Abstract
Escherichia coli modulates its porin expression through a histidine kinase, EnvZ, and its cognate response regulator, OmpR. EnvZ is a bifunctional enzyme that possesses both OmpR kinase and phosphorylated OmpR (OmpR-P) phosphatase activities and thus controls the cellular level of OmpR-P. In an in vitro-assay system, the addition of OmpR to the reaction mixture consisting of the cytoplasmic domain of EnvZ (EnvZc) and ATP produces a barely detectable amount of OmpR-P because of the dual activities of EnvZ. Here we report that DNA fragments containing the upstream promoter regions of the porin genes (ompF and ompC) can shift the equilibrium between OmpR and OmpR-P dramatically toward OmpR-P. Among the four reactions occurring in the mixture, only the EnvZ phosphatase activity was inhibited severely by the specific DNA, in contrast to the previous report by Kenney and her associates that DNA stimulates OmpR phosphorylation by EnvZ [Ames, S. K., Frankema, N. & Kenney, L. J. (1999) Proc. Natl. Acad. Sci. USA 96, 11792–11797]. The autophosphorylation of EnvZc and the phosphotransfer from phosphorylated EnvZc to OmpR were not affected by DNA, whereas the autodephosphorylation of OmpR-P was inhibited slightly. We propose that the apparent inhibitory effect of DNA on the EnvZ phosphatase function is caused by sequestrating OmpR-P from the reaction as a result of OmpR-P binding to DNA.
The EnvZ–OmpR system in Escherichia coli is a member of the histidyl-aspartyl phosphorelay family (also known as two-component signal transduction system). This system serves as the major signal transduction system in prokaryotes to respond to various environmental stresses and growth conditions. The basic components of this system are a histidine protein kinase serving as a signal receptor and its cognate response regulator mediating specific gene expression or cellular locomotion (1).
EnvZ is a transmembrane histidine kinase that monitors environmental osmolarity changes. It is autophosphorylated by using ATP at the highly conserved His residue (His-243) in the cytoplasmic domain. This phosphate group is transferred subsequently to the conserved Asp-55 residue of OmpR forming phosphorylated OmpR (OmpR-P). OmpR-P is a transcription factor that binds upstream promoter regions of genes for outer-membrane porins, ompF and ompC, and differentially modulates their expression. Notably, EnvZ also acts as a phosphatase dephosphorylating OmpR-P to regulate the cellular OmpR-P concentration. In response to low medium osmolarity, the cellular OmpR-P level is reduced as the ratio of EnvZ kinase to phosphatase activity decreases, resulting in the transcription of only the ompF gene. On the other hand, at high osmolarity the kinase/phosphatase ratio of EnvZ increases (mainly because of the decreased phosphatase activity; ref. 2), resulting in higher levels of OmpR-P. Under this condition, OmpR-P functions as a repressor for ompF, and as a transcription activator for ompC. Thus, ompF and ompC are regulated reciprocally by the cellular level of OmpR-P (for a review of the EnvZ–OmpR signal transduction pathway, see refs. 3–5).
EnvZc, the cytoplasmic region of EnvZ (residues 180–450), exists as a dimer and consists of linker region (residues 180–222), domain A (or DHp domain, residues 223–289), and domain B (or CA domain, residues 290–450) (ref. 6; for a review see ref. 7). It possesses both kinase and phosphatase activities similar to the intact EnvZ. The linker region is considered to be important for transducing a signal from the periplasmic receptor domain to the cytoplasmic catalytic domain (8). Three-dimensional structures of both domains A and B have been solved by NMR (9, 10). Recently we demonstrated that domain A by itself serves as a phosphatase for OmpR-P and proposed that the phosphatase of EnvZ is regulated by the spatial relationship between domain A and B (11).
The response regulator OmpR is a two-domain protein. The N-terminal half of the CheY-like receiver domain contains the phosphorylation site (Asp-55) and its C-terminal half contains the DNA-binding function (12). These two domains are connected by a flexible linker peptide (13). The structure of the DNA-binding domain has been determined by x-ray crystallography (14, 15). It contains a winged helix-turn-helix motif. The phosphorylation of OmpR at Asp-55 enhances its ability to bind to the regulatory sequences upstream of the ompC and ompF promoters (16, 17). In vitro and in vivo footprinting studies have shown that OmpR-P binds to the −380 to −361 region (F4 site), the −100 to −39 region (F1, F2, and F3 sites) of ompF, and the −100 to −38 region (C1, C2, and C3 sites) of ompC (18–21). These binding sites consist of 20 base pairs each and share a consensus sequence (22). Two OmpR-P molecules bind to each site in a head-to-tail manner (23). The binding affinities of OmpR-P to these sites are in a hierarchical order such that OmpR-P binds independently to F1 and C1 sites and bind to other sites only after the upstream F1 or C1 site is occupied (17, 21).
