Abstract
Extensive sequence data and structural sampling of expressed proteins from different species lead to the idea that entire molecules or specific domain folds belong to large super-families of proteins. A subset of G protein-coupled receptors, one of the largest families involved in cellular signaling, rod and cone opsins are involved in phototransduction in photoreceptor cells. Here, the evolutionary analysis of opsin sequences and structures predicts key residues involved in the transmission of the signal from the binding site of the chromophore to the cytoplasmic surface and residues that are involved in the spectral tuning of opsins to short wavelengths of light.
Keywords: Rhodopsin, Cone pigment, Spectral tuning, G protein-coupled receptor, Retinal, Color vision
1. Introduction
Rhodopsin is the photoreceptor membrane protein involved in absorption of photons and signaling in phototransduction in retinal photoreceptor cells [1-6]. It is also a member of the superfamily of G protein-coupled receptors (GPCRs) that comprises ∼6% of the human genome [7,8]. Because almost all physiological processes are modulated by GPCRs, the members of this family are the critical targets for interventions by agonists and antagonists [9,10]. Therefore, it is of enormous interest to consider rhodopsin’s mechanism of signaling and its interactions with other proteins in the G protein transduction cascade. Rhodopsin and cone pigments of rod and cone photoreceptors are composed of a hydrophobic protein core of seven transmembrane helices, opsin, and a covalently tethered chromophore, 11-cis-retinal. Specific interaction of the core of residues in the binding sites and the chromophore leads to specific λmax of absorption of these pigments and provides the basis of color vision [11]. The proteins are activated when light strikes these photoreceptors, leading to photoisomerization of the chromophore. The protein part of these receptors changes its conformation, allowing interaction with rod- and cone-type-specific G protein transducins (Gt), receptor kinases and arrestins [12].
Bovine rhodopsin is the first and only member of the family of GPCRs whose three-dimensional structure has been determined crystallographically (see [3] for the summary on the rhodopsin structure). At this time only the dark state of rhodopsin has been crystallized and determined [13]. Structural changes in the protein caused by absorption of a photon transmit the signal from the covalently linked retinal to the cytoplasmic surface where coupling to the G protein (transducin, Gt) occurs. The nature of these changes is not well characterized.
The phototransduction cycle is presented in cartoon form in Fig. 1 [2-5]. Upon reception of a photon, the 11-cis-retinal is converted to all-trans-retinal within a few hundred femtoseconds. Several spectroscopically characterized states occur in the following milliseconds until metarhodopsin II (Meta II), the signaling state, is reached. At this point, Gt in its GDP form binds to rhodopsin, exchanges GTP for the GDP, and dissociates. This binding, exchange, and dissociation reaction occurs multiple times prior to hydrolysis of the Schiff base between all-trans-retinal [2-4] and Lys296 (7.43) (296 is the amino acid number in bovine rhodopsin, 7.43 is a residue numbering system after [14]). Because the hydrolysis is slow on the time scale of phototransduction, the amplification step is attenuated rapidly when rhodopsin kinase binds to photoactivated rhodopsin and phosphorylates several residues in the £exible C-terminal region of the protein [15,16]. Next, arrestin binds to the phosphorylated, photoactivated rhodopsin to completely shut down the interaction of photoactivated rhodopsin with Gt. Finally, retinal dissociates from the receptor and is regenerated in a complex series of reactions occurring in the retinal pigment epithelium (RPE) [17]. Rhodopsin is regenerated by accepting 11-cis-retinal generated in the RPE and transported to the rod outer segment [17].
Fig. 1.
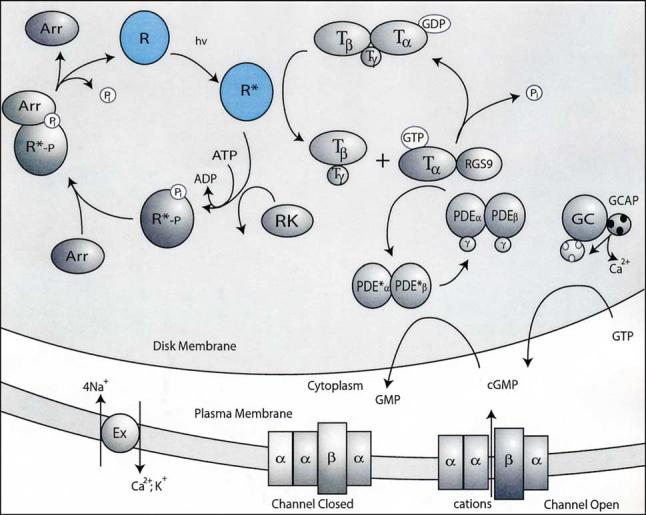
Phototransduction. Phototransduction in the rod photoreceptor cell is initiated when a photon of light is absorbed by rhodopsin causing conformation changes of the receptor. Photoisomerization of the chromophore induces a sequence of conformational changes that culminates in the formation of quasi-stable Meta II, which catalyzes exchange of GDP to GTP in Gt molecules, before rhodopsin is phosphorylated. Gt activation is prevented by binding of arrestin to phosphorylated Meta II [3,4]. The other steps of phototransduction proceed, where the GTP K-subunit of Gt activates phosphodiesterase (PDE), and cGMP is hydrolyzed faster than it is replenished by guanylate cyclase (GC). As a consequence of reduced concentrations of cGMP, the plasma membrane cGMP-gated cation channels close, leading to the hyperpolarization of the cell. Another consequence of phototransduction is lower levels of Ca2+ that trigger a feedback mechanism of the enhanced GC activity through Ca2+ binding protein GCAP1/2 and restoration of the dark levels of cGMP [5].
Rhodopsin of the rod cells and pigments of cone photoreceptor cells all undergo a similar reaction cycle upon the absorption of a photon by the 11-cis-retinylidene chromophore [18]. The three-dimensional models based on the rhodopsin structure of red, green and blue human pigments [19] and many other GPCRs [20] were recently generated. It is expected that a common set of structural changes is involved in the transmission of the conformational changes through the protein. Particular amino acid residues involved in the conformational change might form a pathway within the protein conducting the signal from the chromophore to the protein surface allowing a variety of proteins to associate with photoactivated rhodopsin (Gt, rhodopsin kinase, arrestin, rhodopsin itself in its dimer form) [3,4,21,22].
