations among loci.
Acknowledgments
We thank Marissa Baskett, Andy Clark, and Joanna Mountain for comments. This work was supported by a National Defense Science and Engineering Graduate fellowship to N.A.R. and by National Institutes of Health Grant GM28428 to M.W.F.
References
- 1.De Felice R. Jews in an Arab Land: Libya, 1835–1970. Austin: Univ. of Texas Press; 1985. [Google Scholar]
- 2.Gubbay L, Levy A. The Sephardim: Their Glorious Tradition from the Babylonian Exile to the Present Day. London: Carnell Limited; 1992. [Google Scholar]
- 3.Chouraqui A N. Between East and West: A History of the Jews of North Africa. Philadelphia: Jewish Publication Society of America; 1968. [Google Scholar]
- 4.Simon R. Change Within Tradition Among Jewish Women in Libya. Seattle: Univ. of Washington Press; 1992. [Google Scholar]
- 5.Wexler P. The Non-Jewish Origins of the Sephardic Jews. New York: State Univ. of New York Press; 1996. [Google Scholar]
- 6.Bonné-Tamir B, Ashbel S, Modai J. Hum Genet. 1977;37:319–328. doi: 10.1007/BF00393615. [DOI] [PubMed] [Google Scholar]
- 7.Roth C, Wigoder G, editors. Encyclopedia Judaica. Jerusalem: Keter Publishing House; 1972. pp. 198–206. [Google Scholar]
- 8.Mourant A E, Kopec A C, Domaniewska-Sobczak K. The Genetics of the Jews. Oxford: Clarendon; 1978. [Google Scholar]
- 9.Kobyliansky E, Micle S, Goldschmidt-Nathan M, Arensburg B, Nathan H. Ann Hum Biol. 1982;9:1–34. doi: 10.1080/03014468200005461. [DOI] [PubMed] [Google Scholar]
- 10.Tikochinski Y, Ritte U, Gross S R, Prager E M, Wilson A C. Am J Hum Genet. 1991;48:129–136. [PMC free article] [PubMed] [Google Scholar]
- 11.Ritte U, Neufeld E, Prager E M, Gross M, Hakim I, Khatib A, Bonné-Tamir B. Hum Biol. 1993;65:359–385. [PubMed] [Google Scholar]
- 12.Santachiara-Benerecetti A S, Semino O, Passarino G, Torroni A, Brdicka R, Fellous M, Modiano G. Ann Hum Genet. 1993;57:55–64. doi: 10.1111/j.1469-1809.1993.tb00886.x. [DOI] [PubMed] [Google Scholar]
- 13.Lucotte G, Smets P, Ruffie J. Hum Biol. 1993;65:835–840. [PubMed] [Google Scholar]
- 14.Lucotte G, David F, Berriche S. Hum Biol. 1996;68:467–471. [PubMed] [Google Scholar]
- 15.Ritte U, Neufeld E, Broit M, Shavit D, Motro U. J Mol Evol. 1993;37:435–440. doi: 10.1007/BF00178873. [DOI] [PubMed] [Google Scholar]
- 16.Hammer M F, Redd A J, Wood E T, Bonner M R, Jarjanzi H, Karafet T, Santachiara-Benerecetti S, Oppenheim A, Jobling M A, Jenkins T, Ostrer H, Bonné-Tamir B. Proc Natl Acad Sci USA. 2000;97:6769–6774. doi: 10.1073/pnas.100115997. . (First Published May 9, 2000; 10.1073/pnas.100115997) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomas M G, Parfitt T, Weiss D A, Skorecki K, Wilson J F, le Roux M, Bradman N, Goldstein D B. Am J Hum Genet. 2000;66:674–686. doi: 10.1086/302749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodman R M. Genetic Disorders Among the Jewish People. Baltimore: The Johns Hopkins Univ. Press; 1979. [Google Scholar]
- 19.Bonné-Tamir B, Ashbel S, Bar-Shani S. Am J Phys Anthropol. 1978a;49:465–472. doi: 10.1002/ajpa.1330490406. [DOI] [PubMed] [Google Scholar]
- 20.Bonné-Tamir B, Bodmer J G, Bodmer W F, Pickbourne P, Brautbar C, Gazit E, Nevo S, Zamir R. Tissue Antigens. 1978b;11:235–250. doi: 10.1111/j.1399-0039.1978.tb01255.x. [DOI] [PubMed] [Google Scholar]
- 21.Carmelli D, Cavalli-Sforza L L. Hum Biol. 1979;51:41–61. [PubMed] [Google Scholar]
- 22.Karlin S, Kenett R, Bonné-Tamir B. Am J Hum Genet. 1979;31:341–365. [PMC free article] [PubMed] [Google Scholar]
- 23.Livshits G, Sokal R R, Kobyliansky E. Am J Hum Genet. 1991;49:131–146. [PMC free article] [PubMed] [Google Scholar]
- 24.Amar A, Kwon O J, Motro U, Witt C S, Bonné-Tamir B, Gabison R, Brautbar C. Hum Immunol. 1999;60:723–730. doi: 10.1016/s0198-8859(99)00043-9. [DOI] [PubMed] [Google Scholar]
