Abstract
Self-protection in the mitomycin C (MC)-producing microorganism Streptomyces lavendulae includes MRD, a protein that binds MC in the presence of NADH and functions as a component of a unique drug binding-export system. Characterization of MRD revealed that it reductively transforms MC into 1,2-cis-1-hydroxy-2,7-diaminomitosene, a compound that is produced in the reductive MC activation cascade. However, the reductive reaction catalyzed by native MRD is slow, and both MC and the reduced product are bound to MRD for a relatively prolonged period. Gene shuffling experiments generated a mutant protein (MRDE55G) that conferred a 2-fold increase in MC resistance when expressed in Escherichia coli. Purified MRDE55G reduces MC twice as fast as native MRD, generating three compounds that are identical to those produced in the reductive activation of MC. Detailed amino acid sequence analysis revealed that the region around E55 in MRD strongly resembles the second active site of prokaryotic catalase-peroxidases. However, native MRD has an aspartic acid (D52) and a glutamic acid (E55) residue at the positions corresponding to the catalytic histidine and a nearby glycine residue in the catalase-peroxidases. Mutational analysis demonstrated that MRDD52H and MRDD52H/E55G conferred only marginal resistance to MC in E. coli. These findings suggest that MRD has descended from a previously unidentified quinone reductase, and mutations at the active site of MRD have greatly attenuated its catalytic activity while preserving substrate-binding capability. This presumed evolutionary process might have switched MRD from a potential drug-activating enzyme into the drug-binding component of the MC export system.
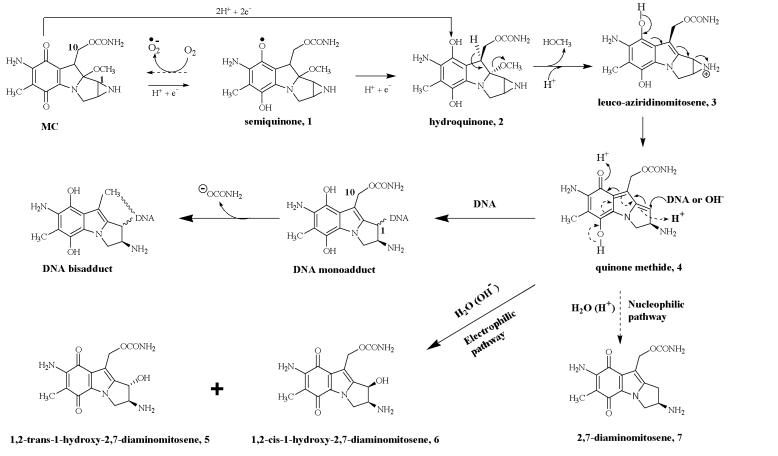
Produced by soil bacterium Streptomyces lavendulae, mitomycin C (MC) is a highly effective antitumor agent commonly used in combination chemotherapy regimens for the treatment of various carcinomas (1, 2). As a prodrug, MC does not have significant cytotoxity; however, upon enzymatic or chemical reductive activation, it will generate highly reactive DNA-alkylating agents that cause lethal intrastrand and interstrand DNA cross-links (3). The intracellular activation of MC is catalyzed by endogenous flavoreductases and proceeds by either anaerobic one electron reduction or oxygen-independent two-electron reduction (Fig. 1). A key step in both routes is the enzymatic reduction of the quinone moiety of MC, which initiates a cascade of spontaneous transformations that result in the production of a reactive quinone methide intermediate (4) and a group of reduced MC metabolites (5, 6, and 7) called mitosenes (1, 2).
Figure 1.
Metabolic activation of MC through one or two electron quinone reduction (modified from refs. 1 and 12). The two reaction pathways of quinone methide 4, are represented by solid arrows (electrophilic pathway) and dashed arrows (nucleophilic pathway).
Activated MC maintains a strong propensity to interact covalently with DNA at 5′-CpG-3′ sequences. Despite the high percentage of G + C in its chromosome, S. lavendulae exhibits extraordinary resistance against MC (4). Previously, three genetic loci have been identified to be involved in the cellular self-protection against MC in this microorganism. One locus (mcrA), which is located outside of the MC biosynthetic gene cluster, encodes a protein (MCRA) that reoxidizes the reductively activated MC species to the prodrug through a redox relay mechanism (5, 6). Two other loci (mrd and mct) were found within the MC biosynthetic gene cluster and encode a small soluble protein (MRD) and a membrane-associated protein (MCT), respectively (7, 8). Whereas MCT has extensive amino acid sequence similarity with several actinomycete antibiotic export proteins, MRD does not significantly resemble any previously characterized protein. DNA protection studies demonstrated that MRD can protect high G + C DNA from MC-mediated cross-linking through a drug-binding activity that depends on the presence of NADH (7). Heterologous expression experiments also indicated that MRD can work together with MCT to form a highly efficient drug binding-export system (8). Together, the functions of MCRA, MRD, and MCT ensure an efficient self-protection network in S. lavendulae, through which most of the produced drug would be exported to the outside of the cell, whereas a few molecules left inside the cell are maintained in the inactive (prodrug) form.
