Abstract
SopB is an inositol phosphate phosphatase that is a virulence factor in Salmonella species. We have overexpressed SopB cDNA in a tetracycline-dependent system in human embryonic 293 cells, and used this model system to directly analyze the role of SopB in altering inositol metabolite levels in vivo. Addition of tetracycline to these cells resulted in the rapid induction of SopB expression, which was coincident with perturbations in the cellular levels of multiple soluble inositol phosphates. All of the changes induced by SopB expression were reversed within 24 h on removal of tetracycline from media. Specifically, cellular inositol 1,3,4,5,6-pentakisphosphate (InsP5) and inositol hexakisphosphate (InsP6) levels were depleted within 4 to 6 h after inducing SopB expression. A transient rise in cellular inositol 1,4,5,6-tetrakisphosphate was also observed and was accompanied by increased chloride channel activity. This indicates that SopB alone is sufficient for changes in chloride channel function in cells infected with Salmonella organisms. Depletion of inositol phosphates, including InsP5 and InsP6 metabolites, was coincident with the accumulation of polyadenylated RNA in the nucleus. This suggested that a defect in nuclear export had occurred. Moreover, the penetrance of the export defect required localization of SopB to the nucleus. These results provide evidence that inositol phosphate productions may be required for efficient mRNA export in mammalian cells.
The inositol signaling pathway is involved in a host of intracellular processes. Previous studies have shown that perturbations of cell signaling can be achieved by molecular genetic manipulation or mutations of the enzymes involved in inositol phosphate metabolism. Type I inositol polyphosphate 5-phosphatase is the enzyme that hydrolyzes the calcium-mobilizing messenger inositol 1,4,5-trisphosphate (Ins-1,4,5-P3). Cells stably transfected with an antisense cDNA for this enzyme have elevated levels of Ins-1,4,5-P3 and display a transformed phenotype (1). Drosophila mutants lacking inositol polyphosphate 1-phosphatase, a lithium-inhibited phosphatase, have a “shaker” phenotype (2). Mice with deletion of SHIP, a hematopoietic cell-specific inositol polyphosphate 5-phosphatase, have excessive cytokine signaling and die from myeloid proliferation (3). Patients with Lowe syndrome, a deficiency of another inositol polyphosphate 5-phosphatase designated OCRL, have decreased phosphatidylinositol 4,5-bisphosphate (PtdIns-4,5-P2) hydrolyzing activity and accumulate the PtdIns-4,5-P2 metabolite in cells. These and other examples not cited show that perturbation of inositol signaling can uncover potential cellular functions for particular metabolites.
Inositol 1,3,4,5,6-pentakisphosphate (InsP5) and inositol hexakisphosphate (InsP6) are soluble inositol polyphosphates that are very abundant in cells, but until recently their cellular functions were unknown (4). Recent studies in the budding yeast Saccharomyces cerevisiae have suggested that these molecules function in the nucleus to stimulate transcription by the ArgR-MCM1 complex and to facilitate nuclear export of mRNA (5, 6). Conserved InsP5 2-kinases have been identified in several fungi (7); however, a mammalian homologue of an InsP5 2-kinase has not yet been reported. Moreover, in vivo roles for InsP5 and InsP6 in higher eukaryotic cells have not been demonstrated. To analyze the roles of such inositol polyphosphates in mammalian cells, we have devised a strategy to deplete these metabolites and to assay for perturbations of nuclear and cellular functions.