Recently it was reported that phosphorylation of OmpR by either acetyl phosphate or EnvZ was enhanced in the presence of a DNA fragment containing OmpR-P-binding regions (24). On the basis of acetyl phosphate being only a phosphor donor for OmpR but not a phosphatase for OmpR-P and that the half-life of OmpR-P in the presence of DNA is slightly longer than that in the absence of DNA, it was concluded that DNA enhances the rate of phosphorylation of OmpR but not the rate of dephosphorylation of OmpR-P. Although acetyl phosphate can phosphorylate OmpR in vivo, EnvZ controls the cellular OmpR-P concentration and mediates osmoregulation (3–5). We reexamined the effect of OmpR-P-bound DNA on EnvZ function and found that the addition of a DNA fragment has a negligible effect on OmpR phosphorylation by EnvZc but dramatically reduces OmpR-P dephosphorylation by EnvZc. The substantial stabilization of OmpR-P in the presence of a DNA fragment was found to be due to the sequestration of OmpR-P from the dephosphorylation reaction by its binding to DNA. As a result a significant increase of the amount of OmpR-P in the reaction was observed. Possible biological significance of our findings will be discussed.
Materials and Methods
Oligonucleotides.
All of the oligonucleotides used in this paper were synthesized on an Applied Biosystems DNA synthesizer. Complementary strands were annealed to create double-stranded DNA fragments, the upper-strand sequences of which (from 5′ to 3′ end) were as follows: F1–F2–F3, 5′-GGGGTT-TACTTTTGGTTACATATTTTTTCTTTTTGAAACC-AAATCTTTATCTTTGTAGCACTTTCAGGGG-3′; C1–C2–C3, 5′-GGGGTTTACATTTTGAAACATCTATAGCGATA-AATGAAACATCTTAAAAGTTTTAGTATCATATTGG-GG-3′; F1–F2, 5′-GGGGTTTACTTTTGGTTACATATTT-TTTCTTTTTGAAACCAAATGGGG-3′; F1, 5′-GGGGTTTACTTTTGGTTACATATTGGGG-3′; F2; 5′-GGGGTTTTCTTTTTGAAACCAAATGGGG-3′; F3, 5′-GGGGTTATCTTTGTAGCACTTTCAGGGG-3′; F4, 5′-GGGGGTTACGGAATATTACATTGCGGGG-3′; C1, 5′-GGGGTTTACATTTTGAAACATCTAGGGG-3′; C1(C5G), 5′-GGGGTTTAGATTTTGAAACATCTAGGGG-3′ and; control, 5′-ATAAAGATATCGCAGCGTGCAACGCCATCAT-3′.
Purification of Proteins.
EnvZc, OmpR, and EnvZcT247Y were purified as described (25, 26).
Phosphorylation and Dephosphorylation Reactions.
All reactions were carried in buffer A (50 mM Tris⋅HCl, pH 8.0/50 mM KCl/5% (vol/vol) glycerol) containing 5 mM MgCl2. To make phosphorylated EnvZc (EnvZc-P), 50 μg EnvZc was phosphorylated by 50 μM [γ-32P]ATP in 200 μl of buffer A containing 5 mM Ca2+ at room temperature for 1 h. Free ATP was removed by exchanging the reaction mixture with a total of 5 ml of buffer A containing 1 mM EDTA through repeating centrifugation using a Biomax-10 ultrafree-4 centrifugal filter (Millipore). The final volume of the EnvZc-P preparation was reduced to 50 μl. No [γ-32P]ATP or [32P]Pi was detected in the preparation as judged by TLC analysis (data not shown). To make OmpR-P, 100 μg OmpR was phosphorylated with 50 μM [γ-32P]ATP by 60 μg glutathione S-transferase (GST)-EnvZc11, a GST fusion of a superkinase mutant, EnvZcT247R, in 200 μl of buffer A containing 5 mM Ca2+ at room temperature for 1 h. HPLC C4 column analysis (24) demonstrated that more than 95% OmpR in the reaction has been converted into OmpR-P (data not shown). The reaction mixture was stopped by addition of 10 mM EDTA and then applied to a Sephacryl S-100 HR gel-filtration column (0.8 × 10 cm; Amersham Pharmacia) to separate GST-EnvZc11 from OmpR-P and to remove free [γ-32P]ATP and inorganic phosphate. The buffer used for the gel-filtration column contains 2 mM EDTA to inhibit the autophosphatase of OmpR-P. The fractions containing only OmpR-P as analyzed by SDS/PAGE were pooled, and the total OmpR concentration was measured by the Bio-Rad protein assay (Bio-Rad).
Native Gel Assay.
Nonlabeled OmpR-P was generated and purified in the same way as the 32P-labeled OmpR-P except that the amount of proteins and the volume of the reaction increased 2-fold and cold ATP instead of [γ-32P]ATP was used. Then the pooled OmpR-P fractions were concentrated into 160 μl by a Biomax-10 ultrafree-4 centrifugal filter (Millipore). To observe interactions between OmpR and EnvZc and between OmpR-P and a DNA fragment, 5 μM OmpR-P purified as described above was mixed with 16 μM C1 DNA fragment and 5 μM EnvZc in buffer A containing 5 mM MgCl2 for 5 min. Then the mixture was analyzed by native PAGE. The composition of the stacking gel was 5% acrylamide/bis (29:1) in 62.5 mM Tris⋅HCl (pH 7.5) and 1 mM EDTA and the composition of the separation gel was 10% acrylamide/bis (29:1) in 187.5 mM Tris⋅HCl (pH 8.9) and 1 mM EDTA. After being run for 4 h at a constant voltage (100 V) at 4°C, the gel was stained first by ethidium bromide to detect DNA bands and then by Coomassie brilliant-blue R to detect protein bands. The desired protein bands were then cut out from the gel and placed vertically on the top of a 17.5% SDS/PAGE gel for a second-dimensional electrophoresis.