A mystery has remained regarding the NADH requirement for binding of MC to MRD, because this cofactor would only be expected for the flavoreductase-mediated bioactivation of MC. In an effort to resolve this question, we re-examined the in vitro activity of MRD toward MC and found that MRD can indeed reductively transform MC into 1,2-cis-1-hydroxy-2,7-diaminomitosene (compound 6, Fig. 1), one of the three mitosene compounds that normally are generated in the bioreductive MC activation pathway. This discovery suggested that MRD might have a potential drug-activating activity, which appears paradoxical with the previously identified drug-resistance activity of MRD. Here we report details of the unique quinone reductase activity of MRD and propose a model to explain the evolutionary switching of MRD from a potential drug-activating enzyme to the drug-binding component of the MC export system.
Materials and Methods
Chemicals and Reagents.
MC was generously provided by Kyowa Hakko Kogyo (Tokyo) and Bristol-Myers Squibb Pharmaceutical Research Institute. Authentic mitosenes (5, 6, and 7) were kindly provided by Maria Tomasz (Hunter College, City University of New York). Dimethyl-d6 sulfoxide (DMSO-d6, D, 99.9%) was purchased from Cambridge Isotope Laboratories, Cambridge, MA.
Protein Overexpression and Purification.
MRD was overexpressed and purified as described (7).
MC Metabolism by MRD.
A 100-μl reaction mixture consisting of 1.25 mM MC, 2.5 mM NADH, and 100 μM purified MRD in 10 mM sodium phosphate buffer (pH 7.0) was incubated at 37°C for 1 h, then added to a Microcon centrifugal filter device (molecular mass cutoff at 10,000 Da, Millipore) and centrifuged at 14,000 × g for 15 min to separate MRD from the mixture. The filtrate was directly loaded onto the HPLC system for analysis. MRD was recollected from the filter, and the protein-bound metabolites were extracted twice with 500 μl of solvent containing chloroform-isopropanol-ethyl acetate (2:2:1). Extracts were combined, dried under vacuum, and reconstituted in a small amount of methanol for HPLC or TLC analysis. For analysis of the prolonged binding of MC and its reduced derivatives to MRD, the reaction mixture was stored at 4°C for 3 days before filtration and protein extraction.
Absorption Analysis of MC Reduction by MRD.
A Shimadzu 160UV spectrophotometer was used to study the absorption changes during the reductive transformation of MC by MRD.
HPLC Analysis of MC Metabolites.
An Econsil C18 column (250 mm × 4.6 mm, 5 μm) was used with isocratic solvent systems at a flow rate of 1 ml/min and dual detection wavelengths of 313 and 365 nm. Initially, the reaction mixture was separated by using a 30% solution B isocratic program for 30 min (solution A: 10 mM ammonium acetate, pH 5.6; solution B: methanol) (condition I). Under this condition, compound 5 migrated along with MC. To resolve 5 from MC, the MC peak collected under condition I was repurified by the same isocratic program but using water as solution A (condition II). For the large-scale reaction using MRDE55G, a 4-ml reaction mixture was incubated at 37°C for 4 h. Extracts of the whole reaction mixture were separated by using a 25% solution B isocratic program (condition III). Under each condition, the standard retention time for each compound was determined by injecting pure authentic materials.
TLC Analysis of MC Metabolites.
Methanol (100%) was used as the solvent system with Whatman K6F silica gel 60A glass plates.
Analysis of MC Metabolites by MS.
A Finnigan-MAT (San Jose, CA) LCQ Deca instrument was used to obtain the electrospray ionization mass spectra of MC metabolites.
NMR Analysis of MC Metabolites.
1H-NMR, 13C-heteronuclear multiple quantum correlation, proton one-dimensional correlated spectroscopy, and heteronuclear multiple bond correlation data of compound 6 were obtained from an Inova8001 NMR machine, and the 13C NMR data were obtained from an Inova6002 NMR machine (Varian). Sample was dissolved in DMSO-d6 and contained in a 5-mm Shigemi NMR tube for all analyses.
Spectrophotometric Analysis of MRD.
