Abstract
Bacterial DNA-dependent RNA polymerase (RNAP) has subunit composition β′βαIαIIω. The role of ω has been unclear. We show that ω is homologous in sequence and structure to RPB6, an essential subunit shared in eukaryotic RNAP I, II, and III. In Escherichia coli, overproduction of ω suppresses the assembly defect caused by substitution of residue 1362 of the largest subunit of RNAP, β′. In yeast, overproduction of RPB6 suppresses the assembly defect caused by the equivalent substitution in the largest subunit of RNAP II, RPB1. High-resolution structural analysis of the ω–β′ interface in bacterial RNAP, and comparison with the RPB6–RPB1 interface in yeast RNAP II, confirms the structural relationship and suggests a “latching” mechanism for the role of ω and RPB6 in promoting RNAP assembly.
Keywords: transcription, β′ subunit, RPB1 subunit
The DNA-dependent RNA polymerase (RNAP) is central to all steps of the transcription cycle (1–5). Bacterial, archaeal, and eukaryotic cellular RNAPs are large, multisubunit enzymes. Bacterial RNAP core enzyme consists of five subunits (β′, β, αΙ, αII, and ω) and has a molecular mass of ≈0.35 MDa (1, 6). Archaeal and eukaryotic RNAP core enzymes consist of 10 to 20 subunits and have molecular masses of 0.4–0.8 MDa (3–5, 7).
It has been shown previously that four subunits of bacterial RNAP core enzyme have sequence, structural, and functional homologs in archaeal and eukaryotic RNAP (2–16). Bacterial RNAP subunit β′, which is the largest subunit and which is involved in catalysis, corresponds to archaeal RNAP subunit RpoA′/RpoA" and eukaryotic RNAP I, II, and III subunits RPA1, RPB1, and RPC1. Bacterial RNAP subunit β, which is the second-largest subunit and which also is involved in catalysis, corresponds to archaeal RNAP subunit RpoB (or RpoB′/RpoB") and eukaryotic RNAP I, II, and III subunits RPA2, RPB2, and RPC2. Bacterial RNAP subunits αI and αII, which are identical in sequence but different in location within RNAP (with αI interacting with β, and αII interacting with β′) and which are involved in RNAP assembly and transcriptional regulation, correspond to archaeal RNAP subunits RpoD and RpoL, eukaryotic RNAP I and III subunits RPC5 and RPC9 (also known as RPAC40 and RPAC19), and eukaryotic RNAP II subunits RPB3 and RPB11.
The role of the fifth subunit of bacterial RNAP core enzyme, ω (17), has been unclear. On the one hand, the Escherichia coli rpoZ gene, which encodes ω, is not essential for viability under standard laboratory conditions (18), and reconstituted RNAP lacking ω is indistinguishable from RNAP containing ω in in vitro transcription assays (refs. 19 and 20; K.S., unpublished data). On the other hand, ω homologs are present in all sequenced genomes of free-living bacteria, suggesting an important, conserved function (Fig. 1), deletion of ω results in a slow-growth phenotype (21), deletion of ω results in association of RNAP with the molecular chaperone GroEL in vivo (22), and ω significantly increases the yield of correctly assembled, active RNAP during in vitro reconstitution of RNAP (ref. 21; D. Markov and K.S., unpublished data).
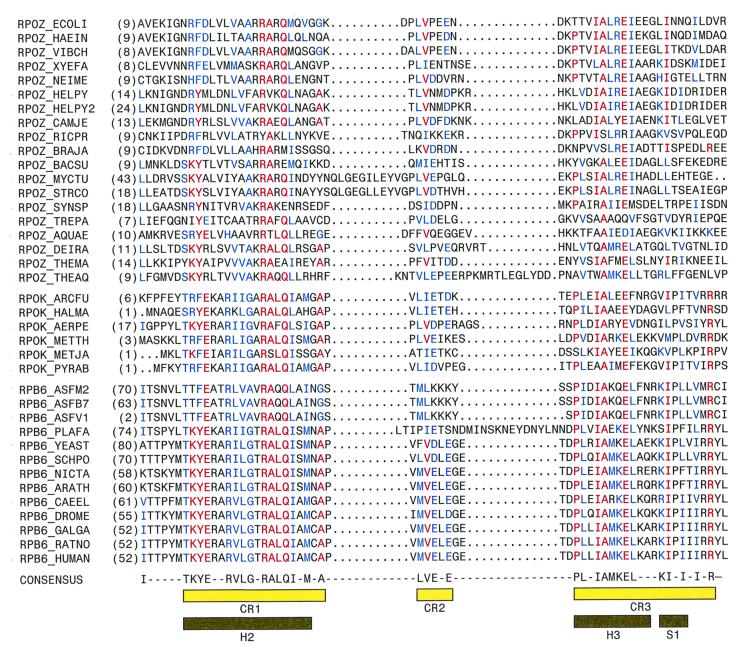
Figure 1.