Results
The Effect of DNA on the Overall Phosphorylation Reaction of OmpR by EnvZc.
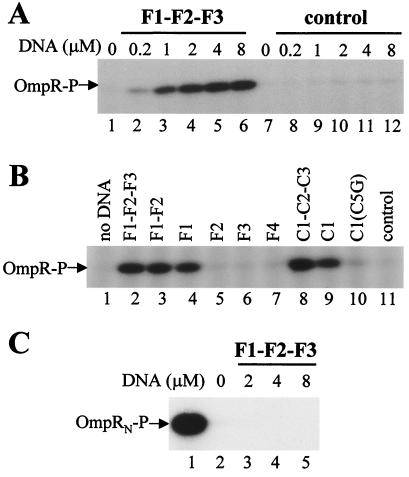
When EnvZc and OmpR were mixed together in the presence of 50 μM ATP and 5 μM Mg2+, the production of OmpR-P was barely detectable (Fig. 1A, lane 1). This result is due to the bifunctional feature of EnvZc, which not only phosphorylates OmpR but also dephosphorylates the resulting OmpR-P. However, when a DNA fragment consisting of F1, F2, and F3, the upstream OmpR-P binding sites of the ompF promoter, is added to the reaction mixture, the apparent production of OmpR-P became increased dramatically (Fig. 1A, lanes 2–6). The amount of OmpR-P thus produced was DNA concentration-dependent: the more F1–F2–F3 DNA fragment was added, the more OmpR-P accumulated in the reaction mixture. A similar result was obtained also with C1–C2–C3 DNA, the OmpR-P binding sites of the ompC promoter (Fig. 1B, lane 8). We subsequently estimated the amount of OmpR-P by SDS-gel electrophoresis by using OmpR-P phosphorylated by GST–EnvZc11 as a standard. As described in Materials and Methods, GST–EnvZc11 phosphorylates more than 95% OmpR under the condition used in the experiment. In the presence of 8 μM F1–F2–F3 (note that one DNA fragment contains six OmpR-P binding sites, and at the start of the reaction the molar ratio between OmpR and DNA is 1:2), approximately 6.4% of OmpR was converted into the phosphorylated form after 35 min incubation at room temperature, as quantified by comparing with serial dilution of OmpR-P produced by GST–EnvZc11 (data not shown). Importantly, the OmpR-P band was barely detectable in the reaction mixture without DNA (Fig. 1A, lane 1). From an overexposed film, the level of OmpR phosphorylation in the absence of DNA was estimated to be approximately 0.12%, 50-fold less than that in the presence of DNA (data not shown).
Figure 1.
The effect of DNA on the accumulation of OmpR-P in the reaction mixture of OmpR, EnvZc, and ATP. (A) OmpR (4 μM) was mixed with different amounts of the F1–F2–F3 fragment or the control DNA (indicated above the gel) in a buffer containing 50 μM [γ-32P]ATP and 5 mM Mg2+. EnvZc (1 μM) was added to the mixture to initiate the reaction. After 35 min of incubation at room temperature, the reaction was stopped by the addition of 5× SDS loading buffer. The products were analyzed by SDS/PAGE and autoradiography. (B) The reaction was carried out as described in A, but different DNA fragments (4 μM each) were used. (C) The N-terminal half fragment of OmpR (OmpRN; residues 1 to 134) (4 μM) was phosphorylated by either GST–EnvZc11 (1 μM, lane 1) or EnvZc (1 μM) in the presence of different amounts of F1–F2–F3 DNA (lanes 2–5) under the same condition as described in A.
To test whether this accumulation of OmpR-P is DNA-specific, a 31-bp unrelated DNA fragment (originally designed for mutagenesis of EnvZ to create E275A and E276A) was used as a control. Gel-mobility shift experiments demonstrated that OmpR-P could not bind to this control DNA (data not shown). As shown in Fig. 1A, lanes 8–12, the control DNA had no effect on the production of OmpR-P.
To examine further the specificity of DNA sequences for the accumulation of OmpR-P, we next investigated the effect of individual OmpR-P binding sequences. Previously it had been shown that F1 and C1 have the highest binding affinity toward OmpR-P (approximately 7 nM), whereas all the other sites have very low affinity (more than 270 nM; ref. 17). Gel-mobility shift experiments further proved that only F1 or C1 was capable of binding to OmpR-P independently but not all the other sites (ref. 21; L.Q. and M.I., unpublished data). Consistent with these results, the stimulation of OmpR-P production depended on either the F1 or C1 sequence as shown in Fig. 1B. The F1-containing fragments such as F1–F2–F3 (lane 2), F1–F2 (lane 3), and F1 (lane 4) enhanced OmpR-P accumulation, but F2 (lane 5), F3 (lane 6), and F4 (lane 7) were unable to enhance OmpR-P accumulation. Similarly, the C1-containing fragments such as C1–C2–C3 (lane 8) and C1 (lane 9) stimulated OmpR-P production. The ability of DNA fragments to enhance OmpR-P accumulation agrees well with the ability of these DNA fragments to cause gel-mobility shift in the presence of OmpR-P. Note that a single base-substitution mutation at position 5 (C to G) in the C1 sequence, which severely affected its affinity to OmpR-P in vivo (27) and completely abolished its affinity to OmpR-P in vitro (L.Q. and M.I., unpublished data), also lost its ability to enhance OmpR-P accumulation (lane 10).