Several assumptions form the basis for an evolutionary analysis of protein structure [23-25]. First, it is assumed that proteins possessing a similar function will have conserved residues at sequence positions essential to that function. Thus, we assume that all of the vertebrate opsins will utilize a similar set of protein-protein interactions for signaling, and the pathways within the molecular structure will be conserved such as to maintain those functions. If there is divergence of function within subgroups of the phylogeny, then we should be able to detect the residues responsible by using suitable difference measures between evolutionary subgroups. However, if particular residues are involved in two separate functions such as color tuning and signaling, then discerning the separate roles can become ambiguous.
Here, an evolutionary analysis of the protein’s structure from aligned sequences of the vertebrate opsins was carried out. The main analysis is aimed at understanding signal transmission from retinal to the Gt within the metarhodopsin Meta II. For our study of the opsin family, we have developed new methods specifically for the combination of previously defined measures of residue divergence. To further explore our methods, we discuss the use of a difference measure for color tuning and discuss some of the pitfalls of this method applied to the opsin family.
2. Materials and methods
Vertebrate opsin sequences were extracted from the tGRAP Mutant Database, release 10 (http://tgrap.uit.no/fam1asel.html) and supplemented with additional sequences from references in the work of S. Yokoyama and co-workers [26,27]. The sequences were realigned with CLUSTALW ([28], see also [29]), and the alignment was manually edited. Bovine rhodopsin’s secondary structure was used to weight the gap penalties in the alignment. The majority of the realignment was in the C-terminal region of the proteins in the family. A total of 116 vertebrate opsin sequences were used in the subsequent analysis.
For construction of phylogenetic dendrograms, CLUSTALW was employed with the bootstrap option. The bootstrap tree obtained from the CLUSTALW program was used to obtain a matrix containing the evolutionary distances between the opsin sequences. Dendrograms (see Fig. 2) were generated using the phylogenetic tree from the PHYLIP [30] programs, FITCH, DRAWGRAM, and DRAWTREE [31].
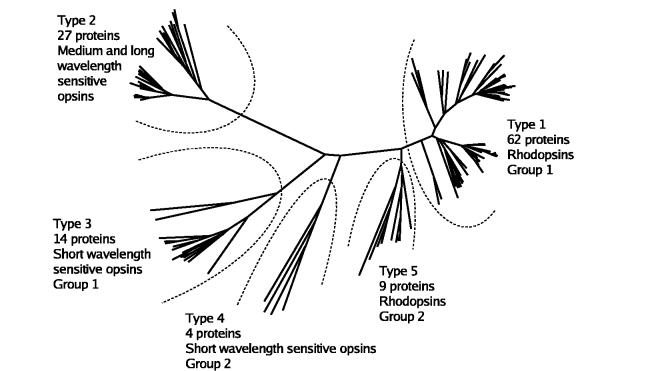
Fig. 2.
Phylogenetic relationships among vertebrate opsins. The names of the 116 proteins have been deleted for clarity. (See supplementary material for details.) Type 1 (RH1) proteins are shown in the upper right. Sixty-two proteins are in this group of rod rhodopsins. The separate branch containing two proteins consists of lamprey rhodopsin sequences. Type 2 opsins (MWS/LWS) are located on the upper left. These are 27 cone opsins which absorb at medium and long wavelengths, (λmax = 508-560 nm). Type 3 opsins (SWS1) include 14 proteins with UV and short wavelength sensitivities (λmax = 367-425 nm). Type 4 opsins (SWS2) include only four proteins from fish and chicken. Type 5 opsins (RH2) are similar to but separate from the RH1 opsins. If lamprey pineal opsin is used as an outgroup in the dendrogram construction, then the apparent root of the dendrogram occurs between the type 2 (MWS/LWS) and type 3 (SWS1) opsins.
Several locally written FORTRAN programs and shell scripts were used to make a sequence profile and map it to the residues present in the coordinate file. The sequence profile for each protein group consists of the bovine rhodopsin sequence number, the bovine rhodopsin sequence residue, the consensus sequence residue of the group, and the three parameters of frequency (Qi), entropy (Si), and evolutionary distance (Pi) given below. Only those residues present in the bovine rhodopsin coordinate file (1HZX, molecule A) [32] were used in the profile. Two separate programs were then used either to analyze an entire family of proteins or to identify the differences between groups of proteins within a family. Finally, a clustering program was used to determine residue clusters and extract coordinates for particular residues from the Protein Data Bank (PDB) file.
For the conservation measures at each residue position, i, in the sequence, three parameters were employed. First, the value of the frequency of occurrence of the majority consensus residue (Qi) was determined. These values were on a scale from 0 to 1.0 and were used as a conservation measure defined as Fi =13Qi. With this measure conserved residues have low values.
For the second measure of amino acid conservation, we used the entropy at each position, i, in the family of sequences, similar to the treatment of Oliveira et al. [33],
| (1) |
where log is the natural logarithm and the exclamation point indicates factorial. N is the number of proteins in the family, 21 is the number of potential residues at a position, i (20 amino acids plus a gap since gaps are allowed in the conservation part of the analysis). Vi is the number of different amino acid residues found at position i, and Wi is the number of occurrences of each amino acid at position i. The sum over this term is taken over the 21 possibilities for residues or gaps at position i. We have inserted the final N to normalize the values for the number of members in a family or subfamily.
The third measure of conservation at each position in the protein was calculated by the PROTPARS program of the PHYLIP package. This program gives the number of mutation steps to a different residue (excluding silent mutations) at each amino acid residue position employed in the construction of the phylogenetic dendrogram. The number at each position thus serves in a sense as a measure of the rate of evolution at that position. The number of steps at each position was divided by the number of members in the family for normalization to give the parameter we denote by Pi.
All three sets of parameter values were normalized to give a minimum value of zero and a mean value of 1.0. The analysis was limited to consider only those residue positions present in the bovine rhodopsin coordinates, 340 positions.
These three measures correlate strongly and it is uncertain how to best combine them into a single measure. To accomplish this combination, we applied singular value decomposition methods [34] on the 340×3 (rows×columns) matrix of values. In performing this operation we take the matrix, [A], and decompose it into three components, [U][w][V]t, where [U] is a 340×3 column-orthogonal matrix, [w] is a 3×3 diagonal matrix and [V]t, the transpose of V, a 3×3 orthogonal matrix. By measuring the length of the three components of [U] at each position, a very good measure of conservation and divergence is achieved. Residues conserved through subgroups in the protein family could then be identified by grouping the degree of conservation from the singular value decomposition into five categories. The above procedure was found to be superior to many other combinations of parameters investigated as a metric for conservation. A second, less restrictive measure of conservation uses together the principal component of [U], and the row vector lengths of [U]. It gives 93 residues in the central cluster rather than the 74 residues found by the [U] row vector lengths alone reported in Table 2.