SopB is one of several proteins secreted by the type III secretion system of the enteric Salmonella dublin species. These proteins are injected into host cells on contact with the bacteria, and the secretion system is required for the pathogenicity of the organisms (8, 9). Deletion of SopB alone does not affect the invasiveness of Salmonella; however, its absence does abrogate diarrhea and inflammation (10). During routine searches of GenBank for homologues of enzymes potentially involved in inositol phosphate metabolism, we noted that SopB contained two motifs that were highly similar to sequence motifs in inositol polyphosphate 4-phosphatases. Notably, one of these motifs (CKSGKDRTGM) contains the active site cysteine residue that is required for 4-phosphatase activity (11–13). We recently showed that SopB indeed encodes an inositol phosphate phosphatase. When SopB was expressed in E. coli and assayed in vitro, it hydrolyzed a variety of inositol phosphates and phospholipids, although it did not hydrolyze InsP6 in vitro (14). In contrast, when S. dublin was added to HeLa cells there was a rapid (1 h) increase in the levels of inositol 1,4,5,6-tetrakisphosphate (Ins-1,4,5,6-P4) without significant changes in other inositol phosphates or phospholipids (14, 15). This finding was significant because Ins-1,4,5,6-P4 has been shown to antagonize the phosphatidylinositol 3,4,5-trisphosphate-mediated closing of cellular chloride channels (15). Thus, we speculated that SopB may enhance chloride and water secretion, thereby inducing diarrhea during infection. We showed that a mutation in the active site cysteine residue of SopB abrogated both inflammation and diarrhea in infected calf intestinal loops. This established the role for SopB in pathogenicity (14).
To further study the function of SopB in vivo in the absence of the many other Salmonella factors, we have cloned a cDNA encoding SopB into a mammalian expression vector that is under the control of a tetracycline-inducible promoter. We have isolated stable cell lines that harbor this SopB construct by using a human embryonic 293 cell line, and have determined the effect of SopB overexpression on cellular inositol phosphate levels and chloride secretion. Coincidentally, the striking depletion of inositol phosphates, including InsP5 and InsP6 metabolite levels, by SopB expression in the nucleus allowed us to test for perturbations of mRNA export from the nucleus to the cytoplasm.
Experimental Procedures
Plasmid Construction.
Full-length cDNA encoding SopB was generated by PCR using chromosomal DNA from S. dublin 2229 as template. The plasmid FLAG-SopB (FS) was constructed by deleting the initiator methionine and inserting a BamHI restriction site at the 5′ end. An EcoRI site was added at the 3′ end. The PCR product was digested with BamHI and EcoRI and subcloned into pcDNA4/TO (Invitrogen), a tetracycline inducible expression vector, to generate pSopB-1. Two complementary oligonucleotides encoding a HindIII site, followed by a Kozak sequence (CACC) (16), and then sequences encoding a methionine residue, a FLAG epitope, and a BamHI site, were synthesized (MDYKDDDK) (17). This was inserted into the HindIII/BamHI site of pSopB-1 to create FS. The oligonucleotides were 5′-AGCTTCACCATGGATTATAAAGATGATGATGATAAAG-3′ and 5′-GATCCTTTATCATCATCATCTTTATAATCCATGGTGA-3′.
The plasmid FLAG-SopB-NLS (FSN) was constructed as described above by using the FLAG oligonucleotides and a PCR product encoding full-length SopB lacking its stop codon. These fragments were inserted into pcDNA4/TO to create pSopB-2. Two complementary oligonucleotides encoding an EcoRI site followed by sequences encoding a nuclear localization sequence (PKKKRKV) (18, 19), two stop codons, and an XbaI site, respectively, were synthesized. The annealed double-stranded oligonucleotides were inserted into the EcoRI/XbaI site of pSopB-2 to create FSN. The oligonucleotides were 5′-AATTCCCGAAAAAGAAACGCAAAGTCTGATGAT-3′ and 5′-CTAGATCATCAGACTTTGCGTTTCTTTTTCGGG-3′.
Stable SopB Mammalian Cell Lines.
FS and FSN constructs were transformed into TOP10 cells (Invitrogen) and plasmid DNA was purified. The isolated plasmids (2 μg DNA) were transfected into 1.5 × 106 T-REx 293 human embryonic cells (Invitrogen) in 2 ml of medium using TransIT-LT1 transfection reagent (Panvera, Madison, WI). Transfected cells were plated at 1000–4000 cells/ml in 15-cm tissue culture plates containing DMEM with 10% FBS (Tet system approved, CLONTECH), 2 mM glutamine, 100 units of penicillin G/ml, 10 μg streptomycin/ml, 0.25 μg amphotericin B/ml, 5 μg blasticidin/ml (selection for tet repressor), and 0.3 mg/ml Zeocin (selection for SopB expression). Colonies were grown from single cells for approximately 2 weeks and then picked and expanded in 96-well plates. SopB expression was confirmed by Western blotting with monoclonal antibodies recognizing the FLAG epitope (Sigma) and by enzyme assay using [3H]Ins-1,3,4,5-P4 as substrate.