The above results indicate that OmpR-P binding to DNA is essential for the observed accumulation of OmpR-P by DNA, consistent with the previous observation (24). We further tested this notion with OmpRN (residues 1–134). OmpRN is known to be able to serve as a substrate for both kinase and phosphatase reactions of EnvZ as well as intact OmpR (28). However, it is unable to bind DNA, because it lacks the C-terminal DNA-binding domain. As shown in Fig. 1C, there was no enhancement of OmpRN-P accumulation even at the highest concentration of the F1–F2–F3 DNA fragment (8 μM; lane 5), which caused a dramatic enhancement of intact OmpR-P accumulation (Fig. 1A, lane 6). Note that this OmpRN can be phosphorylated by GST–EnvZc11 (lane 1). These results clearly suggest that the accumulation of OmpR-P in the presence of DNA requires its ability to bind to DNA.
The Mechanism of DNA Enhancement of OmpR-P Accumulation.
When OmpR and EnvZc are mixed together with ATP, the following four reactions occur simultaneously in solution: autophosphorylation, EnvZc + ATP → EnvZc-P + ADP; phosphotransfer, EnvZc-P + OmpR → EnvZc + OmpR-P; autophosphatase, OmpR-P →OmpR + Pi; and EnvZc phosphatase, OmpR-P → OmpR + Pi. In the absence of DNA, the net result of these reactions is the hydrolysis of ATP, an ATPase reaction, and OmpR mainly exists as unphosphorylated because of the apparent strong phosphatase activity of EnvZc over its kinase activity. However, in the presence of DNA, a significantly higher amount of OmpR is able to remain phosphorylated, indicating one or more of the above reactions is affected by DNA.
Autophosporylation reaction.
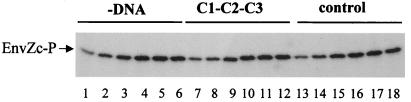
First, we studied whether the autophosphorylation of EnvZc was affected by DNA. EnvZc was mixed with or without DNA, and then [γ-32P]ATP was added. Aliquots were taken at different time points and loaded on an SDS/PAGE to detect the amount of EnvZc-P. As shown in Fig. 2, the autophosphorylation patterns of EnvZc are very similar either in the absence of DNA (lanes 1–6) or in the presence of C1–C2–C3 DNA (lanes 7–12) or control DNA (lanes 13–18), indicating that DNA has no effect on the autophosphorylation reaction.
Figure 2.
The effect of DNA on the autophosphorylation of EnvZc. EnvZc (1 μM) was mixed without DNA or with C1–C2–C3 DNA or control DNA in buffer A containing 50 μM [γ-32P]ATP and 5 mM Mg2+. Aliquots were taken at 0.5 min (lanes 1, 7, and 13), 1 min (lanes 2, 8, and 14), 2 min (lanes 3, 9, and 15), 5 min (lanes 4, 10, and 16), 10 min (lanes 5, 11, and 17), and 35 min (lanes 6, 12, and 18) and were analyzed by SDS/PAGE.
Phosphotransfer reaction.
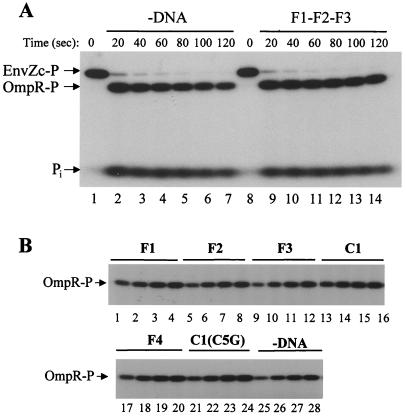
To analyze the phosphotransfer from EnvZc-P to OmpR, EnvZc was phosphorylated first with [γ-32P]ATP and purified free of ATP as described in Materials and Methods. The 32P-labeled EnvZc then was added into a premixed OmpR and DNA solution. Aliquots were taken at several time intervals and analyzed by SDS/PAGE. Fig. 3A indicates that the phosphotransfer reaction was so rapid that only a very small amount of EnvZc-P was left, even at 20 sec after the addition of EnvZc-P, both in the absence (lane 2) or in the presence (lane 9) of F1–F2–F3 DNA. These data strongly suggest that the phosphotransfer reaction is not affected by the addition of DNA.
Figure 3.
The effect of DNA on the EnvZc kinase activity. (A) The effect of DNA on the phosphotransfer reaction from EnvZc-P to OmpR. OmpR (4 μM) was mixed with or without the F1–F2–F3 DNA (4 μM) at room temperature for 5 min before purified 32P-labeled EnvZc-P (1 μM) was added. Aliquots were taken at 20 sec (lanes 2 and 9), 40 sec (lanes 3 and 10), 60 sec (lanes 4 and 11), 80 sec (lanes 5 and 12), 100 sec (lanes 6 and 13), and 120 sec (lanes 7 and 14). Lanes 1 and 8: EnvZc-P at zero time point. Reactions were stopped by adding 5× SDS loading buffer, and products were analyzed by SDS/PAGE and autoradiography. The positions of EnvZc-P, OmpR-P, and inorganic phosphate (Pi) are shown by arrows. (B) The effect of DNA on the OmpR kinase activity of EnvZcT247Y. OmpR (4 μM) was mixed first with different DNA fragments (4 μM), as indicated on the top of the gel. Then EnvZcT247Y (0.2 μM), a Kinase+ Phosphatase− mutant, was added to the mixture. Aliquots were taken at 15 min (lanes 1, 5, 9, 13, 17, 21, and 25), 30 min (lanes 2, 10, 14, 18, 22, and 26), 45 min (lanes 3, 7, 11, 15, 19, 23, and 27), and 60 min (lanes 4, 8, 12, 16, 20, 24, and 28). The products were analyzed by SDS/PAGE and autoradiography.