- 25.Kobyliansky E, Livshits G. Ann Hum Biol. 1983;10:453–464. doi: 10.1080/03014468300006661. [DOI] [PubMed] [Google Scholar]
- 26.Thomas M G, Skorecki K, Ben-Ami H, Parfitt T, Bradman N, Goldstein D B. Nature (London) 1998;394:138. doi: 10.1038/28083. [DOI] [PubMed] [Google Scholar]
- 27.Pritchard J K, Stephens M, Donnelly P J. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pritchard J K, Rosenberg N A. Am J Hum Genet. 1999;65:220–228. doi: 10.1086/302449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sokal R R, Rohlf F J. Biometry. New York: Freeman; 1995. [Google Scholar]
- 30.Cramer H. Mathematical Methods of Statistics. Princeton: Princeton Univ. Press; 1999. [Google Scholar]
- 31.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 32.Mountain J L, Cavalli-Sforza L L. Am J Hum Genet. 1997;61:705–718. doi: 10.1086/515510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minch E, Ruiz-Linares A, Goldstein D B, Feldman M W, Cavalli-Sforza L L. microsat: A Computer Program for Calculating Various Statistics on Microsatellite Allele Data. Stanford Univ., Stanford: Department of Genetics; 1997. , Version 1.5d. [Google Scholar]
- 34.Felsenstein J. phylip (Phylogeny Inference Package) University of Washington, Seattle: Department of Genetics; 1993. , Version 3.5.c. [Google Scholar]
- 35.Bowcock A M, Ruiz-Linares A, Tomfohrde J, Minch E, Cavalli-Sforza L L. Nature (London) 1994;368:455–457. doi: 10.1038/368455a0. [DOI] [PubMed] [Google Scholar]
- 36.Chakraborty R, Jin L. In: DNA Fingerprinting: State of the Science. Pena S D J, Chakraborty R, Epplen J T, Jeffreys A J, editors. Basel: Birkäuser; 1993. pp. 153–175. [Google Scholar]
- 37.Weir B S. Genetic Data Analysis II. Sunderland, MA: Sinauer; 1996. [Google Scholar]
- 38.Bonné-Tamir B, Karlin S, Kenett R. Am J Hum Genet. 1979;31:324–340. [PMC free article] [PubMed] [Google Scholar]
- 39.Quirin J A. The Evolution of the Ethiopian Jews. Philadelphia: Univ. Pennsylvania Press; 1991. [Google Scholar]
- 40.Kessler D F. The Falashas: A Short History of the Ethiopian Jews. London: Frank Cass; 1996. [Google Scholar]
- 41.Cavalli-Sforza L L, Menozzi P, Piazza A. The History and Geography of Human Genes. Princeton: Princeton Univ. Press; 1994. [Google Scholar]
- 42.Tobi J. The Jews of Yemen: Studies in Their History and Culture. Leiden, The Netherlands: E. J. Brill; 1999. [Google Scholar]
- 43.Lucotte G, Smets P. Hum Biol. 1999;71:989–993. [PubMed] [Google Scholar]
- 44.Bonné-Tamir B, Gross Y, Ashbel S, Goldwitch Z. Gene Geogr. 1987;1:1–8. [PubMed] [Google Scholar]
- 45.Zoossmann-Diskin A, Ticher A, Hakim I, Goldwitch Z, Rubinstein A, Bonné-Tamir B. Isr J Med Sci. 1991;27:245–251. [PubMed] [Google Scholar]
- 46.Passarino G, Semino O, Quintana-Murci L, Excoffier L, Hammer M, Santachiara-Benerecetti A S. Am J Hum Genet. 1998;62:420–434. doi: 10.1086/301702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hakim I, Gross B, Bonné-Tamir B. In: Plurisciplinary Approach to Human Isolates. Chaventré A, Roberts D F, editors. Paris: INED; 1990. pp. 43–57. [Google Scholar]
- 48.Jin L, Baskett M L, Cavalli-Sforza L L, Zhivotovsky L A, Feldman M W, Rosenberg N A. Ann Hum Genet. 2000;64:117–134. doi: 10.1017/S0003480000008034. [DOI] [PubMed] [Google Scholar]
- 49.Abu-Izzedin N M. The Druzes: A New Study of Their History, Faith, and Society. Leiden, The Netherlands: E. J. Brill; 1984. [Google Scholar]
- 50.Woolf E. M. Sc. Thesis. Jerusalem: The Hebrew Univ. of Jerusalem; 2000. [Google Scholar]
- 51.Raymond M, Rousset F. J Hered. 1995;86:248–249. [Google Scholar]
- 52.Pérez-Lezaun A, Calafell F, Mateu E, Comas D, Ruiz-Pacheco R, Bertranpetit J. Hum Genet. 1997;99:1–7. doi: 10.1007/s004390050299. [DOI] [PubMed] [Google Scholar]
- 53.Calafell F, Shuster A, Speed W C, Kidd J R, Kidd K K. Eur J Hum Genet. 1998;6:38–49. doi: 10.1038/sj.ejhg.5200151. [DOI] [PubMed] [Google Scholar]