Bacterial ω, archaeal RpoK, and eukaryotic RPB6 are sequence homologs. Aligned sequences of bacterial RNAP ω (Top), archaeal RNAP RpoK (Middle), and poxviral and eukaryotic RNAP RPB6 (Bottom). Residues identical in at least half of the aligned sequences and represented in all three sets of aligned sequences are in red; residues identical or similar in at least half of the aligned sequences and represented in all three sets of aligned sequences are in blue. CR1–CR3 (yellow bars) delineate conserved regions (defined as containing residues identical or similar in at least half of the aligned sequences and represented in all three sets of aligned sequences, and containing no insertions or deletions greater than one residue). Helices 2 and 3 and strand 1 in the crystallographic structure of Thermus aquaticus ω (Fig. 3) are indicated by black bars. Species names and database locus identifiers for the sequences are, in order: Escherichia coli (RPOZ_ECOLI), Haemophilus influenzae (RPOZ_HAEIN), Vibrio cholerae (AAF95849), Xylella fastidiosa (AE003980_3), Neisseria meningitidis (AAF42009), Helicobacter pylori J99 (D71898), Helicobacter pylori 26695 (H64616), Campylobacter jejuni (CAB73527), Rickettsia prowazekii (Y578_RICPR), Bradyrhizobium japonicum (AAF04326), Bacillus subtilis (C69878), Mycobacterium tuberculosis (YD90_MYCTU), Streptomyces coelicolor (CAB93358), Synechocystis sp. (Y61L_SYNY3), Treponema pallidum (A71289), Aquifex aeolicus (F70317), Deinococcus radiodurans (A75266), Thermotoga maritima (B72223), Thermus aquaticus (AJ295839; this work), Archaeoglobus fulgidus (RPOK_ARCFU), Haloarcula marismortui (RPOK_HALMA), Aeropyrum pernix (RPOK_AERPE), Methanobacterium thermoautotrophicum (RPOK_METTH), Methanococcus jannaschii (RPOK_METJA), Pyrococcus abyssi (B75172), African swine fever virus ASF M2 (RPB6_ASFM2), ASF B7 (RPB6_ASFB7), ASF 1 (S35644), Plasmodium falciparum (T18424), Saccharomyces cerevisiae RPB6_YEAST), Schizosaccharomyces pombe (RPB6_SCHPO), Nicotiana tabacum (AF153277_1), Arabidopsis thaliana AC006955_29), Caenorhabditis elegans (RPB6_CAEEL), Drosophila melanogaster (RPB6_DROME), Gallus gallus (CAB62065), Rattus norvegicus (RPB6_RAT), and Homo sapiens (RPB6_HUMAN). Chloroplast RNAP is closely related to bacterial RNAP (39); putative ω homologs are present in chloroplasts from red algae, cryptophyte algae, and low-branching green flagellates [Porphyra purpurea (YC61_PORPU), Cyanidium caldarium (YC61_CYACA), Guillardia theta (YC61_GUITH), and Mesostigma viride (AF166114–93); unpublished data]. Sequences of chloroplast putative ω homologs are highly similar to the sequence of Synechocystis sp. ω and can be aligned analogously.
In this work, we show that ω is homologous in sequence to archaeal RNAP subunit RpoK and is homologous in sequence, structure, and function to eukaryotic RNAP I, II, and III subunit RPB6. In addition, based on structural and genetic analysis, we suggest that ω and RPB6 function in RNAP assembly by “latching” the N- and C-terminal regions of the RNAP largest subunit, thereby facilitating association of the RNAP largest subunit with the βαΙαII assembly intermediate (1) in bacteria and with the corresponding assembly intermediates (12, 23) in eukaryotes.
Materials and Methods
Sequence Analysis.
psi-blast (24) searches were performed (www.ncbi.nlm.nih.gov) with the following specifications: default filtering; substitution matrix = blosum 80; E-value cut-off for iteration 1 = 0.1; E-value cut-off for subsequent iterations = 0.5. Multiple-sequence alignment was performed by ClustalX-based alignment of ω sequences using align-x 5.5 (Informax; substitution matrix = blosum 80; gap-opening penalty = 10; gap-extension penalty = 0.05), followed by manual adjustment, followed by profile-based alignment of RpoK and RPB6 sequences using align-x 5.5 (substitution matrix = blosum 80; gap-opening penalty = 10; gap-extension penalty = 0.05), followed by further manual adjustment (Fig. 1). Multiple-sequence alignment also was performed in fully objective fashion using Ballast 1.0 (ref. 25; Irix 6.5; input = psi-blast report edited to conform to blastp report format) followed by DbClustal 1.0 (ref. 25; Irix 6.5; anchor source = Ballast 1.0 report, with propagation of anchors among all sequences; substitution matrix = Gonnet series; gap-opening penalty = 10; gap-extension penalty for pair-wise comparison = 1; gap-extension penalty for multiple comparison = 0.2; terminal gap penalties = 0) (unpublished data). Results of the two multiple-sequence alignment procedures were nearly identical (identical for CR1 and CR3; nearly identical for CR2, with differences in alignment of CR2 for only 6 of 38 sequences).
Cloning and Sequence Determination.
A ≈90-bp segment of the gene encoding Thermus aquaticus ω was amplified by PCR from T. aquaticus genomic DNA using degenerate primers. The upstream primer, 5′-CCS GGS ATC GAC AAC CTS TTC GG-3′, was designed based on the N-terminal amino acid sequence of the ≈11-kDa polypeptide present in the preparation of T. aquaticus RNAP core enzyme of ref. 6 (XXPGIDLFG; sequence from material isolated by SDS/PAGE and blotted to poly(vinylidene difluoride); sequencing by automated gas-phase Edman analysis at the Rockefeller University Protein-DNA Technology Center). The downstream primer, 5′-SAG CTG CAG SCG GGC CTT SGC-3′, was designed based on the consensus sequence for ω CR1 (Fig. 1). A SacI plasmid library of T. aquaticus genomic DNA was prepared, a clone containing the gene was identified by colony hybridization, plasmid DNA from the clone containing the gene was isolated, and the DNA-nucleotide sequence of the gene was determined (methods as in ref. 26); the sequence of 99-amino acid-long ω was deduced (MAEPGIDKLFGMVDSKYRLTVVVAKRAQQLLRHRFKNTVLEPEERPKMRTLEGLYDDPNAVTWAMKELLTGRLFFGENLVPEDRLQKEMERLYPTEEEA).