Table 2.
Part of the 74 residue cluster which leads from the chromophore to the cytoplasmic surface of rhodopsin
| Level 1 | M44 | E113 | G114 | G121 | S186 | C187 | F261 | W265 | Y268 | A269 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Helix/loop | H-I | H-III | H-III | H-III | E-III | E-III | H-VI | H-VI | H-VI | H-VI | |
| 1:1 contact | X | X | X | X | X | X | X | X | X | ||
| Level 2 | L128 | M257 | P267 | P291 | Y301 | ||||||
| Helix/loop | H-III | H-VI | H-VI | H-VII | H-VII | ||||||
| 2:2 contact | X | X | |||||||||
| Level 3 | I219 | M253 | V254 | N302 | P303 | I305 | |||||
| Helix/loop | H-V | H-VI | H-VI | H-VII | H-VII | H-VII | |||||
| 3:3 contact | X | X | X | ||||||||
| Level 4 | N55 | L72 | L76 | P215 | Y223 | E249 | V250 | Y306 | |||
| Helix loop | H-I | H-II | H-II | H-V | H-V | H-VI | H-VI | H-VII | |||
| 4:4 contact | X | X | X | X | |||||||
| Level 5 | L59 | N73 | I75 | W126 | R135 | A246 | E247 | N310 | F313 | ||
| Helix/loop | H-I | H-II | H-II | H-III | H-III | H-VI | H-VI | H-VII-8 | H-8 | ||
| 5:5 contact | X | X | X | X | |||||||
| Level 6 | T62 | L68 | R69 | N78 | S127 | V138 | V139 | T243 | Q312 | R314 | C316 |
| Helix/loop | H-I | C-I | C-I | H-II | H-III | H-III | H-III | C-III | H-8 | H-8 | H-8 |
| 6:6 contact | X | X | X | X | X | ||||||
| Level 7 | K66 | K67 | E134 | K141 | W161 | ||||||
| Helix/loop | C-I | C-I | H-III | C-II | H-IV | ||||||
| 7:7 contact | none | ||||||||||
| Level 8 | F148 | S343 | |||||||||
| Helix/loop | C-II | C-term | |||||||||
| 8:8 contact | none | ||||||||||
| Level 9 | E341 | ||||||||||
| Helix/loop | C-term | ||||||||||
| 9:9 contact | none |
Lys296 and the retinal (level 0) make direct contacts with the following conserved residues on level 1; level 2 residues contact those of level 1; etc. This table attempts to give the hypothetical signaling pathway from the Schiff base retinal to the cytoplasmic surface of rhodopsin as determined by the contact ranking. Only residues of the central cluster (Fig. 3) which proceed in the direction of the cytoplasmic surface relative to the Schiff base retinal are included in this table. For each level, the residues listed contact those in the next lower level. The rows labeled X:X contact indicate the residues in the shell which contact other residues of that same shell. Residue nomenclature and numbering of loops and helices for bovine rhodopsin based on the 1HZX PDB coordinates: E-I: 1-33, H-I: 34-64, C-I: 65-70, H-II: 71-100, E-II: 101-105, H-III: 106-139, C-II: 140-149, H-IV: 150-172, E-III: 173-199 plug, H-V: 200-225, C-III: 226-243, H-VI: 244-276, E-IV: 277-285, H-VII: 286-309, H-8: 311-323=C-IV, C-term: 324-348.
To determine which of the conserved residues might be associated in transduction pathways in GPCRs, their positions were located in the three-dimensional rhodopsin model (PDB identifier 1HZX) [32] and their inter-residue contacts were calculated. The program ACT of the CCP4 program suite [35] was used to measure the contacts between residues. This program produces all of the hydrogen bond and van der Waals contacts for a protein within specified distance limits. The distance cutoffs used in this study were 3.5 Å for hydrogen bonds and 4.0 Å for van der Waals contacts. Shell scripts and FORTRAN programs were written to select only side chain:side chain contacts (sc:sc) and side chain:main chain (sc:mc) contacts of the conserved residues. Note that this restriction implies the assumption that signal transfer is sc:sc and sc:mc but not mc:mc in character. The reason for this restriction is, in part, that we wished that hypotheses concerning the pathway would be experimentally testable by mutagenesis experiments. The restriction also implicitly assumes that the signaling pathway remains intact in spite of potential rearrangements within helices during signal transmission.
Only those conserved residues in contact with other conserved residues were saved for the consideration of clustering. Clustering was accomplished by considering the residue with the most contacts as the center of the cluster. Generally retinal attached to Lys296 (7.43) was chosen by the program as the center of the main cluster. Residues in contact with the central one were considered members of the first shell. Residues with atoms in contact with these first shell residues were placed into the second shell, etc. That is, the subsequent shells of residues were identified as being in contact with preceding shells. In this way, the cluster of contacting residues is hierarchically ranked. The clustering process thus selects the residues from the atomic coordinate file in a ranked order. Each ranking is separated into individual coordinate files to display the ranked contacts using XtalView [36] and Raster3d [37].
3. Results and discussion
3.1. Evolutionary trace differences between groups
Determining the amino acid residues associated with spectral tuning in opsins has been addressed experimentally by several authors (for example [38-48]; see review by Ebrey and Takahashi [49]). In addition to special packing of the chromophore and its interaction with surrounding side chains, the UV-Vis spectra could to a small degree be in£uenced by temperature [50], and more profoundly, by utilization of modified retinals [51] or, for some pigments, by the binding of anions [43,52]. Here, we focus on the effect of residues in the transmembrane bundle on spectral tuning. The difference method identifies the shells of residues that contact the retinal Schiff base and are conserved within the phylogenetic subgroups. Table 1 shows the cluster of residues which contact retinal, are conserved within the five phylogenetic subgroups shown in Fig. 2, and differ between the SWS1 group and the others. The analysis has been done by combining the difference profiles of each of the phylogenetic groups in comparison with the SWS1 group of proteins. The 30 residue positions given in Table 1 form a contacting cluster in bovine rhodopsin. It seems likely that only the shells at levels 1 and 2 (column 4 of Table 1) would be involved in the color tuning. These results are in contrast with those of Yokoyama and co-workers [41,53-55] and with those of Fasick et al. [45,56] in that there is but little overlap in the residues found by our procedure and their mutagenesis results. The central core of rhodopsin is tightly packed for the most part just as with globular, soluble proteins. It would then appear that structural perturbations of the protein by mutations remote from the retinal ligand could strongly affect the spectrum of the pigment due to packing defects. This may be the case for the recent results of Shi and Yokoyama [54] where residues remote from retinal in helices I, II, and III were proposed as the basis of the SWS1 absorption characteristics.