Immunofluorescence and in Situ Hybridization.
SopB expression and subcellular localization in FS and FSN cells were determined by using cells grown on glass coverslips in 6-well tissue culture plates. When cell density reached 60% confluence, 20 ng tetracycline/ml was added to induce SopB expression. After overnight incubation, cells were washed, fixed, and permeabilized as described (20, 21). Cells were then incubated with 10 μg anti-FLAG M2 monoclonal antibody-FITC conjugate for 1 h at room temperature. mRNA in situ hybridization was carried out as described (20). Photographs were taken with the ×60 objective on a Nikon Eclipse E800 fluorescence microscope.
HPLC of Inositol Phosphates.
Cells were grown in complete media containing 10 μCi (1 Ci = 37 GBq) [3H]inositol/ml in 6-well tissue culture plates for 72 h. After various times of SopB induction with tetracycline (20 ng/ml final), cells were lysed in 0.5 ml methanol/0.5 M HCl (2/1) and extracted with 1 ml chloroform. The aqueous phase was dried, suspended in water, and applied to a 250 × 4.6 mm AdsorboSphere SAX column (Alltech/Applied Science, Deerfield, IL). Inositol phosphates were eluted as described (22). 32P-labeled Ins-1,3,4,5-P4 (22), Ins-1,3,4,5,6-P5 (14), and InsP6 (our unpublished data) were synthesized and used as internal standards. Radiolabeled inositol phosphates were detected by using a Beta-RAM flow-through detector (IN/US System, Tampa, FL). The inositol lipid fraction was deacylated (23) and separated on a PartiSphere SAX column (Whatman) as described (24).
Measurement of Chloride Influx.
Chloride influx was measured by modification of a previously described method (25). Cells were cultured for various times in the presence of 20 ng tetracycline/ml and then trypsinized, washed, and resuspended at 2 × 107 cells/ml. Na36Cl (ICN), 2 μCi/ml, was added for varying times and 30-μl samples were removed and added to Microfuge (Beckman Coulter) tubes containing “stop solution” (330 μl of complete medium containing 140 mM potassium gluconate and 1 mM 4,4′-diisothiocyanatostilbene-2,2′-disulfonic acid (DIDS) layered over 30 μl of dibutyl phthalate). Tubes were immediately spun at top speed in a Microfuge for 30 sec, frozen in a dry ice bath, and the tips of the tubes containing the cell pellets were cut off and analyzed in a liquid scintillation counter.
Protein Synthesis Assay.
Cells grown in six-well plates were treated with 20 ng tetracycline/ml for varying times before addition of Tran35S-label (ICN), 5 μCi, and further incubation for 15 min at 37°C. Protein was precipitated in 10% trichloroacetic acid, filtered onto Whatman glass microfiber filters, and samples were analyzed in a liquid scintillation counter (26).
Results and Discussion
Expression of SopB.