Structure Determination.
In the published structure of T. aquaticus core RNAP, electron density corresponding to a ≈90-residue polypeptide tentatively was identified as ω and was modeled as a polyalanine chain (6). We have modeled the sequence of T. aquaticus ω, determined as described above, into this density. The sequence matched the electron density features well (Fig. 2), and all internal Met residues in the sequence (residues 12, 48, 65, and 89) corresponded to peaks in the selenomethionine (SeMet) Fourier difference map obtained using SeMet-substituted RNAP (ref. 6 and Fig. 2). Refinement of the model, in conjunction with rebuilding and refinement of the entire structure of RNAP core enzyme, was performed (methods as in ref. 6). The current structure has an R factor of 28% (Rfree = 36%) and includes residues 2–96 of ω (of 99 total residues). Coordinates for the current structure have been deposited in the Protein Data Bank. dali (27) structure comparisons were performed (www2.ebi.ac.uk/dali).
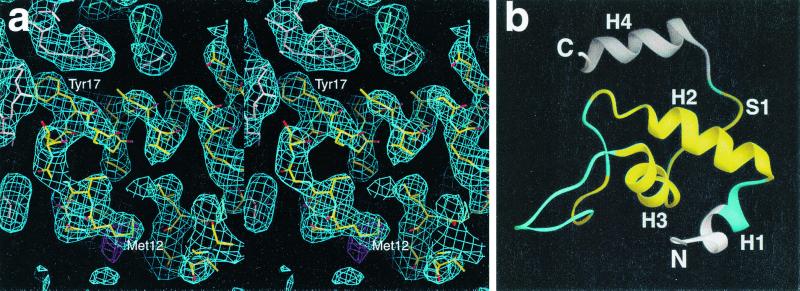
Figure 2.
Structure determination. (a) Stereo view of a portion of the 2|Fo| − |Fc| electron density map (3.2 Å, 1σ, shown in blue) calculated from the T. aquaticus core RNAP structure, showing a region corresponding to the ω subunit (including the N-terminal part of CR1 α-helix; at center, oriented horizontally) and nearby parts of β and β′. Atoms of ω are colored by atom type (C, yellow; O, red; N, blue; S, green). Atoms of β′ and β are colored pink and light blue, respectively. The SeMet difference Fourier peak (3σ) that corresponds to Met12 of ω is shown in magenta. Selected residues of ω are labeled. The figure was generated by using the program o (40). (b) Structure of the ω subunit in T. aquaticus RNAP core enzyme. A ribbon representation of T. aquaticus ω residues (residues 2–96) is shown. Residues of ω not included in the sequence alignment in Fig. 1 are illustrated in white; conserved regions CR1–CR3 are in yellow; nonconserved regions are in cyan. S1 is part of intersubunit β-sheet (two-strand antiparallel β-sheet with residues 1483–1487 of the C-terminal tail of β′).
Plasmids and Strains.
E. coli strains harboring rpoCtsX, rpoCts4, and rpoC397c mutations were described previously (28). For overproduction of ω, plasmids pE3C2 and pGP1–2 (29) were used. Yeast strain H27 (ref. 30; MATa URA3-52 HIS3,4 TRP1 LEU2-3,112 RPB1-1) was provided by M. Hampsey (UMDNJ). RPB6 plasmid pSN316 and vector-only control plasmid pRS424 (ref. 31; 2μ, TRP1) were kindly provided by J. Friesen (University of Toronto).
Results
Bacterial ω, Archaeal RpoK, and Eukaryotic RPB6 Are Sequence Homologs.
In the course of systematic psi-blast searches using bacterial, archaeal, and eukaryotic RNAP subunit sequences as queries, we have encountered a previously unrecognized, but unequivocal, sequence relationship among bacterial RNAP subunit ω, archaeal RNAP subunit RpoK, and eukaryotic RNAP I, II, and III subunit RPB6. Thus, psi-blast searches using ω sequences as queries (E. coli ω residues 10–65, Vibrio cholerae ω, Bacillus subtilis ω, or Xylella fastidiosa ω) retrieve RpoK and RPB6 sequences; psi-blast searches using RpoK sequences as queries (Archaeoglobus fulgidus RpoK, Haloarcula marismortui RpoK, Methanobacterium thermoautotrophicum RpoK, or Pyrococcus abyssi RpoK) retrieve ω and RPB6 sequences; and psi-blast searches using RPB6 sequences as queries (Arabidopsis thaliana RPB6, Caenorhabditis elegans RPB6, Drosophila melanogaster RPB6, or Homo sapiens RPB6) retrieve ω and RpoK sequences. In each case, E scores are ≪1 × 10−4.