The absorbance spectrum (200–800 nm) for MRD (1.0 mg/ml in 0.05 M Tris buffer) was obtained by using a Shimadzu 160UV spectrophotometer.
Metal Analysis of MRD.
MRD (0.5 mg) was added to 5.0 ml of 1% HNO3, and the resulting solution was used for quantification of metal content by inductively coupled plasma photometry.
MRD Fluorescence Quenching Assay.
Binding of NADH and MC to native MRD and MRDE55G was monitored by following the quenching of tryptophan fluorescence intensity induced by the binding of NADH or MC (Fexcitation = 295 nm, Femission = 339 nm), using a Perkin–Elmer LS50 fluorometer. The experiment involved successive titration of 10 μM MRD with 20–200 μM NADH or MC in 20 mM Tris buffer (pH 7.0). Binding affinities were calculated as described (9).
DNA Shuffling of mrd.
A ≈700-bp XbaI–HindIII DNA fragment containing mrd (390 bp) was excised from pDHS7024 (8) and ligated into XbaI–HindIII-digested pUC18. The resulting construct was used as the template for DNA shuffling (10). The reassembled DNA fragments were digested with XbaI and HindIII, ligated back into pUC18, and transformed into Escherichia coli DH5α. The resulting transformants were screened on LB agar plates containing a gradient concentration of MC from 5 μg/ml to 50 μg/ml.
Site-Directed Mutagenesis of mrd.
Site-directed mutagenesis of mrd was performed by using the QuickChange site-directed mutagenesis kit (Stratagene).
MC Resistance Level of E. coli Expressing Various mrd Genes.
E. coli cells expressing various mrd genes were grown overnight at 37°C in LB medium containing 100 μg/ml of ampicillin. The cultures were diluted into fresh LB medium and grown to an OD600 of 0.6. Cells were diluted again and aliquoted (50 μl) into 96-well microplates containing an equal volume of LB medium supplemented with various concentrations of MC. Plates were incubated on an orbital shaker at 37°C, and cell density was monitored by detecting the absorption at 600 nm every 4 h by using a microplate autoreader (Bio-Tek, Burlington, VT).
Western Blot Analysis.
Western blot analysis was performed by using standard protocols (11). Polycolonal anti-MRD antibody was developed in rabbits (HTI Bio-Products, Ramona, CA) using native MRD protein purified from E. coli.
Results
Characterization of the Enzymatic Activity of MRD.
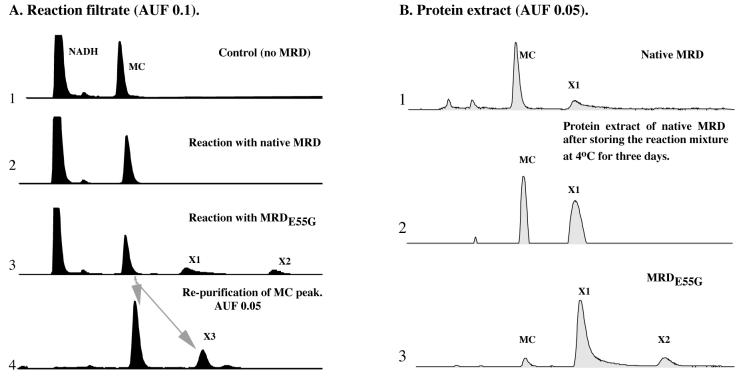
In previous work that led to the identification of MRD as a MC-binding protein, relatively small amounts of protein and substrates (MC, NADH) were used in the in vitro DNA protection assay (7). Although this assay was sensitive enough to detect the protection of DNA from MC-mediated cross-linking, it was not suitable for the detection of potential enzymatic reaction products. We re-examined the in vitro activity of MRD on MC under conditions considered optimal for enzymatic reaction, including higher protein and substrate concentrations and incubation at 37°C for 1 h. Monitoring the UV absorbency of the reaction mixture clearly showed a decrease at 365 nm and 340 nm, suggesting the transformation of MC and oxidation of NADH, respectively. Such changes were not detected in the control reaction containing only MC and NADH (data not shown). To identify the reaction products, the reaction mixture was filtered to remove MRD protein, and the filtrate was analyzed by HPLC with detection at dual wavelengths of 365 nm and 313 nm (characteristic absorptive wavelengths for MC and mitosene compounds, respectively). Interestingly, no specific signals could be detected at either wavelength that are unique to the reaction mixture containing MRD, as compared against the control reaction (Fig. 2A, lanes 1 and 2). However, the MRD protein remaining on the filter exhibited a pink color that is quite different from the original purple color of MC, but typical of its reduced mitosene derivatives. After recovery from the filter, the proteins were extracted with solvent. HPLC analysis of the extract at 365 nm revealed MC and NADH only (data not shown), whereas detection at 313 nm revealed a unique signal (X1) that appeared after the MC peak (Fig. 2B, lane 1). After purification, X1 was identified as a single compound, 1,2-cis-1-hydroxy-2,7-diaminomitosene (compound 6, Fig. 1), by comparison with an authentic standard in TLC, HPLC, and MS analyses.