Table 1.
Cluster of 30 residues which differ between four types of opsins and the SWS1 typea
| 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|
| MWS/LWS:SWS1 | 12 | F | N | 6 |
| MWS/LWS:SWS1 | 28 | N | Q | 4 |
| MWS/LWS:SWS1 | 37 | Y | F | 3 |
| RH1:SWS1 | 43 | Y | F | 1 |
| MWS/LWS:SWS1 | 43 | W | F | 1 |
| RH2:SWS1 | 43 | Y | F | 1 |
| RH1:SWS1 | 47 | L | V | 1 |
| RH2:SWS1 | 47 | L | V | 1 |
| MWS/LWS:SWS1 | 80 | A | S | 3 |
| MWS/LWS:SWS1 | 83 | D | G | 2 |
| SWS2:SWS1 | 83 | N | G | 2 |
| MWS/LWS:SWS1 | 90 | A | S | 3 |
| MWS/LWS:SWS1 | 94 | S | V | 2 |
| MWS/LWS:SWS1 | 98 | Q | S | 2 |
| RH1:SWS1 | 117 | A | G | 1 |
| MWS/LWS:SWS1 | 117 | V | G | 1 |
| SWS2:SWS1 | 117 | A | G | 1 |
| RH2:SWS1 | 117 | A | G | 1 |
| MWS/LWS:SWS1 | 122 | I | L | 1 |
| SWS2:SWS1 | 122 | M | L | 1 |
| RH1:SWS1 | 125 | L | G | 2 |
| MWS/LWS:SWS1 | 125 | L | G | 2 |
| SWS2:SWS1 | 125 | L | G | 2 |
| RH2:SWS1 | 125 | L | G | 2 |
| RH1:SWS1 | 129 | V | A | 3 |
| RH2:SWS1 | 129 | V | A | 3 |
| RH2:SWS1 | 133 | I | F | 4 |
| RH1:SWS1 | 163 | M | I | 2 |
| RH2:SWS1 | 163 | M | I | 2 |
| MWS/LWS:SWS1 | 184 | K | Q | 5 |
| RH1:SWS1 | 191 | Y | W | 1 |
| MWS/LWS:SWS1 | 191 | V | W | 1 |
| RH2:SWS1 | 191 | Y | W | 1 |
| RH1:SWS1 | 203 | R | Y | 2 |
| RH1:SWS1 | 204 | V | T | 2 |
| RH2:SWS1 | 204 | V | T | 2 |
| RH2:SWS1 | 206 | Y | F | 2 |
| RH1:SWS1 | 207 | M | L | 1 |
| RH1:SWS1 | 211 | H | C | 1 |
| RH2:SWS1 | 211 | H | C | 1 |
| MWS/LWS:SWS1 | 212 | C | F | 1 |
| RH1:SWS1 | 265 | W | Y | 1 |
| MWS/LWS:SWS1 | 265 | W | Y | 1 |
| RH2:SWS1 | 265 | W | Y | 1 |
| MWS/LWS:SWS1 | 284 | H | D | 7 |
| SWS2:SWS1 | 292 | S | A | 1 |
| MWS/LWS:SWS1 | 293 | Y | F | 1 |
| RH1:SWS1 | 296 | K | K | 0 |
| MWS/LWS:SWS1 | 296 | K | K | 0 |
| SWS2:SWS1 | 296 | K | K | 0 |
| RH2:SWS1 | 296 | K | K | 0 |
| MWS/LWS:SWS1 | 299 | T | C | 1 |
The objective is to and the likely residues responsible for the shift to low wavelength opsins using the residues conserved within each phylogenetic type. This table gives the conserved residues of each phylogenetic group which change to a different conserved residue in the SWS1 group. The numbers in the columns following the residue changes are the level of interaction with the Lys-retinal Schiff base. K296 is given a level of 0. The columns are as follows: (1) phylogenetic groups, (2) residue number, (3) residue in the SWSI group, (4) residue in SWS1, (5) level of contact with retinal. For example, the MWS/LWS group has residue 28 Asn (bovine rhodopsin numbering) while it converts to Gln in the SWS1 phylogenetic group. This residue is at level 4 with respect to contact with retinal.
Using the difference in conserved residues between phylogenetic groups is not a good method to determine the residues causing particular wavelength shifts. The reason is that different taxonomic groups such as birds and mammals have used differing mechanisms to affect the wavelength shifts over the course of evolution. This aspect of the spectral tuning problem is discussed by Cowing et al. [57]. Spectral tuning is further complicated because there appear to be multiple ways to produce similar absorption spectra. For example, Fasick et al. [45] demonstrated that the single mutation F86Y (2.53) in the mouse blue opsin sequence (SWS1 class, λmax = 358 nm) red shifted the absorption maximum to 424 nm. The bovine blue cone pigment which has Y86 in the wild type could be shifted from λmax = 438 nm of the wild type to λmax = 367 nm by the Y86F mutation. The human pigment has Leu at position 86 and the L86Y mutation in this protein caused only a 2 nm shift (λmax = 423C425 nm). Table 1 does not even include residue 86 because it is not conserved within the subgroups analyzed. Moreover, it is unclear how the intermolecular packing [21,22], interactions with phospholipids, and disruption or modification of the interface between individual molecules/lipids by natural or introduced mutations will affect repacking of the visual pigment’s side chains and therefore their spectral properties.
Some of the factors which affect spectral tuning are discussed by Lin et al. [47] and Kochendoerfer et al. [46]. The dipolar and electrostatic environment of the retinal due to contacting residues as well as the solvent cavities which are presumably occupied by water [32] will strongly in£uence the spectrum. Non-polar residues may also play a role by constraining the torsion angles of the retinal due to steric factors. Conclusions from our studies and those in the literature are that color shifting can be accomplished in a variety of ways by residues that are not conserved within a subfamily.