Multiple-sequence alignment of bacterial ω, archaeal RpoK, and poxviral and eukaryotic RPB6 sequences retrieved from psi-blast searches using bacterial ω sequences as queries confirms the sequence relationship and defines three regions of sequence similarity (Fig. 1): conserved region 1 (CR1, corresponding to residues 15–34 of E. coli ω), conserved region 2 (CR2, corresponding to residues 38–42 of E. coli ω), and conserved region 3 (CR3, corresponding to residues 46–65 of E. coli ω). CR1 and CR3 exhibit high sequence similarity, containing, respectively, eight positions and six positions with residues identical in at least half of aligned sequences and represented in all three sets of aligned sequences (ω, RpoK, and RPB6); CR2 exhibits modest sequence similarity (Fig. 1).
Bacterial ω and Eukaryotic RPB6 Exhibit Structural Similarity.
The structure of RNAP core enzyme from the thermophilic bacterium T. aquaticus revealed a molecule with a “crab-claw” shape, with a central mass formed by residues of β′, β, αΙ, and αΙΙ, and two prominent pincer-like projections formed by residues of β′ and β (6). In addition, the structure contained a ≈90-amino acid polypeptide of unknown sequence, corresponding to an 11-kDa polypeptide present in the preparation of T. aquaticus RNAP core enzyme, tentatively identified as T. aquaticus ω (based on the correspondence of the length and predicted secondary structure of the polypeptide to the length and predicted secondary structure of E. coli ω; ref. 6).
We have determined the N-terminal amino acid sequence of the 11-kDa polypeptide present in the preparation of T. aquaticus RNAP core enzyme and cloned and sequenced the corresponding gene (see Materials and Methods). The inferred amino acid sequence of the 11-kDa polypeptide exhibits high similarity to bacterial ω sequences, confirming the identity of the 11-kDa polypeptide as T. aquaticus ω (Fig. 1). We have modeled the amino acid sequence into the structure of T. aquaticus RNAP core. The sequence matches experimental electron density well (Fig. 2), and each internal Met residue in the sequence corresponds to a peak in the SeMet Fourier difference map obtained by using SeMet-substituted RNAP (ref. 6; Fig. 2).
The residues of ω shown in the sequence alignment in Fig. 1 form a discrete, compact, globular protein domain (Figs. 1, 2b, and 3a). Conserved regions CR1–CR3 form the core of the protein domain in ω (yellow in Figs. 2b and 3a). The nonconserved regions between CR1 and CR2 and between CR2 and CR3 form loops that make excursions from the protein domain (cyan in Figs. 2b and 3a). Conserved regions CR1 and CR3 correspond to specific secondary-structure elements (Figs. 1, 2b, and 3a). CR1 corresponds, almost precisely, to α-helix 2; the first half of CR3 corresponds to α-helix 3; the second half of CR3 corresponds to β-strand 2 (Fig. 2b).
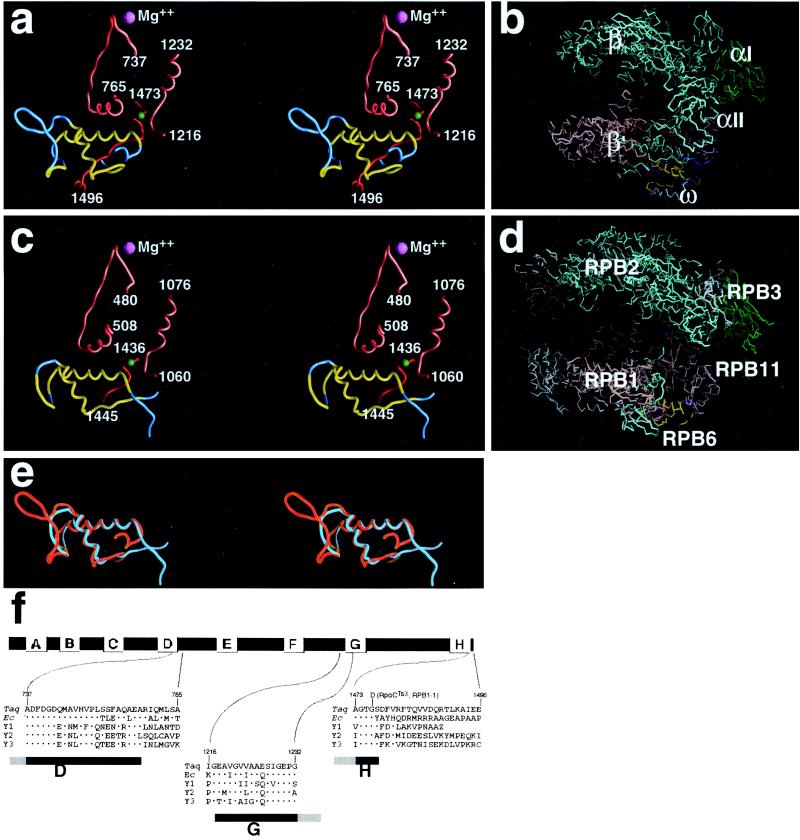
Figure 3.