Figure 2.
HPLC analysis (313 nm) of MC reduction by native MRD or MRDE55G. (A) Reaction filtrate. (B) Protein extract.
Previous studies on the reductive activation of MC by various flavoreductases have established that only the initial reduction of the quinone moiety of MC is enzyme-catalyzed and that mitosenes (5, 6, and 7) arise from the reactions between the ambivalent intermediate 4 and water. Specifically, the electrophilic form of 4 is attacked by water to produce 5 and 6, and the nucleophilic form of 4 abstracts the water-derived proton to form 7 (see Fig. 1) (12). Detection of 6 in the reaction containing MRD, MC, and NADH clearly suggested that MRD can use NADH to reduce the quinone moiety of MC.
Previously, only flavoreductases have been found to reductively activate MC. We, therefore, examined whether MRD contains cofactors that may facilitate electron transfer. The spectrophotometric analysis of purified MRD failed to reveal absorption peaks characteristic of flavoproteins, and the inductively coupled plasma spectrometry analysis suggested that MRD does not coordinate a metal center sufficient for redox chemistry (data not shown). Therefore, MRD appears to be a unique reductase that independently catalyzes the direct transfer of hydride from NADH to the quinone moiety of MC. In fact, comparison of the reactions conducted under aerobic or anaerobic conditions demonstrated that there was no significant difference in reaction rate, product profile or product distribution (data not shown). Together, these findings provide evidence that the reaction catalyzed by MRD entails an oxygen-independent two-electron reduction.
A striking characteristic of the action of MRD on MC is the slow reaction rate (Table 1). Coincidentally, both the substrate (MC) and the reduced product (6) were found to be MRD-bound for a prolonged time (Fig. 2B, lane 2). Further analysis of the reactions conducted under various pH conditions demonstrated that the reaction rate (measured by monitoring the decrease of absorption at 363 nm for the initial 5 min) increased as pH increased. The optimal reaction rate was found at a pH of approximately 7.5 (data not shown). It is possible that the reductase activity of MRD was inhibited by the binding of 6 to MRD. However, the mechanism of this inhibition appears different from what was observed previously in the NAD(P)H:quinone reductase (DT-diaphorase)-catalyzed MC reduction, which could be eliminated by lowering the pH (13, 14).
Table 1.
Comparison of MC reduction catalyzed by native MRD and MRDE55G, and the ligand binding constants of these two proteins
| MRD (native) | MRDE55G | |
|---|---|---|
| Specific activity (nmol MC/min per nmol protein) | 0.22 | 0.53 |
| Product profile | 6 | 5, 6, 7 (1:16.8:3.3) |
| Product distribution | Protein bound | Free (∼58%) and protein bound (∼42%)* |
| Molar ratio of bound MC to MRD | 0.47 | 0.22 |
| Binding constant for NADH | 166.48 ± 50.12 μM | 169.56 ± 56.89 μM |
| Binding constant for MC | 40.31 ± 7.49 μM | 54.58 ± 4.62 μM |
Estimated based on the total peak areas of mitosenes in the HPLC analyses.
Construction and Characterization of MRDE55G.