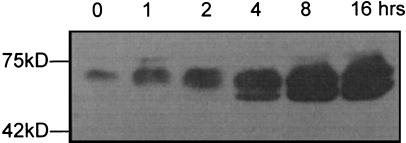
Our previous studies have shown that SopB is an inositol phosphate phosphatase (14), and the recombinant protein assayed in vitro hydrolyzes a wide range of inositol phosphates and phospholipids (not including InsP6). However, when a virulent strain of S. dublin is added to HeLa cells, it appears that Ins-1,3,4,5,6-P5 is the major, if not only, phosphatase substrate with cellular levels of Ins-1,4,5,6-P4 rising 14-fold in 30 min (14, 15). Thus, other bacterial proteins besides SopB may be affecting intracellular inositol metabolites. To investigate the effect of SopB itself on mammalian cells, a cDNA encoding full-length SopB fused to a sequence encoding the FLAG epitope was cloned into a mammalian expression vector and placed under the control of a tetracycline-inducible promoter. Stable cell lines expressing FS were generated in the human embryonic 293 cell line (see Experimental Procedures). The expression of FS was induced with tetracycline for varying times, and we estimated by western blotting with an antibody recognizing the FLAG epitope that FS was induced at least 50- to 100-fold after 16 h, as shown in Fig. 1. At the zero time point in the absence of tetracycline, a low level of FS was expressed, indicating that the promoter was leaky. The highest expression level was achieved 20 h after the addition of 20 ng/ml tetracycline in the media (data not shown).
Figure 1.
Western blotting of FSN in lysates from stably transfected human 293 cells. Cells were induced with 20 ng/ml tetracycline for varying times. Total protein (50 μg) was loaded on SDS/PAGE and then transferred to a poly(vinylidene difluoride) (PVDF) membrane. Monoclonal antibody against the FLAG epitope was used to visualize the overexpressed SopB protein.
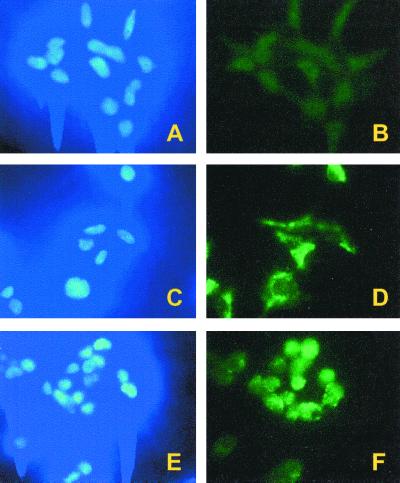
To examine the subcellular localization of FS, indirect immunofluorescence microscopy was conducted with an anti-FLAG monoclonal antibody. Without tetracycline induction, only background staining was observed (Fig. 2B). After induction overnight, the majority of the anti-FS staining was present in the cytoplasm (Fig. 2D). Significant levels of FS were not observed in the nuclei of most cells, as indicated by the absence of staining in the areas corresponding to DAPI (4′,6-diamidino-2-phenylindole) staining (Fig. 2 C vs. D). The lack of nuclear localization is likely due to the fact that the predicted molecular mass of FS (≈62 kDa) is just above the predicted diffusion limit for nuclear entry.
Figure 2.
Immunofluorescence of SopB expression in human 293 cells. (A and B) FS cells without tetracycline induction. (C and D) FS cells induced overnight. (E and F) FSN cells induced overnight. Cell nuclei were stained by using DAPI (Left), and SopB was visualized by FITC conjugated anti-FLAG monoclonal antibody (Right).
To be able to perturb inositol phosphate pools in both the nuclear and cytoplasmic compartments, we inserted a nuclear localization signal at the carboxyl terminus of FS to form FSN. Stable tetracycline-inducible FSN cell lines were isolated and characterized. As for FS, the expression levels of FSN were analyzed by Western blotting and nearly identical results were obtained as in Fig. 1. Based on the signal increase, at least a 50- to 100-fold induction was observed over the basal level in the absence of tetracycline. Indirect immunofluorescence microscopy showed that FSN was present throughout the cell with a notable concentration in the nucleus (Fig. 2F). We also noted that after induction of either FS or FSN expression, the cells rounded up and tended to detach from the dishes. Removal of the tetracycline and overnight incubation resulted in restoration of normal cell morphology and growth.
SopB Expression Alters Cellular Inositol Phosphate Levels.