Bacterial ω and eukaryotic RPB6 are structural homologs. (a) Structure of ω and the ω–β′ interface in T. aquaticus RNAP core enzyme. Residues of ω included in the sequence alignment in Fig. 1 are illustrated in a ribbon representation (residues 9–81); conserved regions CR1–CR3 are in yellow; nonconserved regions are in cyan. Residues of β′ conserved regions D and G are in pink; residues of the β′ C-terminal tail are in red, with the residue corresponding to the residue substituted in the E. coli rpoCtsx mutant (panel f and Fig. 4a) indicated in green. (b) Location of ω within T. aquaticus RNAP core enzyme. The structure of T. aquaticus RNAP core enzyme is illustrated in a Cα representation. Conserved regions CR1–CR3 of ω are in yellow; nonconserved regions of ω are in blue; β′ is in pink, with the β′ C-terminal tail in red; β is in cyan, αΙ and αΙΙ are in green; the active-center Mg2+ is in magenta. (c) Structure of RPB6 and the RPB6–RPB1 interface in yeast RNAP II (atomic coordinates from ref. 2, PDB accession code 1EN0, with reassignment of 79 C-terminal residues of RPB1 based on refined atomic coordinates) (P. Cramer, D. Bushnell, and R. Kornberg, personal communication). Residues of RPB6 included in the sequence alignment in Fig. 1 are illustrated in a ribbon representation (residues 80–138); conserved regions CR1–CR3 are in yellow; nonconserved regions are in cyan. Residues of RPB1 conserved regions D and G are in pink; residues of the RPB1 C-terminal tail are in red, with the residue substituted in RPB1-1 (panel f and Fig. 4b) indicated in green. (Residues of the C-terminal tail following residue Ile1445 are not defined in the available structure.) Residue numbers in structural elements of RPB6 and RPB1 were inferred by reference to residue numbers in structurally equivalent elements of ω and β′ (a) and to sequence alignments (Fig. 1; also panel f herein), and are expected to be correct within ≈1 residue. (d) Location of RPB6 within yeast RNAP II (atomic coordinates as in c). The structure of yeast RNAP II is illustrated in a Cα representation. Conserved regions CR1–CR3 of RPB6 are in yellow; nonconserved regions of RPB6 are in blue; RPB1 is in pink, with the RPB1 C-terminal tail in red; RPB2 is in cyan, RPB3 and RPB11 are in green; subunits of RNAP II without counterparts in bacterial RNAP (RPB5, RPB8, RPB9, RPB10, and RPB12) are in gray; the active-center Mg2+ is in magenta. (e) Structural alignment of ω (cyan; residues 9–81) and RPB6 (red; residues 80–138). (f) Sequences of segments of the RNAP largest subunit that interact with ω (a) and RBP6 (c). Conserved regions of the RNAP largest subunit are indicated by lettered boxes (8). The amino acid substitutions in E. coli rpoCtsX and yeast RPB1-1 are indicated above the aligned sequences. Sequences shown are, in order: T. aquaticus β′ (CAB65466), E. coli β′ (RPOC_ECOLI), S. cerevisiae RPA1 (RPA1_YEAST), S. cerevisiae RPB1 (RPB1_YEAST), and S. cerevisiae RPC1 (RPC1_YEAST). The dots indicate amino acid identities.
One face of the α-helix corresponding to CR1 of ω interacts with an α-helix within conserved region D of β′ (residues 755–762; pink in Fig. 3a). Another face of the α-helix corresponding to CR1 interacts with conserved region G of β′ (residues 1216–1220; pink in Fig. 3a) and with residues 1476–1486 of the C-terminal tail of β′ (red in Fig. 3a). The α-helix corresponding to the first half of CR3 makes additional interactions with the C-terminal tail of β′. The β-strand corresponding to the second half of CR3 wraps over and around the C-terminal tail of β′, with residues 1483–1489 of the C-terminal tail of β′ literally being threaded through the narrow gap between CR1 and the second half of CR3 (Fig. 3a). Nonconserved residues C-terminal to CR3 form an α-helix (α-helix 4; residues 83–91, DRLQKEMER, Fig. 2b) that further secures the C-terminal tail of β′ (not shown). (The presence of α-helix 4, and interactions by α-helix 4, may be unique to T. aquaticus ω, which is longer than most examples of ω.) The extensive contacts between the ω subunit and β′ are in agreement with published crosslinking studies (32).
The α-carbon backbone of yeast RNAP II has been determined by x-ray analysis at 3.0-Å resolution (7). If the sequence similarities between ω and RPB6 described above are meaningful, one would expect that (i) ω and RPB6 would be structurally similar, and (ii) ω and RPB6 would interact with the remainder of RNAP in a structurally similar fashion. The structural analysis presented in Fig. 3 reveals that these expectations are fulfilled.
First, the conserved regions of ω and RPB6 are structurally equivalent (Fig. 3e). The structures of ω and RPB6 are superimposible, with a rms deviation of 2.2 Å for 47 superimposed Cα atoms (Fig. 3e). As in ω, in RPB6, CR1 corresponds to an α-helix, the N-terminal half of CR3 corresponds to an α-helix, and the C-terminal half of CR3 corresponds to a β-strand.
Second, the conserved regions of ω and RPB6 are positioned equivalently relative to the remainder of RNAP, each being located at the base of the pincer-like projection formed by the largest RNAP subunit (compare Fig. 3 b and d), and each making interactions with conserved region D, conserved region G, and the C-terminal tail of the largest RNAP subunit (compare Fig. 3 a and c).
Bacterial ω and Eukaryotic RPB6 Exhibit Functional Similarity.
To determine whether ω and RPB6 exhibit functional similarity, we made use of the serendipitous observation that the E. coli rpoCtsX mutation and the yeast RPB1-1 mutation result in amino acid substitutions that are equivalent, that are located in the ω/β′ and RPB6/RPB1 interfaces, that destabilize RNAP, and that result in temperature-sensitive phenotypes (28, 30, 33).
