Abstract
We have established a model system using the caspase-2 pre-mRNA and initiated a study on the role of alternative splicing in regulation of programmed cell death. A caspase-2 minigene construct has been made that can be alternatively spliced in transfected cells and in nuclear extracts. Using this system, we have identified a 100-nt region in downstream intron 9 that inhibits the inclusion of the 61-bp alternative exon. This element (In100) can facilitate exon skipping in the context of competing 3′ or 5′ splice sites, but not in single-intron splicing units. The In100 element is also active in certain heterologous pre-mRNAs, although in a highly context-dependent manner. Interestingly, we found that In100 contains a sequence that highly resembles a bona fide 3′ splice site. We provide evidence that this sequence acts as a “decoy” acceptor site that engages in U2 snRNP-dependent but nonproductive splicing complexes with the 5′ splice site of exon 9, hence conferring competitive advantage to the exon-skipping splicing event (E8-E10). These results reveal a mechanism of action for a negative intronic regulatory element and uncover a role for U2 snRNP in the regulation of alternative splicing.
Alternative pre-mRNA splicing is a fundamental mechanism for regulating the expression of a multitude of eukaryotic genes (for reviews, see refs. 1 and 2). The basic splicing signals, which include the 5′ splice site, branch site, and polypyrimidine track-AG, are initially recognized by the U1 snRNP, U2 snRNP, and U2 snRNP auxiliary factor (U2AF), respectively. These basic splicing signals tend to be degenerate in higher eukaryotes and cannot alone confer the specificity required to achieve accurate splice site selection. Various types of exonic and intronic elements that can modulate the use of nearby splice sites have now been identified. Among the best known examples of such elements are the exonic splicing enhancers, which were initially identified as purine-rich sequences that could promote the use of an upstream 3′ splice site (3–5). Intronic enhancer elements, usually residing downstream of alternative exons, are generally diverse in sequence (e.g., refs. 6–8). Although the factors interacting with these elements are beginning to be identified (ref. 9 and references therein), mechanisms involved in splicing stimulation are still largely unknown. Splicing silencers have also been found in exons (ref. 10 and references therein) or introns (e.g., refs. 11–14). However, in most cases, it is still not clear how splicing inhibition is achieved.
We have been using the caspase (casp)-2 (also known as Ich-1, interleukin-1-β converting enzyme homologue 1, or Nedd2) gene as a model system to study the importance of alternative splicing regulation in programmed cell death. Interestingly, alternative splicing of a small 61-bp cassette-exon in the casp-2 pre-mRNA leads to the formation of two mRNAs, encoding protein isoforms with antagonistic activities in apoptosis (15). CASP-2L is derived from the skipping of alternative exon 9 and can induce cell death. CASP-2S is a truncated version of the protein produced because of a premature termination codon created by the inclusion of exon 9, and it prevents apoptotic death in certain cell types (16). Casp-2L is the predominant casp-2 transcript present in most tissues, including the ovaries, thymus, and spleen, whereas casp-2S expression is restricted to the adult brain and skeletal muscles (15–18). We have previously reported that splicing factors SC35, ASF/SF2, and hnRNP A1 could modulate casp-2 alternative splicing (17). We report here the identification of an independent intronic element located in the intron downstream of exon 9. Our results show that this element represses the inclusion of the alternative exon through a mechanism involving usage of a decoy 3′ acceptor site that forms nonproductive splicing complexes containing U2 snRNP.
Materials and Methods
Plasmid Constructions and Splicing Substrates.
Sequences of oligonucleotides as well as details of cloning procedures for constructs used in this study can be obtained upon request.
Transfections and Reverse Transcription (RT)–PCR.
Transfection was done using a standard calcium phosphate precipitation procedure with 3 μg of DNA. Cells were harvested after 48 h, and the RNA was extracted using Trizol reagent (GIBCO/BRL). Spliced products derived from the expressed minigenes were detected using RT-PCR as described (17) and resolved on 6% polyacrylamide/1× TBE gels.
In Vitro Splicing Assays.
RNA substrates were synthesized and the full-length transcripts gel purified as described (19). HeLa cell nuclear extracts were prepared according to previously established protocols and contained ≈20 mg/ml total proteins (20). Splicing reactions were set up and processed as described (21). Splicing products were resolved on 8% polyacrylamide/8 M urea gels.
U1 snRNP RNase H Protection Assay.
Two femtomoles of 553 RNA substrates were added to a 12.5-μl splicing reaction containing MOCK- or U1 snRNP-depleted HeLa nuclear extracts (22) and incubated for 0, 5, or 20 min at 30°C. Oligonucleotides (75 pmol) directed against 5′ splice sites of exon 8 and of exon 9 (UP and ALT, respectively) were then added along with 0.2 unit of RNase H (GIBCO/BRL), and the incubation continued for 15 min at 37°C. Resulting RNA fragments were resolved on 8% polyacrylamide/8 M urea gels. Relative levels of protection were quantified as described (23).
Electrophoretic Separation of Splicing Complexes.
This procedure was adapted from refs. 24 and 25. ATP depletion of the nuclear extracts was achieved as in ref. 26. U2 snRNP or U7 snRNP depletion was achieved by preincubation of the nuclear extract with 2 μM or 7 μM of the respective 2′-O-methyl oligoribonucleotide.
UV Cross-Linking and Immunoprecipitation Assays.