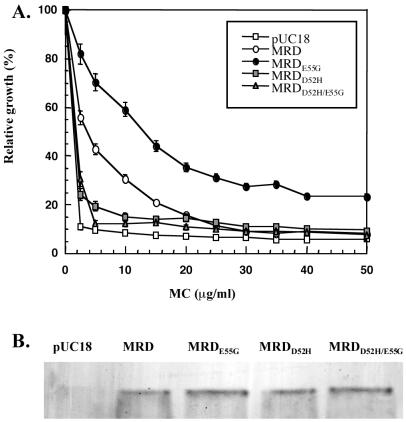
Because MRD catalyzed such a slow reduction of MC coupled with the fact that the protein fails to resemble any known reductase, we were compelled to generate a mutant version of MRD with enhanced catalytic activity. Such a mutant would help to identify the active site and perhaps elucidate the unique functional mechanism of MRD. Toward this end, DNA shuffling technology was used. After one round of shuffling, one recombinant E. coli clone was identified to exhibit increased MC resistance, as compared with the clone harboring wild-type mrd (Fig. 3A). Sequence analysis of the mutated gene identified several nucleotide changes but only one amino acid replacement, E55G. Site-directed mutagenesis confirmed that the improved MC resistance was indeed due to the single E55G replacement. Western blot analysis showed that MRDE55G was produced at a similar level compared with native MRD in E. coli (Fig. 3B), ruling out the possibility that the improved resistance was a result of enhanced protein expression or stabilization. Incubation of purified MRDE55G with MC and NADH resulted in a color change of the reaction mixture from light purple to deep purple-pink after 10 min (no significant color change was observed in the original reaction using native MRD). HPLC analysis of this reaction mixture revealed a significant amount of 6 (peak X1), as well as a new peak (X2), in both the reaction filtrate (Fig. 2A, lane 3) and the protein extract (Fig. 2B, lane 3). In addition, collection and repurification of the MC peak revealed an additional unique product (X3) (Fig. 2A, lane 4). A large-scale reaction (4 ml) then was conducted to generate enough of these metabolites for further characterization. By comparison with authentic standards in TLC, HPLC, and MS analyses, X2 was identified as 7, and X3 was identified as 5. A suitable amount of the major metabolite (6) also was collected for various NMR analyses (1H-NMR, 13C-NMR, 13C-heteronuclear multiple quantum correlation, proton one-dimensional correlated spectroscopy, heteronuclear multiple bond correlation), confirming its structure and the cis configuration (data not shown). Therefore, all three mitosene compounds that have been previously observed in the bioreductive activation of MC by various flavoreductases were detected in the MRDE55G-catalyzed MC reduction.
Figure 3.
(A) MC resistance of E. coli cells expressing various forms of MRD. (B) Western blot of E. coli cells expressing various forms of MRD.
Comparison of the reactions catalyzed by MRDE55G and native MRD are summarized in Table 1. In conclusion, MRDE55G appears to be an efficient quinone reductase, whereas native MRD functions primarily as a drug-binding protein due to its marginal enzymatic activity. Interestingly, the binding affinities of MC and NADH to MRDE55G were similar to native MRD, as measured by fluorescent quenching analysis (Table 1). Therefore, the improved enzymatic activity of this protein was not caused by enhanced substrate or cofactor binding, but more likely resulted from the rapid release of the reduced mitosene metabolites from MRDE55G, as indicated by the detection of 5, 6, and 7 in the filtrate of the reaction mixture. The small amount of these compounds detected in the solvent extract of MRDE55G could be the result of the random and noninhibitory binding to the protein, as observed before in the brewers' yeast NADPH oxidoreductase-catalyzed reduction of MC (12).
Protein Sequence Analysis and Identification of the Active Site of MRD.
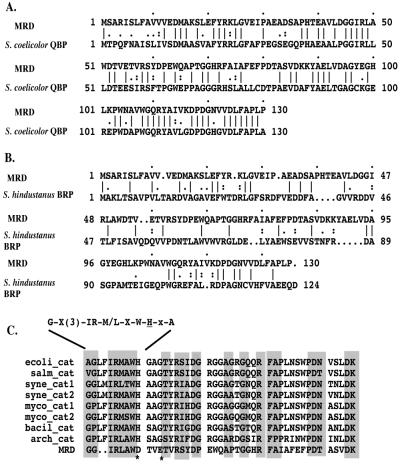
Discovery of the reductase activity of MRD prompted us to reiterate a search of the most recent databases. Two MRD homologues are now apparent. One is a putative quinone binding protein (QBP, GenBank accession no. Q9Z549) that has been recently identified through the genome sequencing project of Streptomyces coelicolor (15), and the other is the bleomycin binding protein (BRP, GenBank accession no. X52869) from Staphylococcus hindustanus (16). The putative QBP of S. coelicolor shows extensive homology with MRD (55% similarity and 49% identity) (Fig. 4A). Interestingly, the gene encoding this putative protein is located near the 6 o'clock position on the S. coelicolor chromosomal map, which encompasses a 1-Mb range of DNA that includes the act, whiE, and red antibiotic biosynthetic gene clusters. On the other hand, amino acid sequence alignment between MRD and BRP displayed only limited homology (35% similarity, 26% identify) (Fig. 4B). Overall, two striking similarities are shared by all three proteins: small size (containing about 130 amino acid residues with deduced molecular mass less than 15 kDa), and acidic nature (pI below 5).
Figure 4.