The E. coli rpoCtsX mutation results in replacement of Gly1360, within conserved region H of β′, by Asp (ref. 28; Fig. 3f). In the structure of T. aquaticus RNAP, Gly1474, which corresponds to E. coli Gly1360, is located close to the point where the C-terminal tail of β′ emerges from the structure of RNAP and is in direct van der Waals contact with the third residue of CR1 of ω (Fig. 3a, green sphere). RpoCtsX mutants are temperature-sensitive for growth (28). The mutant RNAP is temperature-sensitive for activity, is so unstable that it can be purified only by use of high concentrations of glycerol in chromatographic buffers, and exhibits a high tendency to dissociate into β′and βαΙαΙΙ subassembly (ref. 34; E. Nedea, D. Markov, and K.S., unpublished results).
The yeast RPB1-1 mutation results in replacement of the corresponding residue of conserved region H of RPB1 (residue Gly1437) by Asp (ref. 33; Fig. 3f). In the structure of RNAP II (7), this residue is expected to be located close to the point where the C-terminal tail of RPB1 emerges from the structure of RNAP and to be in direct van der Waals contact with the third residue of CR1 of RPB6 (Fig. 3c, green sphere). RPB1-1 mutants are temperature-sensitive for growth and cease transcription immediately upon temperature upshift, suggesting that the mutant RNAP is temperature-sensitive for activity (30). The mutant RNAP is so unstable that it cannot be purified by conventional procedures (30). Overexpression of RPB1-1 suppresses the temperature-sensitive phenotype (35), suggesting that RPB1-1 is partially defective in interaction with the rest of RNAP.
We have compared effects of overproduction of ω on the temperature-sensitive phenotype of E. coli rpoCtsX cells and of overproduction of RPB6 on the temperature-sensitive phenotype of yeast RPB1-1 cells. To assess effects of overproduction of ω on the temperature-sensitive phenotype of E. coli rpoCtsX cells, we introduced a pair of plasmids encoding ω under control of the λ PR promoter (29), or a vector-only control plasmid (29), into rpoCtsX cells and plated at permissive and nonpermissive temperatures (37°C and 42°C). As a control, to assess allele specificity of suppression, we also introduced the same pair of plasmids into rpoCts4 cells (which produce a β′ derivative having a replacement of Gly181 by Asp, and which exhibit a temperature-sensitive defect in RNAP assembly) (ref. 28; E. Nedea and K.S., unpublished results) and rpoC397C cells (which produce a β′ derivative lacking residues 1359–1407, and which exhibit a temperature-sensitive defect in RNAP assembly) (ref. 36; E. Nedea and K.S., unpublished results).
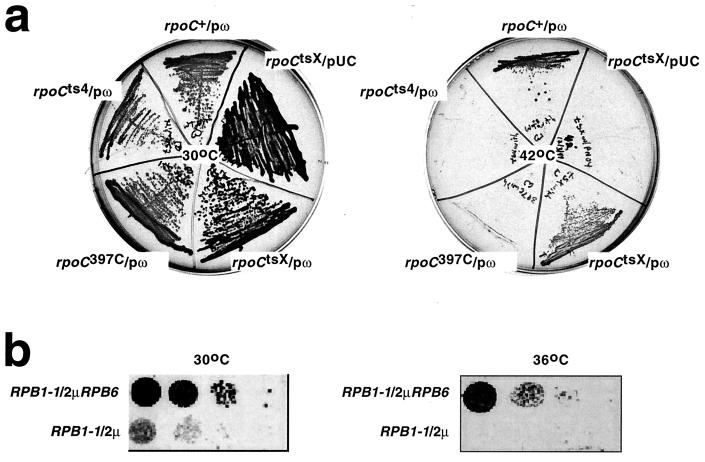
Overproduction of ω suppressed the temperature-sensitive phenotype of rpoCtsX cells but did not suppress the temperature-sensitive phenotypes of rpoCts4 cells and rpoC397C cells (Fig. 4a). To assess effects of overproduction of RPB6 on the temperature-sensitive phenotype of yeast RPB1-1 cells, we introduced a high-copy number plasmid encoding RPB6 (31), or a vector-only control plasmid (31), into RPB1-1 cells and plated at permissive and nonpermissive temperatures (30°C and 36°C). Overproduction of RPB6 suppressed the temperature-sensitive phenotype of RPB1-1 (Fig. 4b). Thus, overproduction of ω and overproduction of RPB6 have equivalent (and allele-specific) suppressing effects on temperature-sensitive phenotypes of equivalent substitutions within β′ and RPB1.
Figure 4.
Bacterial ω and eukaryotic RPB6 are functional homologs. (a) Effects of overproduction of ω on the temperature-sensitive phenotypes of rpoCtsX. The figure shows 15-h growth on LB/ampicillin plates of rpoCtsX cells, rpoCts4 cells, and rpoC397C cells, transformed with plasmids overproducing ω (pω), or a control plasmid (pUC). (Left) Permissive temperature (30°C). (Right) Nonpermissive temperature (42°C). (b) Effects of overproduction of RPB6 on the temperature-sensitive phenotypes of RPB1-1. The figure shows 48-h growth on YEPD plates of RPB1-1 cells transformed with a plasmid overproducing RPB6 (2μRPB6), or a control plasmid (2μ). (Left) Permissive temperature (30°C). (Right) Nonpermissive temperature (36°C).