3.2. G protein signaling by vertebrate visual opsins
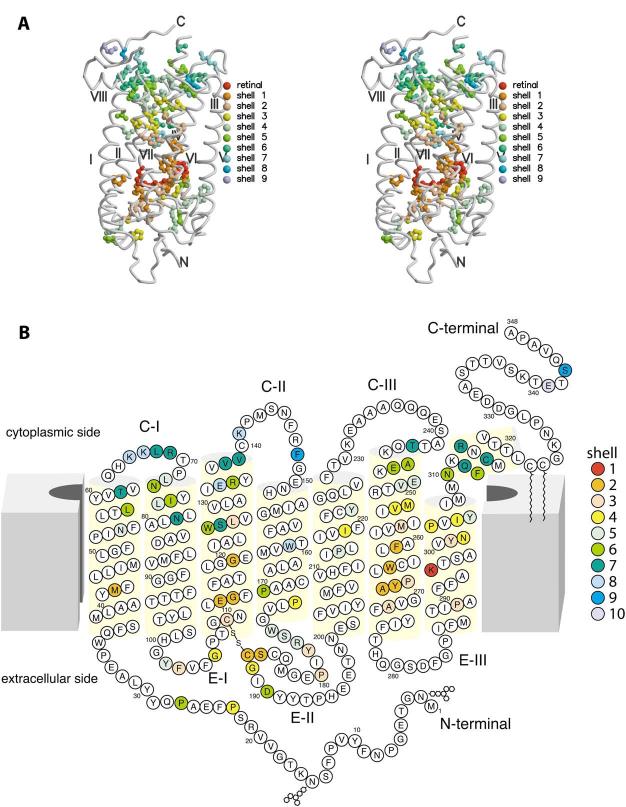
The principal reason for developing the evolutionary trace method used in this study was to determine whether a potential pathway for G protein signaling within the rhodopsin molecule could be hypothesized from application of the evolutionary trace method to the entire opsin family. One must realize that all functions of the protein that require conservation of residues from folding of the polypeptide to the signaling step and beyond may be included in the results. We have found a central cluster of 74 residues that leads from retinal to other parts of the protein. These residues are conserved among the 116 protein sequences considered and have sc:sc or sc:mc contacts when the bovine rhodopsin structure is used as the template. In Table 2, the ranked levels of the cluster are considered in the direction of the cytoplasmic surface, a subset of 55 residues. Fig. 3A,B presents all 74 residues of the cluster.
Fig. 3.
Three- and two-dimensional models of rhodopsin with indicated conserved residues. A: Conserved residues among opsins on the three-dimensional model of bovine rhodopsin. This figure is a stereo image of the rhodopsin molecule with the 74 residue cluster surrounding retinal given as colored shells for the successive levels of contact. For this figure, the helices have been smoothed for clarity. B: Two-dimensional model of rhodopsin. In this diagram we have used the same coloring as in A for the successive shells of contacting residues. The diagram indicates that the contacting residues are not at all sequentially related. The model is after Hargrave and McDowell [69] and modified based on the three-dimensional structure of rhodopsin [13].
The hypothesized signal transfer within the protein begins with the perturbation of F261 (6.44) and W265 (6.48) and propagates along helix H-VI, jumping to H-VII at Y301 (7.48) and N302 (7.49). See Table 2 for the progression of this wave of contacting residues. Tyrosyl 301 makes contact with F261 and C264 (6.47). C264 is conserved except in the RH2 phylogenetic group, so it does not appear in the cluster determined by the restrictive conservation measure for data reported here, but we hypothesize that it is involved in the signaling pathway (Fig. 4). From Y301, contacts proceed along helices VII and VIII via the NPxxY_F (residues 302-306T313) motif as well as through contacts with other conserved residues near the cytoplasmic surface (Table 2). M257 (6.40) contacts Phe261 at level 1 and also provides a link to helix VII via the NPxxY_F motif. N302 (7.49) and I305 (7.52) make van der Waals contacts with M257. Additionally, the signal proceeds along helices V and VI to reach the R135 (3.50) TE247 (6.30) contact of helices III and VI. This R135 (3.50) makes an ion pair with E134 (3.49) in the dark state of rhodopsin. Upon activation E134 is proposed to become protonated [58,59]. These residues are part of the ERY(W) motif which is part of the Gt:Meta II interface (reviewed in[3,60,61]). The ranking of successive shells of Table 2 shows that the interactions behave like a wave traveling towards the cytoplasmic surface and spread to surrounding helices and loops as it progresses (Fig. 3). We conjecture that the signal transfer proceeds along this wave by a succession of small motions of the residues which possibly amplifies as it progresses.
Fig. 4.
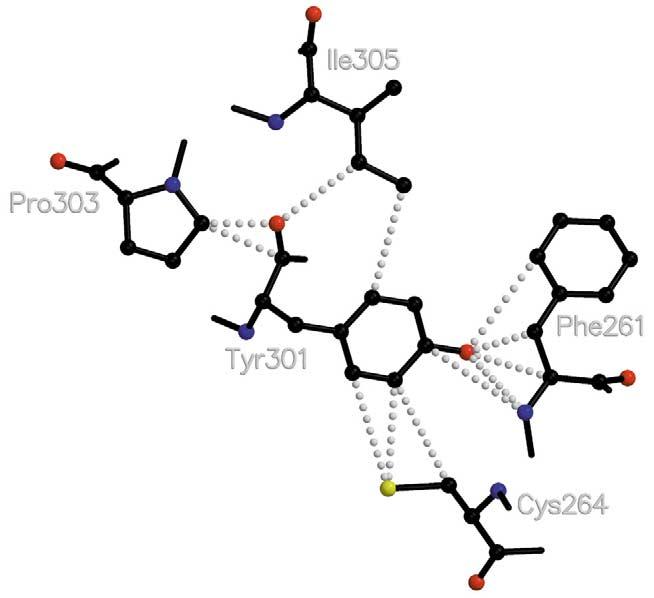
Schematic diagram of contacts between Y301 (7.48) indicating contacting residues between helix VI and helix VII. Helix VI residues F261 (6.44) and C264 (6.47) make substantial contact with Y301 (7.48) which, in turn, contacts P303 (7.50) and I305 (7.52). Numbering of amino acid residues after Ballesteros and Weinstein [14].