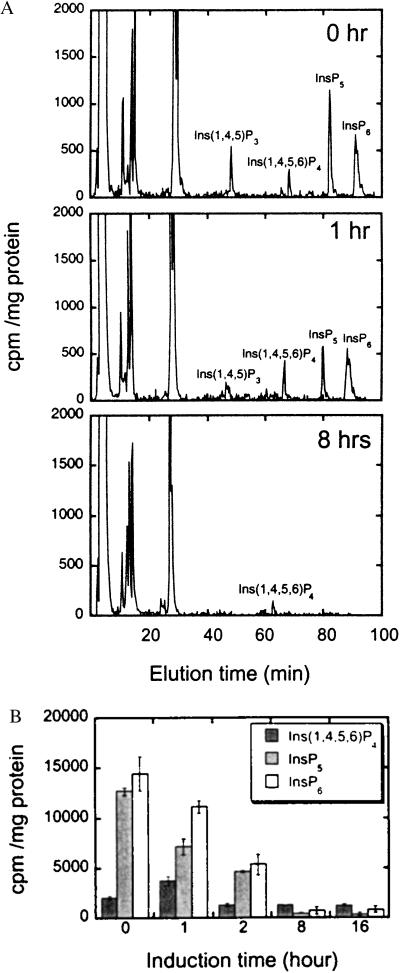
To analyze perturbations on the cellular inositol phosphates at various times after induction of either FS or FSN expression, we measured the amounts of soluble inositol phosphates by steady-state radiolabeling and HPLC analysis (see Experimental Procedures) (Fig. 3). We also analyzed the effects on inositol lipids by HPLC of glyceryl phosphoryl inositol derivatives and saw no significant changes in any of these molecules. The results were very similar for expression of both FS and FSN, and a summary of the changes averaged from three independent experiments is shown in Fig. 3B. Compared with cells before induction, at least three major effects were noted. First, the levels of Ins-1,4,5,6-P4 increased at 1 h and subsequently decreased. The elevation of Ins-1,4,5,6-P4 levels that is observed without tetracycline presumably results from the low basal expression level of SopB without tetracycline (see Fig. 1). Second, Ins-1,3,4,5,6-P5 levels decreased at 1 h and were absent by 8 h. Finally, InsP6 was also surprisingly depleted by 8 h (Fig. 3). In some experiments, Ins-1,3,4,5,6-P5 and InsP6 were largely depleted within 2 hours. The decrease in InsP6 levels was also unexpected because InsP6 is not an in vitro substrate for SopB. The depletion of InsP6 indicates that this metabolite can turn over rapidly, perhaps in an effort to maintain cellular Ins-1,3,4,5,6-P5 levels. The perturbations in inositol phosphate metabolite levels were reversed within 24 h by removal of the tetracycline (data not shown).
Figure 3.
HPLC of inositol phosphates in FS-overexpressing cells. (A) Inositol phosphates in human 293 cells without induction, or after FSN expression was induced for 1 and 8 h. (B) Changes in Ins-1,4,5,6-P4, InsP5, and InsP6 in SopB-overexpressing 293 cells after different times of induction. The averages of three independent experiments are shown ± SD (two trials of FS and 1 trial of FSN).
Chloride Influx Is Stimulated in SopB-Expressing Cells.
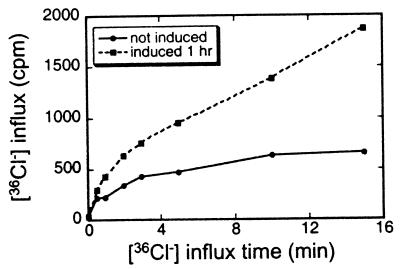
Because Salmonella induces a rapid rise in Ins-1,4,5,6-P4 in cells exposed to the organism and SopB expression alone also induced a transient rise in this metabolite, we measured chloride channel activity 1 h after induction (Fig. 4). Influx in uninduced cells reached a steady state by 15 min corresponding to an intracellular chloride concentration of 25 mM. The cells induced for SopB overexpression took up 36Cl 2- to 3-fold faster and did not reach a steady state by 15 min. This experiment was repeated several times with similar results. We also measured uptake 2 h after induction of SopB expression, at which time Ins-1,4,5,6-P4 levels had returned to baseline. At this point, 36Cl uptake was the same as that in uninduced cells (data not shown). These results clarify the role of SopB alone in altering cell functions in the absence of any other proteins from Salmonella. Thus, increased levels of Ins-1,4,5,6-P4, with its resulting increased chloride channel activity, are due to SopB alone.