Discussion
The principal result of this work is the demonstration that bacterial RNAP subunit ω is homologous in sequence to archaeal RNAP subunit RpoK and homologous in sequence, structure, and function to eukaryotic RNAP I, II, and III subunit RPB6. Previous work had demonstrated that four subunits of bacterial RNAP (β′, β, αΙ, and αΙΙ) have counterparts in archaeal and eukaryotic RNAP. It is now clear that the similarity extends further than had been anticipated and that, in fact, all five subunits of bacterial RNAP have counterparts in archaeal and eukaryotic RNAP.
A further result of this work is the demonstration that ω and RPB6 promote RNAP assembly, and/or increase RNAP stability, through specific interactions with the RNAP largest subunit (β′ in bacteria, RPB1 in eukaryotic RNAP II). A precedent for this result comes from the observation that overproduction of RPB6 suppresses the temperature-sensitive phenotype of RPB1-rpo21-4 (37, 38). The RPB1-rpo21-4 mutation results in a four-residue insertion between residues 594 and 595 of yeast RPB1 (38)—residues that, based on the structure of yeast RNAP II (7), are expected to be close to RPB6. A further precedent for this result comes from the observation that mutation of RPB6 results in a decrease in the steady-state level of RPB1 in vivo (31). The observation that mutation of RPB6 likewise results in a decrease in the steady-state level of the largest subunit of eukaryotic RNAP I in vivo (31) supports the reasonable inference that RPB6 likewise promotes RNAP assembly, and/or increases RNAP assembly, through interactions with the largest subunits of eukaryotic RNAP I and RNAP III.
The structures of ω and RBP6, and the structures of the ω/β′ and RPB6/RPB1 interfaces, suggest a molecular mechanism for the function of ω and RPB6 in promoting RNAP assembly and/or stability. The conserved regions of ω and RPB6 form a compact structural domain that interacts simultaneously with conserved regions D and G of the largest RNAP subunit and with the C-terminal tail following conserved region H of the largest RNAP subunit (Fig. 3 a and c). The second half of CR3 of ω and RPB6 forms an arc that projects away from the remainder of the structural domain and wraps over and around the C-terminal tail of the largest RNAP subunit, clamping it in a crevice formed by CR1 and the first half of CR3, and threading the C-terminal tail of the largest RNAP subunit through the narrow gap between CR1 and the second half of CR3 of ω and RPB6 (Fig. 3 a and c). We propose that ω and RPB6 promote RNAP assembly by acting as molecular “hook-fasteners” or “latches” (with the C-terminal residues of ω and RPB6 corresponding to the hook or hasp). Specifically, we propose that ω and RPB6 latch conserved regions D and G of the largest RNAP subunit to the C-terminal tail of the largest RNAP subunit, thereby conformationally constraining N- and C-terminal regions of the largest subunit in a manner that reduces the configurational entropy of the largest subunit and that facilitates interaction of the largest subunit with the βαΙαΙΙ assembly intermediate (1) in bacterial RNAP and with the corresponding assembly intermediates (12, 23) in eukaryotic RNAP. Consistent with this proposal, deletion of the C-terminal tail of β′ does not abrogate binding of ω to β′ but does abrogate function of ω in promoting RNAP assembly and stability (mutant rpoC397C) (ref. 36; Fig. 4a; L.M. and K.S., unpublished data).
Acknowledgments
We thank F. Plewniak for Irix 6.5 executables for Ballast 1.0; J. Friesen and M. Hampsey for strains and plasmids; and R. Kornberg and P. Cramer for discussion. This work was supported by National Institutes of Health Grant GM53759 (to S.A.D.); Howard Hughes Medical Investigatorship and National Institutes of Health Grants GM41376 and GM53665 (to R.H.E.); and a Career Award in Biomedical Sciences from the Borroughs Welcome Fund for Biomedical Research, a March of Dimes Research Grant, and National Institutes of Health Grant GM59295 (to K.S.). L.M. is a recipient of a Charles and Johanna Busch Postdoctoral Fellowship.
Abbreviations
- RNAP
RNA polymerase
- SeMet
selenomethionine
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AJ295839).
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1HQM).