The analysis shows that many of the expected G protein interacting residues are involved in this propagating wave, including the ERY(W) motif at residues 134-136 (3.49-3.51) and the NPxxY_F motif of helices VII and 8 (NPxxY are residues 7.49-7.53). For the opsin family, Y301 (7.48) is a key residue in signal propagation from H-VI to H-VII. Y301 has not been modified in rhodopsin mutagenesis studies, to our knowledge. However, when M257 is converted to any residue except Leu, then a constitutively active rhodopsin derivative is produced [62]. Fritze et al. [63] recently observed that the ERY and NPxxY_F motifs are functionally connected by means of studies conducted with 11-cis-9-demethyl-retinal. We postulate that Y301, like N302, is the branch point for propagation of the signal to these two separated regions of the molecule.
3.3. Cluster of residues leading from retinal to the extracellular surface
Among the other members in the cluster of 74 conserved, contacting residues surrounding the retinal Schiff base is the R177TD190 (E2, or ‘plug’ region) contact recently found to be essential for the thermal stability of the dark state of rhodopsin by Janz et al. [64]. The intriguing aspect of this interaction is that it appears to be part of a pathway leading to the extracellular surface (Fig. 3). Potential residues of this pathway include C187 (contact with retinal) and residues P170, P171, W175, S176, R177, Y178, G188, and D190. How this pathway might be involved in rhodopsin’s function is unclear. It may be the case that these structurally conserved residues are necessary for correct protein folding and stability rather than an active physiological function. To date only the R177TD190 set of contacts have been examined [64]. A recent paper may shed some light on this residue conservation of the plug (E2) residues. Yan et al. [65] have examined the role of E181 in the photoactivation process of rhodopsin by mutagenesis and spectroscopy. Their conclusion is that E181 is the counterion in Meta I, implying a counterion shift from E113 in the dark state to E181 in Meta I due to a proton shift. Their model of activation of rhodopsin then proposes that the reorganization of E2 propagates the signal to H-III via the C187-C110 disulfide bridge. This proposal of the counterion shift would nicely fit with the conservation of the plug residues that we observed.
4. Conclusions
In this evolutionary analysis we have used new methods to examine possible pathways for signal transfer within the rhodopsin molecule. The methods differ from those previously used by others in that we have combined several measures of conservation by means of the singular value decomposition technique. Also, rather than using side chains with variable arbitrary radii to measure contacts within clusters of conserved residues, we have used the coordinate contacts themselves. The basis for this is that proteins are compactly folded and we anticipate that this compactness will be preserved over evolutionary time. Conserved residues will contact others closely and non-conserved residues will adapt to retain the compactness of the protein. One singular advantage (and danger) of the method is that a relatively small and homogeneous family of proteins can be used for the analysis. The method gives similar results to those of Ranganathan and co-workers ([66,67]), but both methods appear to have deficiencies. In our method, critical residues which are not sufficiently conserved are missed such as Y136 (3.51) which is either Y or W among the opsins. The coupled pathway found by Süel et al. [66] does not include R135 (3.50), a residue essential for coupling to Gt. As with the method developed in Ranganathan’s laboratory, the recently published study of the GPCRs from Vriend’s laboratory [68] uses large numbers of proteins for the analysis, relying on statistical analyses for the validation of a pathway. Our method, in contrast, is designed as a hypothesis generator and relies on a careful examination of the results for interpretation.
From the evolutionary difference analysis we performed, we conclude that the analysis of spectral tuning is not readily accessible by this means at the phylogenetic resolution we have employed. We feel that the spectral tuning of the SWS1 class has occurred by multiple pathways to achieve the current wavelengths of absorption of the cone opsins in this class. Birds, reptiles, amphibians, fish, and mammals have arrived at short wavelength opsin spectral tuning by multiple routes [57], and mutagenesis experiments have demonstrated that it is possible to obtain similar absorption spectra by multiple routes [45-48,54]. Possibly by making three-dimensional models such as that for the human cone pigments [19], and analyzing the conformation of the chromophore, electrostatic potential of the binding cavity and delocalization of the positive charge if the Schiff base between the chromophore and opsin is protonated, one may gain insight into this problem of spectral tuning.
We have proposed a hypothetical signaling pathway for the propagation of the information within the rhodopsin molecule which should correspond to the activation of the molecule (Table 2). At this point, the pathway is conjectural and may be di/cult to test experimentally because it relies on contacts of side chains within the core of the molecule. It may well be the case that the same residues proposed for the pathway are necessary to correctly fold rhodopsin to a compact molecule so that mutagenesis experiments could be complicated. However, by expanding the analysis to include other GPCRs and observing their contact pathways, it may be possible to suggest the design of experiments to test the pathway and its generality among the GPCRs.
Finally, we have found a potential pathway of unknown significance leading from retinal to the extracellular surface of the molecule. The two residues of the pathway that have been examined to date have been implicated in the thermal stability of rhodopsin. At this point we may speculate the pathway is part of the counterion shift recently proposed for E181 [65]. It remains to be examined what other physiological functions of rhodopsin and cone pigments are suggested by our observations.
Acknowledgments
Acknowledgements: This research was supported by NIH Grants GM63020, EY09339, EY13385, a grant from Research to Prevent Blindness, Inc. (RPB) to the Department of Ophthalmology at the University of Washington, and a grant from the E.K. Bishop Foundation. K.P. is an RPB Senior Investigator. We thank Drs. Kevin Ridge, Tom Ebrey, and Jeremy Nathans for helpful comments during manuscript preparation.
Footnotes
Supplementary data associated with this article can be found at doi:10.1016/S0014-5793(03)01152-9.