Figure 4.
[36Cl−] uptake in FS-overexpressing 293 cells that were not induced with tetracycline (●) and induced for 1 h (■).
Expression of SopB Inhibits the Nuclear Export of Polyadenylated RNA.
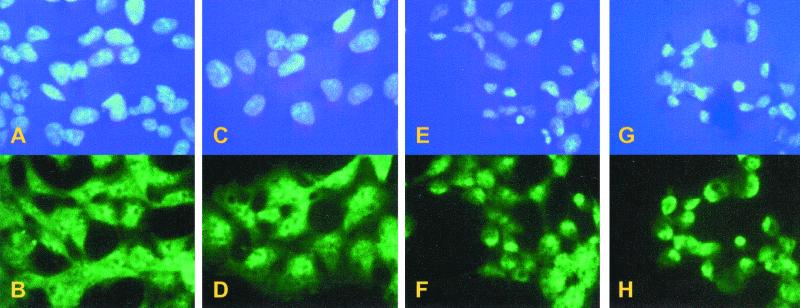
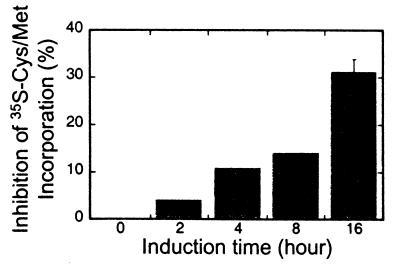
Recent studies in S. cerevisiae have demonstrated that InsP6 is required for the efficient nuclear export of mRNA. Mutant yeast strains that are deficient in any of the enzymes required for the production of InsP6 from PtdIns-4,5-P2 each individually have mRNA export defects at a restrictive growth temperature (5). This includes phospholipase C (Plc1), a dual specificity Ins-1,4,5-P3 6-kinase, and Ins-1,4,5,6-P4 3-kinase (Ipk2) and an Ins-1,3,4,5,6-P5 2-kinase (Ipk1) (5, 6). Thus, a loss of InsP6 production correlates with a decrease in mRNA nuclear export. If the export of mRNA in mammalian cells is also dependent on InsP6, we predicted that the SopB expressing cells should have a nuclear mRNA export defect. To test for this defect, after the induction of FS or FSN expression, the localization of polyadenylated RNA was visualized by in situ hybridization. Before induction, staining was detected in both the cytoplasm and nucleus (Fig. 5B). After induction of FSN for 4 h, polyadenylated RNA staining disappeared from the cytoplasm and the intranuclear staining intensity significantly increased (Fig. 5D). The accumulation of polyadenylated RNA in the nuclei became more prominent after 8 and 16 h of induction (Fig. 5 F and H, respectively). As an alternative indicator of mRNA export capacity, we measured [35S]methionine/cysteine incorporation into total cellular protein at various times after induction of SopB (Fig. 6). We reasoned that if mRNA is trapped in the nucleus, protein synthesis should decrease as cytosolic mRNA levels are depleted. By 16 h after induction of SopB expression, there was a marked 30% inhibition of total protein synthesis. Both the mRNA export and protein synthesis defects were reversed to the uninduced status after overnight incubation of the cells in the absence of tetracycline.
Figure 5.
In situ hybridization of mRNA in FSN cells. Cells were not induced (A and B) or induced for 4 h (C and D), 8 h (E and F), and 16 h (G and H). Cell nuclei were stained by using DAPI (Upper). mRNA was hybridized with a digoxigenin-coupled oligo(dT) and then visualized by using FITC-conjugated anti-digoxigenin Fab fragments.
Figure 6.
Inhibition of protein synthesis in FSN cells after tetracycline induction. Protein synthesis was measured as the incorporation of [35S]cysteine/methionine. The inhibition at 16 h was from three independent experiments and the standard deviations are shown.