(A) Protein sequence alignment between MRD and the putative QBP from S. coelicolor. (B) Protein sequence alignment between MRD and the BRP from S. hindustanus. (C) Amino acid sequence alignment between the central region of MRD and the region around the second active site of various prokaryotic catalase-peroxidases. Motif of the second active site of catalase-peroxidases is shown above the sequences, with the catalytic histidine residues underlined. The two amino acid residues in MRD discussed in the text are indicated by *. ecoli_cat: E. coli catalase-peroxidase; salm_cat: Salmonella typhimurium catalase; syne_cat: Synechococcus sp. (PCC 7942) catalase-peroxidase; syne_cat2: Synechocystis sp. (PCC 6803) catalase-peroxidase; myco_cat1: Mycobacterium tuberculosis catalase-peroxidase; myco_cat2: Mycobacterium intracellulare catalase-peroxidase; bacil_cat: Bacillus stearothermophilus catalase-peroxidase; arch_cat: Archaeoglobus fulgidus catalase-peroxidase.
Similarity to QBP or BRP failed to reveal any insight regarding the enzymatic activity of MRD. However, the fact that the E55G replacement in MRD could significantly facilitate the reductive reaction suggested that E55 is located near the active site of MRD. We further scrutinized the protein sequence of MRD around E55 and found that this segment of amino acid sequence strongly resembles the second active site of prokaryotic catalase-peroxidases (Fig. 4C), a group of heme-binding enzymes exhibiting both catalase and broad-spectrum peroxidase activity (17). In general, these enzymes have two active sites, each containing a conserved histidine residue. The first one serves as the fifth ligand of the heme iron, and the second one functions as an acid-base catalyst in the reaction between the enzyme and hydrogen peroxide (18, 19). The amino acid alignment between MRD and the second active site of prokaryotic catalase-peroxidases can extend to 42 amino acid residues with several mismatches, including an aspartic acid residue (D52) and a glutamic acid residue (E55) in MRD corresponding to the catalytic histidine residue and a nearby glycine residue in catalase-peroxidases, respectively. Therefore, the E55G replacement in MRD has actually made this region more similar to a catalase-peroxidase. Because this replacement resulted in a significant increase in MRD reductase activity, we propose that the active site of native MRD was derived from the second active site of prokaryotic catalase-peroxidases, and that replacements of the catalytic residue and several coordinate residues have served to attenuate the reductase activity. This modification of the active site of MRD is presumed to be critical, because a fully functional reductase would activate MC, generating cytotoxic intermediates that could pose a serious threat to S. lavendulae cells.
To test this hypothesis, two MRD mutants (MRDD52H and MRDD52H/E55G) were generated by site-directed mutagenesis. Expression of either mutant protein in E. coli resulted in greater sensitivity of the cells to MC compared with cells expressing native MRD (Fig. 3A), implying that cytotoxic intermediates might have been generated from the action of the mutated MRD on MC. The cells expressing these two mutant MRD proteins still exhibited marginal MC resistance, as compared with cells harboring vector alone. This result suggests that the two mutant proteins might still maintain the capability to bind and degrade MC. Furthermore, compared with native protein, neither the single nor the double mutant forms of MRD has altered expression level or stability, as shown by Western blot analysis (Fig. 3B).
Discussion
The work presented here has revealed that MRD possesses an inherent, albeit low, level of quinone reductase activity that slowly transforms MC into a mitosene product. This finding, however, does not conflict with the previous conclusion that MRD is simply a drug-binding protein. It is shown that the slow rate of the reductive reaction catalyzed by native MRD actually resulted in a prolonged association of intact MC with the protein. If a drug transport protein is coupled with MRD, it appears likely that the rate of export might exceed the rate of reductive activation, resulting in rapid removal of MC from the cell instead of transformation of MC into a cytotoxic species. The existence of this process is evident in the producing strain, because MC (not mitosenes) is the major product generated and secreted by S. lavendulae. Indeed, a drug transport protein (MCT) already has been identified in S. lavendulae, which cooperates with MRD to form an efficient drug binding and export system that delivers MC to the outside of the cell (8). Furthermore, attempts to disrupt mrd in wild-type S. lavendulae cells were not successful, suggesting that MRD is a key component of the predominant cellular self-protection mechanism (M.H. and D.H.S., unpublished work).
An interesting issue arising from this work is how MRD has evolved from a quinone reductase to a drug-binding protein. Cells typically use quinone reductases to perform two-electron reduction in the detoxification and degradation of quinones generated in primary or secondary metabolism. Compared with the one-electron reduction that is inhibited by oxygen, two-electron reduction is oxygen-independent and bypasses the generation of a semiquinone radical, which, when reoxidized, causes the production of a reactive oxygen species (14). It is not surprising that S. lavendulae, as a highly aerobic microorganism, may produce MRD as a quinone reductase to carry out two-electron reductions. However, the emergence of MC as a secondary metabolite must have coincided with selective pressure for the evolution of MRD. During this process, modification of MRD was essential to minimize the potential damage caused by toxic MC metabolites.