To confirm the above conclusions further, we took advantage of an EnvZc mutant, T247Y, which has kinase activity but lacks phosphatase activity (26). As shown in Fig. 3B, the patterns of OmpR-P formation with DNA fragments that cannot bind OmpR-P independently, including F2 (lanes 5–8), F3 (lanes 9–12), F4 (lanes 17–20), and C1(C5G) (lanes 21–24), are almost identical to those with F1 (lanes 1–4) and C1 (lanes 13–16), which bind OmpR-P. These results indicate that DNA has no effect on the EnvZcT247Y's OmpR kinase activity that consists of autophosphorylation and phosphotransfer reactions. Note that OmpR-P-bound DNA has very little effect on the autophosphatase of OmpR-P, as shown later. There is slightly nonspecific enhancement of OmpR-P formation with DNA [approximately 1.5-fold, compared the experiment without DNA (lanes 25–28) with all the other experiments], the reason for which is not known at present.
Autophosphatase reaction.
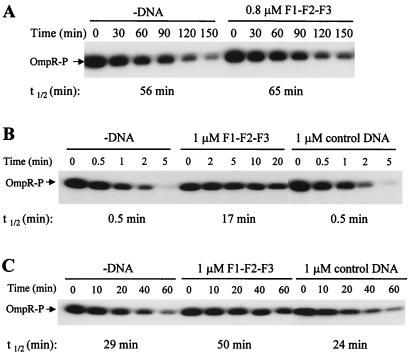
OmpR-P has a detectable autophosphatase activity with a half-life of 56 min (Fig. 4A). This activity was inhibited slightly by the addition of F1–F2–F3, increasing the half-life of OmpR-P to 65 min (Fig. 4A). This fact is consistent with a previous report (24) and suggests that OmpR-P can be stabilized slightly when it binds to DNA. However, this inhibitory effect of DNA on the autophosphatase reaction is too little to account for the dramatic enhancement of OmpR-P accumulation caused by DNA as shown in Fig. 1A.
Figure 4.
The effect of DNA on the autophosphatase activity of OmpR-P (A) and the phosphatase activity of EnvZc (B) and domain A (C). Purified OmpR-P (0.4 μM) was mixed first with or without DNA fragments. After 5 min, 0.2 μM EnvZc and 100 μM ADP (B) or 0.2 μM domain A (C) was added to the mixture. Aliquots were taken at the time points indicated above the gel, and reactions were stopped by adding 5× SDS loading buffer. The products were analyzed by SDS/PAGE and autoradiography. The amounts of OmpR-P were quantified by a PhosphorImager to calculate the half-lives (t1/2) of OmpR-P, which are shown below the gel.
Phosphatase reaction.
In the presence of Mg2+ and cofactor ADP, EnvZc displays a strong phosphatase activity, reducing the half-life of OmpR-P to less than 30 sec. When the F1–F2–F3 DNA was added to the reaction mixture, the phosphatase activity was reduced approximately 34-fold, and the half-life of OmpR-P increased to 17 min (Fig. 4B). This inhibition is DNA sequence-dependent, because the control DNA did not have any effect on the dephosphorylation of OmpR-P by EnvZc (Fig. 4B). These results clearly indicate that the enhancement of OmpR-P accumulation by DNA was caused mainly from the inhibitory effect of DNA on the OmpR-P dephosphorylation by EnvZc.
This inhibition of EnvZc phosphatase activity by DNA was observed also in the phosphotransfer experiment (Fig. 3A). After completely transferring its phosphoryl group to OmpR within 40 sec, EnvZc functions as phosphatase toward OmpR-P. It is apparent that there was more OmpR-P left at 120 sec in the presence of F1–F2–F3 than in the absence of DNA (lane 14 vs. lane 7), suggesting that the phosphatase activity of EnvZc was inhibited by DNA.
Recently, we demonstrated that domain A by itself possesses OmpR-P phosphatase activity (11). Therefore, we also tested the effect of DNA on the phosphatase activity of domain A. The half-life of OmpR-P was approximately 29 min in the absence of DNA and 24 min in the presence of control DNA, while in the presence of F1–F2–F3 it increased to 50 min (Fig. 4C), indicating that domain A's phosphatase activity is inhibited also by specific DNA.
Inhibition of OmpR-P Dephosphorylation by Its DNA Binding.