Interestingly, although mRNA export defects were also observed with the expression of SopB that lacks a nuclear localization signal (FS), the results were not as penetrant or consistent as those observed with the FSN. This suggests that specific depletion of a nuclear inositol phosphate pool is required for the effective inhibition of export. It follows that a major site of inositol phosphate function may be in the nucleus. Finally, the fact that all of the perturbations noted in SopB-overexpressing cells are reversed on removal of tetracycline suggests that no permanent disruption of cell function is induced by depletion of the higher inositol phosphates. This is consistent with regulatory roles for such inositol molecules. Because FS did not enter the nucleus and had no consistent effect on mRNA export, we do not believe that perturbation of mRNA export is caused by SopB in Salmonella.
These results provide the first evidence that inositol phosphates are also required for the efficient export of mRNA in mammalian cells. Based on recent results in S. cerevisiae, we speculate that the production of InsP6 will be specifically required for mRNA export in mammalian cells. In addition, induction of SopB expression resulted in the coincident decrease in InsP6 levels. However, it is possible that depletion of Ins-1,3,4,5,6-P5 or other unmeasured changes in inositol phosphate metabolism caused by SopB expression play a role in the phenotype of these cells. Overall, these results support the hypothesis that the mechanism of inositol phosphate action in mRNA export will be conserved across species.
A recent study (27) has described an additional distinct nuclear inositol phosphate function by showing that in vitro InsP6 is required for efficient nonhomologous end-joining DNA repair. This recent emergence of functions for inositol phosphates in nuclear physiology expands further the panoply of intracellular processes that are carried out by such inositol signaling molecules. At present, there is no way to restore InsP5 or InsP6 levels in cells without altering other metabolites. Further studies pinpointing the mammalian inositol phosphates that are required for mRNA export will likely await the identification of mammalian enzymes that mediate InsP5 and InsP6 production and turnover.
Acknowledgments
We thank Li Cao and Cecil Buchanan for technical assistance and Monita Wilson, Kathryn Ryan, Lisa Strawn, and Mythili Suntharalingam for comments on the manuascript. This work was supported by Grants GM51219 (to S.R.W.), HL55772, and HL16634 (to P.W.M.) from the National Institutes of Health.
Abbreviations
- InsP5
inositol 1,3,4,5,6-pentakisphosphate
- InsP6
inositol hexakisphosphate
- Ins-1,4,5-P3
inositol 1,4,5-trisphosphate
- PtdIns-4,5-P2, phosphatidylinositol 4,5-bisphosphate
Ins-1,4,5,6-P4, inositol 1,4,5,6-tetrakisphosphate
- PtdIns
phosphatidylinositol
- DAPI
4′,6-diamidino-2-phenylindole
- FS
FLAG-SopB
- FSN
FLAG-SopB-NLS
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.021558098.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.021558098
References
- 1.Speed C J, Little P J, Hayman J A, Mitchell C A. EMBO J. 1996;15:4852–4861. [PMC free article] [PubMed] [Google Scholar]
- 2.Acharya J K, Labarca P, Delgado R, Jalink K, Zuker C S. Neuron. 1998;20:1219–1229. doi: 10.1016/s0896-6273(00)80502-4. [DOI] [PubMed] [Google Scholar]