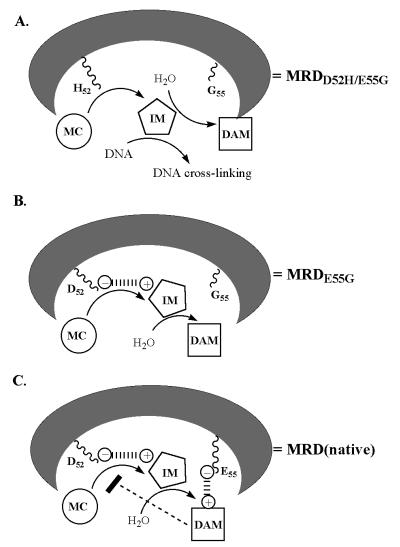
Based on the experimental results presented above, we propose an evolutionary model to explain the functional switching of MRD, which might be achieved primarily through the replacements of two amino acid residues at the active site of MRD (Fig. 5). Presumably, the original amino acid sequence (residues 40–80) in MRD might be quite similar to that of the second active site of prokaryotic catalase-peroxidases, with a histidine residue at position 52 and a glycine residue at position 55. With such an active site, MRD might function as an efficient quinone reductase (Fig. 5A). However, as the MC biosynthetic pathway evolved, the original catalytic residue (H52) was replaced by an aspartic acid residue, resulting in a protein equivalent to MRDE55G. This replacement should not significantly reduce the catalytic activity of MRD, as shown by the fact that MRDE55G can still efficiently reduce MC. Instead, this replacement might prevent the release of the reactive intermediate 4 from the active site, possibly through electrostatic interaction between the positively charged 4 and the deprotonated, negatively charged D52 residue. The sequestered 4 might not be susceptible to the nucleophilic attack of macromolecules like DNA, but should still be available for the nucleophilic or electrophilic attack of water to form various mitosenes (Fig. 5B). Therefore, MRDE55G might confer resistance to MC through fast binding and transformation of MC into three mitosenes (5, 6, and 7). The fact that 6 was the major product of MRDE55G catalyzed reaction also implies that 4 might be in an unusual configuration during the reaction, because in the normal reductive MC activation catalyzed by various flavoreductases, the trans (5) and cis (6) 1-hydroxy mitosenes were produced in almost equal amounts (12, 20, 21).
Figure 5.
Schematic representation of the proposed evolutionary model for the active site of MRD (details are described in the text). IM: reaction intermediate; DAM: diaminomitosenes.
The second significant replacement within the active site of MRD might be the switching of G55 to a glutamic acid (E55). The bulky and negative charged side chain of E55 might interact with the basic mitosene products (mainly, 6), preventing their release from the active site. The accumulation of these mitosenes at the active site, in turn, might interfere with the electron transfer catalyzed by the D52 residue and thus greatly reduce the reductive reaction rate (Fig. 5C). Replacements of several other amino acid residues near the active site also might be necessary to ensure optimal binding of the reaction intermediate or mitosene products. Therefore, the final choice of the amino acid residues at the active site, as found in native MRD, fulfills dual goals: preventing the exposure of cytotoxic reaction intermediates to DNA and minimizing the reductive transformation of MC.
It is noteworthy that a gene encoding a protein (QBP) very similar to MRD also is found in the S. coelicolor genome. Localization of this gene in the chromosome implies that QBP might also be involved in the binding and transport of secondary metabolites. The homology between MRD and the BRP is intriguing. Unlike MC, bleomycin belongs to a family of glycopeptide antibiotics that lack the quinone moiety. However, this compound is also a DNA damaging agent that requires bioreductive activation (21). Previous analysis of BRP also has raised the possibility that this protein may be involved in drug export (16). It is possible that antibiotic-producing bacteria might have evolved a general drug-binding-transport system for drug secretion and self-protection, and a common structural fold might be shared by the drug-binding proteins.
In summary, a previous puzzle regarding the requirement of a NADH cofactor for binding of MC to MRD has been resolved through the work presented here. The capability of MRD to provide resistance to MC apparently is derived from a remnant enzymatic activity that could potentially activate MC. The evolutionary change of the active site of MRD, as demonstrated in the proposed model, might represent a simple, yet efficient, mechanism for protein functional switching and could have a general significance for protein engineering and enzyme development.