References
- 1.Chamberlin M. In: RNA Polymerase. Losick R, Chamberlin M, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1977. pp. 17–67. [Google Scholar]
- 2.Young R A. Annu Rev Biochem. 1991;60:689–715. doi: 10.1146/annurev.bi.60.070191.003353. [DOI] [PubMed] [Google Scholar]
- 3.Thurioux P, Sentenac A. In: The Molecular and Cellular Biology of the Yeast Saccharomyces: Gene Expression. Jones E W, Pringle J R, Broach J R, editors. Vol. 2. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. pp. 1–48. [Google Scholar]
- 4.Archambault J, Friesen J D. Microbiol Rev. 1993;57:703–724. doi: 10.1128/mr.57.3.703-724.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bell S D, Jackson S P. Cold Spring Harbor Symp Quant Biol. 1998;63:41–51. doi: 10.1101/sqb.1998.63.41. [DOI] [PubMed] [Google Scholar]
- 6.Zhang G, Campbell L, Minakhin L, Richter C, Severinov K, Darst S A. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]
- 7.Cramer P, Bushnell D A, Fu J, Gnatt A L, Maier-Davis B, Thompson N E, Burgess R R, Edwards A M, David P R, Kornberg R D. Science. 2000;288:640–649. doi: 10.1126/science.288.5466.640. [DOI] [PubMed] [Google Scholar]
- 8.Allison L A, Moyle M, Shales M, Ingles C J. Cell. 1985;42:599–610. doi: 10.1016/0092-8674(85)90117-5. [DOI] [PubMed] [Google Scholar]
- 9.Sweetser D, Nonet M, Young R A. Proc Natl Acad Sci USA. 1987;84:1192–1196. doi: 10.1073/pnas.84.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang G, Darst S A. Science. 1998;281:262–266. doi: 10.1126/science.281.5374.262. [DOI] [PubMed] [Google Scholar]
- 11.Larkin R M, Guilfoyle T J. J Biol Chem. 1997;272:12824–12830. doi: 10.1074/jbc.272.19.12824. [DOI] [PubMed] [Google Scholar]
- 12.Kimura M, Ishiguro A, Ishihama A. J Biol Chem. 1997;272:25851–25855. doi: 10.1074/jbc.272.41.25851. [DOI] [PubMed] [Google Scholar]
- 13.Svetlov V, Nolan K, Burgess R R. J Biol Chem. 1998;273:10827–10830. doi: 10.1074/jbc.273.18.10827. [DOI] [PubMed] [Google Scholar]
- 14.Eloranta J J, Kato A, Teng M S, Weinzierl R O. Nucleic Acids Res. 1998;26:5562–5567. doi: 10.1093/nar/26.24.5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan Q, Linask K L, Ebright R H, Woychik N A. Genes Dev. 2000;14:339–348. [PMC free article] [PubMed] [Google Scholar]
- 16.Naryshkina T, Rogulja D, Golub L, Severinov K. J Biol Chem. 2000;275:31183–31190. doi: 10.1074/jbc.M003884200. [DOI] [PubMed] [Google Scholar]
- 17.Burgess R R. J Biol Chem. 1969;244:6168–6178. [PubMed] [Google Scholar]
- 18.Gentry D, Xiao H, Burgess R, Cashel M. J Bacteriol. 1991;173:3901–3903. doi: 10.1128/jb.173.12.3901-3903.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zalenskaya K, Lee J, Gujuluva C N, Shin Y K, Slutsky M, Goldfarb A. Gene. 1990;89:7–12. doi: 10.1016/0378-1119(90)90199-2. [DOI] [PubMed] [Google Scholar]
- 20.Tang H, Severinov K, Goldfarb A, Ebright R. Proc Natl Acad Sci USA. 1995;92:4902–4906. doi: 10.1073/pnas.92.11.4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mukherjee K, Chatterji D. Eur J Biochem. 1997;247:884–889. doi: 10.1111/j.1432-1033.1997.00884.x. [DOI] [PubMed] [Google Scholar]
- 22.Mukherjee K, Nagai H, Shimamoto N, Chatterji D. Eur J Biochem. 1999;266:228–235. doi: 10.1046/j.1432-1327.1999.00848.x. [DOI] [PubMed] [Google Scholar]
- 23.Kolodziej P A, Young R A. Mol Cell Biol. 1991;11:4669–4678. doi: 10.1128/mcb.11.9.4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson J D, Plewniak F, Thierry J, Poch O. Nucleic Acids Res. 2000;28:2919–2926. doi: 10.1093/nar/28.15.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 27.Holm L, Sander C. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 28.Nedea E C, Markov D, Naryshkina T, Severinov K. J Bacteriol. 1999;181:2663–2665. doi: 10.1128/jb.181.8.2663-2665.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gentry D R, Burgess R R. Protein Expression Purif. 1990;1:81–86. doi: 10.1016/1046-5928(90)90050-9. [DOI] [PubMed] [Google Scholar]
- 30.Nonet M, Scafe C, Sexton J, Young R. Mol Cell Biol. 1987;7:1602–1611. doi: 10.1128/mcb.7.5.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nouraini S, Archambault J, Friesen J D. Mol Cell Biol. 1996;16:5985–5996. doi: 10.1128/mcb.16.11.5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gentry D R, Burgess R R. Biochemistry. 1993;32:11224–11227. doi: 10.1021/bi00092a036. [DOI] [PubMed] [Google Scholar]
- 33.Scafe C, Martin C, Nonet M, Podos S, Okamura S, Young R A. Mol Cell Biol. 1990;10:1270–1275. doi: 10.1128/mcb.10.3.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Panny S R, Heil A, Mazus B, Palm P, Zillig W, Mindlin S Z, Ilyina T S, Khesin R B. FEBS Lett. 1974;48:241–245. doi: 10.1016/0014-5793(74)80477-1. [DOI] [PubMed] [Google Scholar]
- 35.Nouraini S, Xu D, Nelson S, Lee M, Friesen J D. Nucleic Acids Res. 1997;25:3570–3579. doi: 10.1093/nar/25.18.3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Christie G E, Cale S B, Isaksson L A, Jin D J, Xu M, Sauer B, Calendar R. J Bacteriol. 1996;178:6991–6993. doi: 10.1128/jb.178.23.6991-6993.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Archambault J, Schappert K T, Friesen J D. Mol Cell Biol. 1990;10:6123–6131. doi: 10.1128/mcb.10.12.6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Archambault J, Drebot M A, Stone J C, Friesen J D. Mol Gen Genet. 1992;232:408–414. doi: 10.1007/BF00266244. [DOI] [PubMed] [Google Scholar]
- 39.Hess W R, Borner T. Int Rev Cytol. 1999;190:1–59. doi: 10.1016/s0074-7696(08)62145-2. [DOI] [PubMed] [Google Scholar]
- 40.Jones T A, Zou J-Y, Cowan S, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]