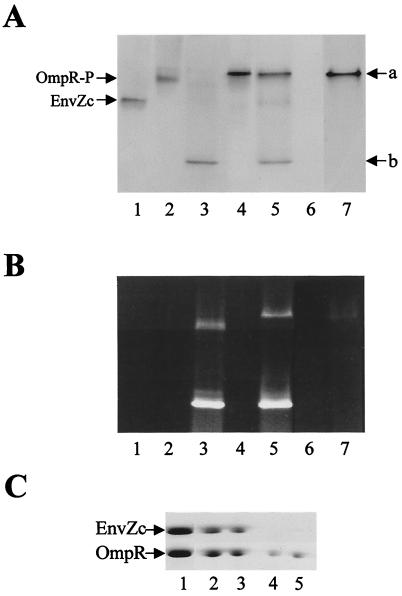
The association of OmpR and EnvZc has been observed by the Ni-nitrilotriacetic acid resin-binding method, either by using His-tagged EnvZc (H. Park & M.I., unpublished data) or His-tagged OmpR (25). Recently, we found that native PAGE is another approach to detecting the interaction between OmpR and EnvZc. As shown in Fig. 5A, EnvZc (lane 1) and OmpR-P (lane 2) migrate at different positions on native PAGE. After they were mixed in an equal-molar ratio, a new band appeared at position “a,” but both OmpR-P and EnvZc bands disappeared (lane 4). Subsequently, this band was extracted from the gel and analyzed by SDS/PAGE (Fig. 5C, lane 2). Clearly, this band is a complex consisting of both OmpR and EnvZc proteins. In this complex, OmpR may exist as both a phosphorylated and unphosphorylated form (because of the phosphatase of EnvZc). Unphosphorylated OmpR can also form the complex band with EnvZc at the same position (data not shown).
Figure 5.
Effect of C1 DNA on the interaction between OmpR and EnvZc. (A and B) Native gel analysis was performed on the following mixtures: EnvZc alone (lane 1); OmpR-P alone (lane 2); OmpR-P and C1 (lane 3); OmpR-P and EnvZc (lane 4); OmpR-P, C1, and EnvZc (lane 5); C1 (lane 6); and OmpR-P, F4, and EnvZc (lane 7). After electrophoresis, the gel was stained first with ethidium bromide (B) and then with Coomassie brilliant-blue R (A). (C) The bands at positions “a” and “b” in lanes 3, 4, and 5 of A were cut out and loaded on an SDS/PAGE gel for second-dimensional electrophoresis. The gel was stained with Coomassie brilliant-blue R. Lane 1, EnvZc and OmpR marker; lane 2, band at position “a” in lane 4 of A; lane 3, band at position “a” in lane 5 of A; lane 4, band at position “b” in lane 5 of A; and lane 5, band at position “b” in lane 3 of A.
In the same gel system, the OmpR-P–DNA complex can be detected also when OmpR-P is mixed with C1 DNA (a new band at position “b” in Fig. 5A, lane 3). This result was confirmed by second-dimensional SDS/PAGE, because it contained OmpR protein (Fig. 5C, lane 5), and by ethidium-bromide staining, because it contained DNA (Fig. 5B, lane 3). Note that free C1 DNA run out of the gel under the condition used (Fig. 5B, lane 6). Next, OmpR-P was added to the mixture of C1 DNA and EnvZc. It was found that both bands at positions “a” and “b” appeared (Fig. 5A, lane 5). The components of both bands were identified by ethidium-bromide staining (Fig. 5B, lane 5) and by second-dimensional SDS/PAGE (Fig. 5C, lanes 3 and 4 for bands at positions “a” and “b,” respectively). Note that the intensity of the OmpR–EnvZc complex band at position “a” decreased (compared with lane 4) with concomitant appearance of a diffused band corresponding to EnvZc. These results indicate that OmpR-P interacted with both EnvZc and C1 DNA, forming two distinct complexes at positions “a” and “b” at the same time. Also note that OmpR-P–DNA complex formation only occurred with OmpR-specific DNA but not with nonspecific DNA like F4 (Fig. 5A and B, lane 7). A small amount of C1 DNA was retarded at the position of OmpR-P (Fig. 5B, lane 3) or the EnvZc–OmpR complex (lane 5), likely because of nonspecific interaction.
Together, the above results suggest that in the presence of specific DNA, a fraction of OmpR-P binds to DNA and sequestrates from interacting with EnvZc. Thus, these OmpR-P proteins may be protected from the EnvZc's phosphatase, resulting in the significant accumulation of OmpR-P observed in Fig. 1A.
Discussion
The previous studies by Kenney and her associates (24) and the present results indicate that the equilibrium between OmpR and OmpR-P in the presence of EnvZc is affected greatly by DNA fragments carrying OmpR-P-binding sites, resulting in accumulation of OmpR-P. In the present paper, we demonstrated that the phosphatase reaction rather than the kinase reaction is affected by OmpR-P-specific DNA.
The mechanism of the reciprocal transcription of ompF and ompC mediated by OmpR-P has been proposed (21). At low osmolarity, OmpR-P cooperatively binds to F1 as well as F2 and somewhat loosely to F3 resulting in ompF expression by direct interaction of bound OmpR-P with the C-terminal domain of the α subunit of RNA polymerase (29, 30), whereas ompC is not induced, because OmpR-P is able to bind only to C1 but not to C2 and C3 sites. At high osmolarity, the OmpR-P concentration increases such that OmpR-P becomes capable of binding to the upstream F4 site by interacting with OmpR-P molecules already bound to F1–F2–F3. As a result, the ompF-promoter region is considered to form a loop blocking the ompF transcription. At the ompC-promoter region, C2 and C3 sites are occupied now by OmpR-P to transcribe ompC. Notably, the unphosphorylated form of OmpR has a very low affinity toward the promoters and thus is incapable of enhancing ompF and ompC transcription (17). In the present report, we demonstrated that OmpR-P bound to ompF- and ompC-promoter regions became protected from being dephosphorylated by EnvZ phosphatase. As a result, the half-life of DNA-bound OmpR-P is prolonged significantly compared with free OmpR-P (more than 30-fold; Fig. 4B), allowing the formation of either stable OmpR-P–RNA polymerase complexes on ompF and ompC promoters to enhance their transcriptions or the multiple OmpR-P complex on F1, F2, F3, and F4 sites to block ompF transcription.