Acknowledgments
We thank Peter W. Villalta at the Cancer Center of University of Minnesota for MS analysis, Beverly Ostrowski at the Department of Biochemistry University of Minnesota for NMR analysis, and Namthip Sitachitta for interpreting the NMR data. We are grateful to Kyowa Hakko Kogyo, Ltd., and Bristol-Myers Squibb for gifts of mitomycin C and Maria Tomasz for mitosene standards. This work was supported by National Institutes of Health Grant CA81172.
Abbreviations
- MC
mitomycin C
- QBP
quinone binding protein
- BRP
bleomycin binding protein
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.031314998.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.031314998
References
- 1.Spanswick V J, Cummings J, Smyth J F. Gen Pharmacol. 1998;31:539–544. doi: 10.1016/s0306-3623(98)00055-x. [DOI] [PubMed] [Google Scholar]
- 2.Cummings J, Spanswick V J, Tomasz M, Smyth J F. Biochem Pharmacol. 1998;56:405–414. doi: 10.1016/s0006-2952(98)00073-2. [DOI] [PubMed] [Google Scholar]
- 3.Tomasz M, Palom Y. Pharmacol Ther. 1997;76:73–87. doi: 10.1016/s0163-7258(97)00088-0. [DOI] [PubMed] [Google Scholar]
- 4.Tomasz M. Chem Biol. 1995;2:575–579. doi: 10.1016/1074-5521(95)90120-5. [DOI] [PubMed] [Google Scholar]
- 5.August P R, Flickinger M C, Sherman D H. J Bacteriol. 1994;176:4448–4454. doi: 10.1128/jb.176.14.4448-4454.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson D A, August P R, Shacketon C, Liu H W, Sherman D H. J Am Chem Soc. 1997;119:2576–2577. [Google Scholar]
- 7.Sheldon P J, Johnson D A, August P R, Liu H W, Sherman D H. J Bacteriol. 1997;179:1796–1804. doi: 10.1128/jb.179.5.1796-1804.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sheldon P J, Mao Y, He M, Sherman D H. J Bacteriol. 1999;181:2507–2512. doi: 10.1128/jb.181.8.2507-2512.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Epps D E, Raub T J, Caiolfa V, Chiari A, Zamai M. J Pharm Pharmacol. 1999;51:41–48. doi: 10.1211/0022357991772079. [DOI] [PubMed] [Google Scholar]
- 10.Stemmer W P. Proc Natl Acad Sci USA. 1994;91:10747–10751. doi: 10.1073/pnas.91.22.10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sambrook J, Fritsch E F, Maniatis R. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 12.Peterson D M, Fisher J. Biochemistry. 1986;25:4077–4084. doi: 10.1021/bi00362a014. [DOI] [PubMed] [Google Scholar]
- 13.Siegel D, Beall H, Kasai M, Arai H, Gibson N W, Ross D. Mol Pharmacol. 1993;44:1128–1134. [PubMed] [Google Scholar]
- 14.Ross D, Siege D, Beall H, Prakash A, Mulcahy R T, Gibson N W. Cancer Metastasis Rev. 1993;12:83–101. doi: 10.1007/BF00689803. [DOI] [PubMed] [Google Scholar]
- 15.Redenbach M, Kieser H M, Denapaite D, Eichner A, Cullum J, Kinashi H, Hopwood D A. Mol Microbiol. 1996;21:77–96. doi: 10.1046/j.1365-2958.1996.6191336.x. [DOI] [PubMed] [Google Scholar]
- 16.Dumas P, Bergdoli M, Cagnon C, Masson J. EMBO J. 1994;13:2483–2492. doi: 10.1002/j.1460-2075.1994.tb06535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Welinder K G. Biochim Biophys Acta. 1991;1080:215–220. doi: 10.1016/0167-4838(91)90004-j. [DOI] [PubMed] [Google Scholar]
- 18.Kimura S, Ikeda-Saito M. Proteins. 1988;3:113–120. doi: 10.1002/prot.340030206. [DOI] [PubMed] [Google Scholar]
- 19.Henrissat B, Saloheimo M, Lavaitte S, Knowles J K C. Proteins. 1990;8:251–257. doi: 10.1002/prot.340080307. [DOI] [PubMed] [Google Scholar]
- 20.Tomasz M, Lipman R. Biochemistry. 1981;20:5056–5061. doi: 10.1021/bi00520a036. [DOI] [PubMed] [Google Scholar]
- 21.Burger R M, Peisach J, Horwitz S B. J Biol Chem. 1981;256:11636–11644. [PubMed] [Google Scholar]