- GPCR
- G protein-coupled receptor
- Gt
- photoreceptor G protein
- Meta
- metarhodopsin
- H-
- helix
- PDB
- Protein Data Bank
- RPE
- retinal pigment epithelium
- sc
- side chain
- mc
- main chain
- RH1
- rhodopsins of group 1 or type 1 rod opsins (λmax = 480-510 nm)
- MWS/LWS
- cone opsins with medium to long wavelength absorption (λmax = 508-560 nm), type 2 opsins
- SWS1
- opsins with λmax = 367-425 nm, type 3 opsins
- SWS2
- opsins with λmax = 470-510 nm absorption, type 4 opsins
- RH2
- rhodopsins of groups 2 or type 5 opsins (λmax = 470-510 nm)
References
- 1.Nathans J. Biochemistry. 1992;31:4923–4931. doi: 10.1021/bi00136a001. [DOI] [PubMed] [Google Scholar]
- 2.Ebrey T, Koutalos Y. Prog. Retin. Eye Res. 2001;20:49–94. doi: 10.1016/s1350-9462(00)00014-8. [DOI] [PubMed] [Google Scholar]
- 3.Filipek S, Stenkamp RE, Teller DC, Palczewski K. Annu. Rev. Physiol. 2003;65:851–879. doi: 10.1146/annurev.physiol.65.092101.142611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okada T, Ernst OP, Palczewski K, Hofmann KP. Trends Biochem. Sci. 2001;26:318–324. doi: 10.1016/s0968-0004(01)01799-6. [DOI] [PubMed] [Google Scholar]
- 5.Polans A, Baehr W, Palczewski K. Trends Neurosci. 1996;19:547–554. doi: 10.1016/s0166-2236(96)10059-x. [DOI] [PubMed] [Google Scholar]
- 6.Sakmar TP, Menon ST, Marin EP, Awad ES. Annu. Rev. Biophys. Biomol. Struct. 2002;31:443–484. doi: 10.1146/annurev.biophys.31.082901.134348. [DOI] [PubMed] [Google Scholar]
- 7.Mirzadegan T, Benko G, Filipek S, Palczewski K. Biochemistry. 2003;42:2759–2767. doi: 10.1021/bi027224+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vassilatis DK, et al. Proc. Natl. Acad. Sci.; USA. 2003. pp. 4903–4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klabunde T, Hessler G. Chembiochem. 2002;3:928–944. doi: 10.1002/1439-7633(20021004)3:10<928::AID-CBIC928>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 10.Ballesteros J, Palczewski K. Curr. Opin. Drug Discov. Dev. 2001;4:561–574. [PMC free article] [PubMed] [Google Scholar]
- 11.Nathans J, Merbs SL, Sung CH, Weitz CJ, Wang Y. Annu. Rev. Genet. 1992;26:403–424. doi: 10.1146/annurev.ge.26.120192.002155. [DOI] [PubMed] [Google Scholar]
- 12.Palczewski K, Polans AS, Baehr W, Ames JB. BioEssays. 2000;22:337–350. doi: 10.1002/(SICI)1521-1878(200004)22:4<337::AID-BIES4>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 13.Palczewski K, et al. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 14.Ballesteros J, Weinstein H. Methods Neurosci. 1985;25:366–428. [Google Scholar]
- 15.Maeda T, Imanishi Y, Palczewski K. Prog. Retin. Eye Res. 2003;22:417–434. doi: 10.1016/s1350-9462(03)00017-x. [DOI] [PubMed] [Google Scholar]
- 16.Ohguro H, Van Hooser JP, Milam AH, Palczewski K. J. Biol. Chem. 1995;270:14259–14262. doi: 10.1074/jbc.270.24.14259. [DOI] [PubMed] [Google Scholar]
- 17.McBee JK, Palczewski K, Baehr W, Pepperberg DR. Prog. Retin. Eye Res. 2001;20:469–529. doi: 10.1016/s1350-9462(01)00002-7. [DOI] [PubMed] [Google Scholar]
- 18.Shichida Y, Imai H. Cell. Mol. Life Sci. 1998;54:1299–1315. doi: 10.1007/s000180050256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stenkamp RE, Filipek S, Driessen CA, Teller DC, Palczewski K. Biochim. Biophys. Acta. 2002;1565:168–182. doi: 10.1016/s0005-2736(02)00567-9. [DOI] [PubMed] [Google Scholar]
- 20.Filipek S, Teller DC, Palczewski K, Stenkamp R. Annu. Rev. Biophys. Biomol. Struct. 2003;32:375–397. doi: 10.1146/annurev.biophys.32.110601.142520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang Y, Fotiadis D, Filipek S, Saperstein DA, Palczewski K, Engel A. J. Biol. Chem. 2003;278:21655–21662. doi: 10.1074/jbc.M302536200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fotiadis D, Liang Y, Filipek S, Saperstein DA, Engel A, Palczewski K. Nature. 2003;421:127–128. doi: 10.1038/421127a. [DOI] [PubMed] [Google Scholar]
- 23.Gribaldo S, Casane D, Lopez P, Philippe H.Mol. Biol. Evol 2003. in press [DOI] [PubMed] [Google Scholar]
- 24.Gough J. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1897–1900. doi: 10.1107/s0907444902015160. [DOI] [PubMed] [Google Scholar]
- 25.Burley SK, Bonanno JB. Annu. Rev. Genomics Hum. Genet. 2002;3:243–262. doi: 10.1146/annurev.genom.3.022502.103227. [DOI] [PubMed] [Google Scholar]
- 26.Yokoyama S. Genes Genet. Syst. 1999;74:189–199. doi: 10.1266/ggs.74.189. [DOI] [PubMed] [Google Scholar]
- 27.Yokoyama S, Radlwimmer FB. Genetics. 1999;153:919–932. doi: 10.1093/genetics/153.2.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson JD, Higgins DG, Gibson TJ. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Felsenstein J. Syst. Biol. 1997;46:101–111. doi: 10.1093/sysbio/46.1.101. [DOI] [PubMed] [Google Scholar]
- 31.Felsenstein J.PHYLIP (Phylogeny Inference Package) 1993version 3.5cDepartment of Genetics, University of Washington; Seattle, WA: distributed by the author [Google Scholar]