E. coli appears to have a number of other OmpR-P-binding sites in addition to a total of 14 OmpR-P molecules binding to the porin promoters (24, see references therein). It is important to note that because E. coli contains OmpR at the level of 103 molecules per cell (4), even at a level of 6.4% phosphorylation of cellular OmpR there are likely to be enough OmpR-P molecules to cover all of the chromosomal OmpR-binding sites. The dissociation half-life of OmpR-P from C1 DNA was estimated to be approximately 7 min in Mg2+ buffer (L.Q. and M.I., unpublished data), indicating that OmpR-P can be stably maintained on specific DNA. These data, together with the fact that OmpR-P bound to DNA is protected from the EnvZ phosphatase reaction, imply DNA is likely an important factor for ompF and ompC regulation. However, the in vivo relevance of our results remains to be clarified, because EnvZ is a cytoplasmic membrane protein.
Previously, Ames et al. reported that OmpR phosphorylation by acetyl phosphate and EnvZ were approximately 2-fold and 2- to 7-fold, respectively, stimulated by C1 DNA fragments (24). Based on the fact that the stability of OmpR-P was affected only slightly by the presence of DNA, these authors concluded that the increased level of OmpR-P in the presence of DNA is caused by the stimulation of the phosphorylation reaction of OmpR by acetyl phosphate and EnvZ. However, the present results revealed that DNA had little effect on the phosphorylation reaction of OmpR. The 2- to 7-fold stimulation of OmpR phosphorylation by acetyl phosphate in the presence of DNA may be a unique feature of OmpR phosphorylation by a small molecule. It has been reported recently that OmpRV203 M was unable to be phosphorylated by acetyl phosphate, but it was still able to be phosphorylated by EnvZ both in vivo and in vitro (31). A similar result has been observed with several chemotaxis response-regulator CheY mutants (32). It should be noted that although it takes 3 h to reach 40% phosphorylation of OmpR in the presence of 3,000-fold excess acetyl phosphate without DNA (24), it takes less than 20 sec to transfer almost all of the phosphoryl group from 1 μM EnvZc-P to 4 μM OmpR (Fig. 3A). It has been shown also that CheY is phosphorylated much more rapidly by CheA-P (1,000-fold) than by acetyl phosphate and that CheY binds much more tightly to CheA-P (1,000-fold) than to acetyl phosphate (33). It has been proposed that CheA accelerates the phosphotransfer to CheY by providing an environment of low ionic strength that cannot be achieved by a small phosphodonor (33).
The present results indicate that the apparent inhibitory effect of DNA on EnvZ phosphatase activity is caused by the interference of OmpR-P interaction with EnvZ by trapping OmpR-P on DNA, because there was no EnvZc in the OmpR-P–DNA complex (Fig. 5). OmpR-P is known to bind to F- or C-site DNA in a head-to-tail manner (23), which may block the interaction between EnvZ and the N-terminal domain of OmpR-P sterically. An alternative possibility is that OmpR-P binding to DNA results in a conformational change in OmpR-P that interferes with its interaction with EnvZ. Further structural characterization of OmpR-P in the presence and in the absence of DNA will provide insights into the precise role of DNA in the osmoregulation of ompF and ompC genes.
Acknowledgments
This work is supported by Grant GM19043 from the National Institutes of Health.