- 3.Helgason C D, Damen J E, Rosten P, Grewal R, Sorensen P, Chappel S M, Borowski A, Jirik F, Krystal G, Humphries R K. Genes Dev. 1998;12:1610–1620. doi: 10.1101/gad.12.11.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shears S B. Biochim Biophys Acta. 1998;1436:49–67. doi: 10.1016/s0005-2760(98)00131-3. [DOI] [PubMed] [Google Scholar]
- 5.York J D, Odom R, Murphy R, Ives E B, Wente S R. Science. 1999;285:96–100. doi: 10.1126/science.285.5424.96. [DOI] [PubMed] [Google Scholar]
- 6.Odom A R, Stahlbert A, Wente S R, York J D. Science. 2000;287:2026–2029. doi: 10.1126/science.287.5460.2026. [DOI] [PubMed] [Google Scholar]
- 7.Ives E B, Nichols J, Wente S R, York J D. J Biol Chem. 2000;275:36575–36583. doi: 10.1074/jbc.M007586200. [DOI] [PubMed] [Google Scholar]
- 8.Galán J E. Proc Natl Acad Sci USA. 1998;95:14006–14008. doi: 10.1073/pnas.95.24.14006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee C A. Trends Microbiol. 1997;5:148–155. doi: 10.1016/S0966-842X(97)01029-9. [DOI] [PubMed] [Google Scholar]
- 10.Galyov E E, Wood M W, Rosqvist R, Mullan P B, Watson P R, Hedges S, Wallis T S. Mol Microbiol. 1997;25:903–912. doi: 10.1111/j.1365-2958.1997.mmi525.x. [DOI] [PubMed] [Google Scholar]
- 11.Norris F A, Majerus P W. J Biol Chem. 1994;269:8716–8720. [PubMed] [Google Scholar]
- 12.Norris F A, Auethavekiat V, Majerus P W. J Biol Chem. 1995;270:16128–16133. doi: 10.1074/jbc.270.27.16128. [DOI] [PubMed] [Google Scholar]
- 13.Norris F A, Atkins R C, Majerus P W. J Biol Chem. 1997;272:23859–23864. doi: 10.1074/jbc.272.38.23859. [DOI] [PubMed] [Google Scholar]
- 14.Norris F A, Wilson M P, Wallis T S, Galyov E E, Majerus P W. Proc Natl Acad Sci USA. 1998;95:14057–14059. doi: 10.1073/pnas.95.24.14057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eckmann L, Rudolf M T, Ptasznik A, Schultz C, Jiang T, Wolfson N, Tsien R, Fierer J, Shears S B, Kagnoff M F, et al. Proc Natl Acad Sci USA. 1997;94:14456–14460. doi: 10.1073/pnas.94.26.14456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozak M. Nucleic Acids Res. 1987;15:8125–8132. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hopp T P, Prickett K S, Price V L, Libby R T, March C J, Cerretti D P, Urdal D L, Conlon P J. Bio/Technology. 1988;6:1204–1210. [Google Scholar]
- 18.Kalderon D, Roberts B L, Richardson W D, Smith A E. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 19.Kalderon D, Richardson W D, Markham A F, Smith A E. Nature (London) 1984;311:33–38. doi: 10.1038/311033a0. [DOI] [PubMed] [Google Scholar]
- 20.Watkins J L, Murphy R, Emtage J L T, Wente S R. Proc Natl Acad Sci USA. 1998;95:6779–6784. doi: 10.1073/pnas.95.12.6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wente S R, Blobel G. J Cell Biol. 1993;123:275–284. doi: 10.1083/jcb.123.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson M P, Majerus P W. J Biol Chem. 1996;271:11904–11901. doi: 10.1074/jbc.271.20.11904. [DOI] [PubMed] [Google Scholar]
- 23.Lips D L, Majerus P W. J Biol Chem. 1989;264:8759–8763. [PubMed] [Google Scholar]
- 24.Kisseleva M V, Wilson M P, Majerus P W. J Biol Chem. 2000;275:20110–20116. doi: 10.1074/jbc.M910119199. [DOI] [PubMed] [Google Scholar]
- 25.Hogg M, Ng L L, Davies J E. Clin Sci. 1991;81:405–412. doi: 10.1042/cs0810405. [DOI] [PubMed] [Google Scholar]
- 26.Bonifacino J S. In: Current Protocols in Molecular Biology. Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. New York: Wiley; 1997. pp. 10.18.1–10.18.9. [Google Scholar]
- 27.Hanakahi L A, Bartlet-Jones M, Chappell C, Pappin D, West S C. Cell. 2000;102:721–729. doi: 10.1016/s0092-8674(00)00061-1. [DOI] [PubMed] [Google Scholar]