- 32.Teller DC, Okada T, Behnke CA, Palczewski K, Stenkamp RE. Biochemistry. 2001;40:7761–7772. doi: 10.1021/bi0155091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oliveira L, Paiva AC, Vriend G. Chembiochem. 2002;3:1010–1017. doi: 10.1002/1439-7633(20021004)3:10<1010::AID-CBIC1010>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 34.Press WH, Teukolsky SA, Vetterling WT, Flannery BP. Numerical Recipes in Fortran. 2nd ed. Vol. 77. Cambridge University Press; New York: 1996. pp. 51–63. [Google Scholar]
- 35.Collaborative Computational Project Acta Crystallogr. D. 1994;50:760–763. [Google Scholar]
- 36.McRee DE. J. Mol. Graphics. 1992;10:44–46. [Google Scholar]
- 37.Merritt EA, Bacon JD. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 38.Kochendoerfer GG, Wang Z, Oprian DD, Mathies RA. Biochemistry. 1997;36:6577–6587. doi: 10.1021/bi970322o. [DOI] [PubMed] [Google Scholar]
- 39.Merbs SL, Nathans J. Photochem. Photobiol. 1993;58:706–710. doi: 10.1111/j.1751-1097.1993.tb04956.x. [DOI] [PubMed] [Google Scholar]
- 40.Nathans J. Biochemistry. 1990;29:937–942. doi: 10.1021/bi00456a013. [DOI] [PubMed] [Google Scholar]
- 41.Yokoyama S. Prog. Retin. Eye Res. 2000;19:385–419. doi: 10.1016/s1350-9462(00)00002-1. [DOI] [PubMed] [Google Scholar]
- 42.Sun H, Macke JP, Nathans J. Proc. Natl. Acad. Sci. USA. 1997;94:8860–8865. doi: 10.1073/pnas.94.16.8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Z, Asenjo AB, Oprian DD. Biochemistry. 1993;32:2125–2130. doi: 10.1021/bi00060a001. [DOI] [PubMed] [Google Scholar]
- 44.Asenjo AB, Rim J, Oprian DD. Neuron. 1994;12:1131–1138. doi: 10.1016/0896-6273(94)90320-4. [DOI] [PubMed] [Google Scholar]
- 45.Fasick JI, Applebury ML, Oprian DD. Biochemistry. 2002;41:6860–6865. doi: 10.1021/bi0200413. [DOI] [PubMed] [Google Scholar]
- 46.Kochendoerfer GG, Lin SW, Sakmar TP, Mathies RA. Trends Biochem. Sci. 1999;24:300–305. doi: 10.1016/s0968-0004(99)01432-2. [DOI] [PubMed] [Google Scholar]
- 47.Lin SW, Kochendoerfer GG, Carroll KS, Wang D, Mathies RA, Sakmar TP. J. Biol. Chem. 1998;273:24583–24591. doi: 10.1074/jbc.273.38.24583. [DOI] [PubMed] [Google Scholar]
- 48.Neitz M, Neitz J, Jacobs GH. Science. 1991;252:971–974. doi: 10.1126/science.1903559. [DOI] [PubMed] [Google Scholar]
- 49.Ebrey TG, Takahashi Y. In: Photobiology for the 21th Century. Coohill TP, Valenzeno DP, editors. Vandenmar Publishing Company; Overland Park, KS: 2003. [Google Scholar]
- 50.Ala-Laurila P, Saarinen P, Albert R, Koskelainen A, Donner K. Vis. Neurosci. 2002;19:781–792. doi: 10.1017/s0952523802196088. [DOI] [PubMed] [Google Scholar]
- 51.Harosi FI. Vis. Res. 1994;34:1359–1367. doi: 10.1016/0042-6989(94)90134-1. [DOI] [PubMed] [Google Scholar]
- 52.Kleinschmidt J, Harosi FI. Proc. Natl. Acad. Sci. USA. 1992;89:9181–9185. doi: 10.1073/pnas.89.19.9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kawamura S, Blow NS, Yokoyama S. Genetics. 1999;153:1839–1850. doi: 10.1093/genetics/153.4.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi Y, Yokoyama S. Proc. Natl. Acad. Sci. USA. 2003;100:8308–8313. doi: 10.1073/pnas.1532535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shi Y, Radlwimmer FB, Yokoyama S. Proc. Natl. Acad. Sci. USA. 2001;98:11731–11736. doi: 10.1073/pnas.201257398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fasick JI, Lee N, Oprian DD. Biochemistry. 1999;38:11593–11596. doi: 10.1021/bi991600h. [DOI] [PubMed] [Google Scholar]
- 57.Cowing JA, Poopalasundaram S, Wilkie SE, Robinson PR, Bowmaker JK, Hunt DM. Biochem. J. 2002;367:129–135. doi: 10.1042/BJ20020483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vogel R, Siebert F. Biopolymers. 2003;72:133–148. doi: 10.1002/bip.10407. [DOI] [PubMed] [Google Scholar]
- 59.Fahmy K, Sakmar TP, Siebert F. Biochemistry. 2000;39:10607–10612. doi: 10.1021/bi000912d. [DOI] [PubMed] [Google Scholar]
- 60.Marshall GR. Biopolymers. 2001;60:246–277. doi: 10.1002/1097-0282(2001)60:3<246::AID-BIP10044>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 61.Hamm HE. Proc. Natl. Acad. Sci. USA. 2001;98:4819–4821. doi: 10.1073/pnas.011099798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Han M, Smith SO, Sakmar TP. Biochemistry. 1998;37:8253–8261. doi: 10.1021/bi980147r. [DOI] [PubMed] [Google Scholar]
- 63.Fritze O, Filipek S, Kuksa V, Palczewski K, Hofmann KP, Ernst OP. Proc. Natl. Acad. Sci. USA. 2003;100:2290–2295. doi: 10.1073/pnas.0435715100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Janz JM, Fay JF, Farrens DL. J. Biol. Chem. 2003;278:16982–16991. doi: 10.1074/jbc.M210567200. [DOI] [PubMed] [Google Scholar]
- 65.Yan EC, Kazmi MA, Ganim Z, Hou JM, Pan D, Chang BS, Sakmar TP, Mathies RA. Proc. Natl. Acad. Sci. USA. 2003;100:9262–9267. doi: 10.1073/pnas.1531970100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Süel GM, Lockless SW, Wall MA, Ranganathan R. Nat. Struct. Biol. 2003;10:59–69. doi: 10.1038/nsb881. [DOI] [PubMed] [Google Scholar]
- 67.Lockless SW, Ranganathan R. Science. 1999;286:295–299. doi: 10.1126/science.286.5438.295. [DOI] [PubMed] [Google Scholar]
- 68.Oliveira L, Paiva PB, Paiva AC, Vriend G. Proteins. 2003;52:553–560. doi: 10.1002/prot.10489. [DOI] [PubMed] [Google Scholar]
- 69.Hargrave PA, McDowell JH. FASEB J. 1992;6:2323–2331. doi: 10.1096/fasebj.6.6.1544542. [DOI] [PubMed] [Google Scholar]