Abbreviations
- OmpR-P
phosphorylated OmpR
- EnvZc-P
phosphorylated EnvZc
- GST
glutathione S-transferase
- OmpRN
N-terminal half fragment of OmpR
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.031383098.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.031383098
References
- 1.Grebe T W, Stock J B. Adv Microb Physiol. 1999;41:139–227. doi: 10.1016/s0065-2911(08)60167-8. [DOI] [PubMed] [Google Scholar]
- 2.Jin T, Inouye M. J Mol Biol. 1993;232:484–492. doi: 10.1006/jmbi.1993.1404. [DOI] [PubMed] [Google Scholar]
- 3.Egger L A, Park H, Inouye M. Genes Cells. 1997;2:167–184. doi: 10.1046/j.1365-2443.1997.d01-311.x. [DOI] [PubMed] [Google Scholar]
- 4.Pratt L, Silhavy T J. In: Two-Component Signal Transduction. Hoch J A, Silhavy T J, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 105–127. [Google Scholar]
- 5.Forst S A, Roberts D L. Res Microbiol. 1994;145:363–373. doi: 10.1016/0923-2508(94)90083-3. [DOI] [PubMed] [Google Scholar]
- 6.Park H, Saha S K, Inouye M. Proc Natl Acad Sci USA. 1998;95:6728–6732. doi: 10.1073/pnas.95.12.6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dutta R, Qin L, Inouye M. Mol Microbiol. 1999;34:633–640. doi: 10.1046/j.1365-2958.1999.01646.x. [DOI] [PubMed] [Google Scholar]
- 8.Park H, Inouye M. J Bacteriol. 1997;179:4382–4390. doi: 10.1128/jb.179.13.4382-4390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomomori C, Tanaka T, Dutta R, Park H, Saha S K, Zhu Y, Ishima R, Liu D, Tong K I, Kurokawa H, et al. Nat Struct Biol. 1999;6:729–734. doi: 10.1038/11495. [DOI] [PubMed] [Google Scholar]
- 10.Tanaka T, Saha S K, Tomomori C, Ishima R, Liu D, Tong K I, Park H, Dutta R, Qin L, Swindells M B, et al. Nature (London) 1998;396:88–92. doi: 10.1038/23968. [DOI] [PubMed] [Google Scholar]
- 11.Zhu Y, Qin L, Yoshida T, Inouye M. Proc Natl Acad Sci USA. 2000;97:7808–7813. doi: 10.1073/pnas.97.14.7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kato M, Aiba H, Tate S, Nishimura Y, Mizuno T. FEBS Lett. 1989;249:168–172. doi: 10.1016/0014-5793(89)80617-9. [DOI] [PubMed] [Google Scholar]
- 13.Kenney L J, Bauer M D, Silhavy T J. Proc Natl Acad Sci USA. 1995;92:8866–8870. doi: 10.1073/pnas.92.19.8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinez-Hackert E, Stock A M. Structure (London) 1997;5:109–124. doi: 10.1016/s0969-2126(97)00170-6. [DOI] [PubMed] [Google Scholar]
- 15.Kondo H, Nakagawa A, Nishihira J, Nishimura Y, Mizuno T, Tanaka I. Nat Struct Biol. 1997;4:28–31. doi: 10.1038/nsb0197-28. [DOI] [PubMed] [Google Scholar]
- 16.Forst S, Delgado J, Inouye M. Proc Natl Acad Sci USA. 1989;86:6052–6056. doi: 10.1073/pnas.86.16.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Head C G, Tardy A, Kenney L J. J Mol Biol. 1998;281:857–870. doi: 10.1006/jmbi.1998.1985. [DOI] [PubMed] [Google Scholar]
- 18.Huang K J, Igo M M. J Mol Biol. 1996;262:615–628. doi: 10.1006/jmbi.1996.0540. [DOI] [PubMed] [Google Scholar]
- 19.Rampersaud A, Harlocker S, Inouye M. J Biol Chem. 1994;269:12559–12566. [PubMed] [Google Scholar]
- 20.Tsung K, Brissette R E, Inouye M. J Biol Chem. 1989;264:10104–10109. [PubMed] [Google Scholar]
- 21.Bergstrom L C, Qin L, Harlocker S L, Egger L A, Inouye M. Genes Cells. 1998;3:777–788. doi: 10.1046/j.1365-2443.1998.00228.x. [DOI] [PubMed] [Google Scholar]
- 22.Harlocker S L, Bergstrom L, Inouye M. J Biol Chem. 1995;270:26849–26856. doi: 10.1074/jbc.270.45.26849. [DOI] [PubMed] [Google Scholar]
- 23.Harrison-McMonagle P, Denissova N, Martinez-Hackert E, Ebright R H, Stock A M. J Mol Biol. 1999;285:555–566. doi: 10.1006/jmbi.1998.2375. [DOI] [PubMed] [Google Scholar]
- 24.Ames S K, Frankema N, Kenney L J. Proc Natl Acad Sci USA. 1999;96:11792–11797. doi: 10.1073/pnas.96.21.11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qin L, Dutta R, Kurokawa H, Ikura M, Inouye M. Mol Microbiol. 2000;36:24–32. doi: 10.1046/j.1365-2958.2000.01837.x. [DOI] [PubMed] [Google Scholar]
- 26.Dutta R, Yoshida T, Inouye M. J Biol Chem. 2000;275:38645–38653. doi: 10.1074/jbc.M005872200. [DOI] [PubMed] [Google Scholar]
- 27.Pratt L A, Silhavy T J. Mol Microbiol. 1995;17:565–573. doi: 10.1111/j.1365-2958.1995.mmi_17030565.x. [DOI] [PubMed] [Google Scholar]
- 28.Delgado J, Forst S, Harlocker S, Inouye M. Mol Microbiol. 1993;10:1037–1047. doi: 10.1111/j.1365-2958.1993.tb00974.x. [DOI] [PubMed] [Google Scholar]
- 29.Kato N, Aiba H, Mizuno T. FEMS Microbiol Lett. 1996;139:175–180. doi: 10.1111/j.1574-6968.1996.tb08199.x. [DOI] [PubMed] [Google Scholar]
- 30.Pratt L A, Silhavy T J. J Mol Biol. 1994;243:579–594. doi: 10.1016/0022-2836(94)90033-7. [DOI] [PubMed] [Google Scholar]
- 31.Tran V K, Oropeza R, Kenney L J. J Mol Biol. 2000;299:1257–1270. doi: 10.1006/jmbi.2000.3809. [DOI] [PubMed] [Google Scholar]
- 32.Zhu X, Amsler C D, Volz K, Matsumura P. J Bacteriol. 1996;178:4208–4215. doi: 10.1128/jb.178.14.4208-4215.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mayover T L, Halkides C J, Stewart R C. Biochemistry. 1999;38:2259–2271. doi: 10.1021/bi981707p. [DOI] [PubMed] [Google Scholar]