Splicing reactions were set up as above except that 20 fmol of RNA substrates were used. Six-microliter aliquots were transferred onto a 96-well microtiter plate previously cooled at −20°C and irradiated with ≈1 Joule in a UV Stratalinker 1800 (Stratagene). Samples were then treated for 30 min at 37°C with 1 volume of RNase A (5 mg/ml). Radiolabeled cross-linked proteins were boiled for 5 min in 1× SDS loading buffer and separated on 12.5% SDS/PAGE. For immunoprecipitation, samples were processed as before for cross-linking except that antibodies were added following the RNase A treatment. After incubation for 30–60 min on ice, protein A/G agarose beads were mixed in, and incubation was continued for 2 h at 4°C with gentle shaking. The beads were then thoroughly washed (4×) with NET-2 buffer (22). The proteins retained on the beads were then boiled as before and resolved on 12.5% SDS/PAGE.
Results and Discussion
An Intronic Element Inhibits casp-2 Exon 9 Inclusion.
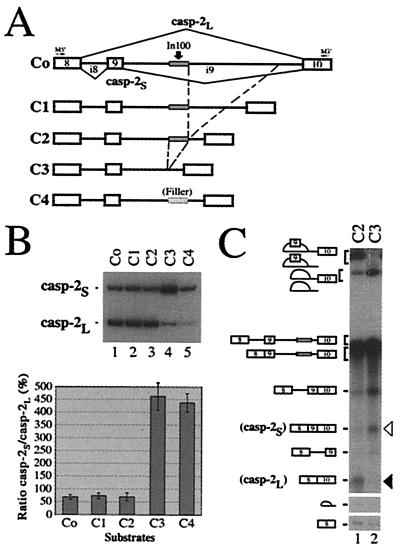
We have constructed a casp-2 minigene containing the common upstream exon 8, the 61-bp alternative exon 9 and flanking introns (84 nt and 2.8 kb, respectively), followed by downstream exon 10 (see Fig. 1A and ref. 17). To characterize the sequences regulating the alternative splicing of exon 9, we used a PCR-based deletion strategy to gradually shorten the downstream 2.8-kb intron. The deletion mutant constructs were expressed transiently in HeLa cells, and the alternative splicing products (casp-2L and casp-2S) were detected using RT-PCR with primers specific for the minigenes. Internal deletions resulting in the shortening of intron 9 from 2.8 kb to 0.3 kb did not significantly affect the ratio of casp-2S to casp-2L (Fig. 1B, C0–C2, lanes 1–3 and graph). In contrast, deletion of an additional 100 nt (C3) produced a drastic change in the splicing profile, strongly promoting alternative exon 9 inclusion (Fig. 1B, lane 4 and graph). Because the distance between splice sites has been reported to affect splicing efficiency (27), we replaced In100 with an unrelated 100-bp fragment so that the resulting construct (C4) had a downstream intron identical in length to that in C2. This C4 construct also gave rise to a higher proportion of the casp-2S isoform (Fig. 1B, lane 5), showing that the shift observed upon deletion from 0.3 kb to 0.2 kb was not due to an increased proximity of the splice sites. To further characterize this intronic element, we set up an in vitro splicing system using HeLa cell nuclear extracts. The casp-2 minigene (C2) was spliced efficiently in vitro, and the ratio of casp-2S to casp-2L mRNAs produced was similar to the one observed in vivo (Fig. 1C, lane 1). More importantly, the C3 substrate lacking the 100-nt region again led to the predominant production of the casp-2S inclusion product (Fig. 1 C, lane 2). Thus, these experiments reveal a 100-nt region (In100) in the intron downstream of the 61-bp alternative exon 9 that inhibits the production of casp-2S mRNA in HeLa cells and nuclear extracts.
Figure 1.
An intronic element inhibits casp-2 exon 9 inclusion. (A) Structure of casp-2 deletion mutant constructs. The gray box in the intron indicates the location of the In100 element. The small cross-hatched rectangle depicts “filler” sequences used as a size control. Positions of oligonucleotides used for RT-PCR are indicated. (B) RT-PCR analysis was carried out using total RNA extracted from cells transfected with the constructs shown in A. Ratio of casp-2S to casp-2L (%) was determined from five independent experiments and plotted in the histogram below. (C) 32P-labeled transcripts were incubated in HeLa cell nuclear extracts under standard splicing conditions. Splicing products were resolved on an 8% acrylamide/8 M urea gel. The black and white arrowheads indicate the position of the casp-2L (skipping) and casp-2S (inclusion) mRNAs, respectively.
The In100 Element Requires the Context of Competing Splice Sites.
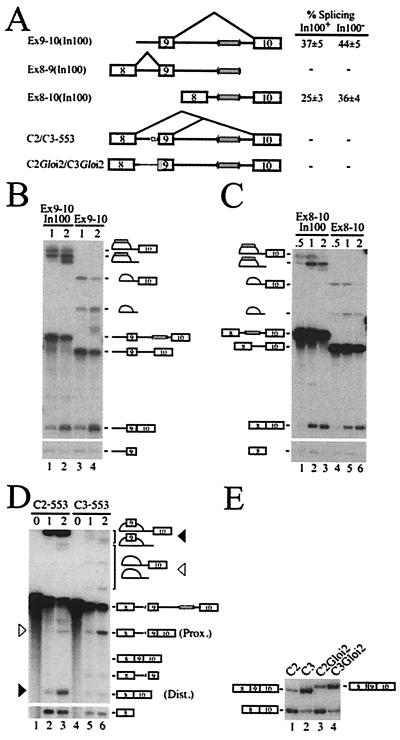
From the above results, it was not clear whether the In100 intronic element was acting by inhibiting exon 9 inclusion or by stimulating the splicing of 8 to 10. To gain insights into the mechanism of action of In100, we constructed a series of minigene derivatives containing or lacking the In100 sequence (Fig. 2A). We first tested if the In100 element affected the joining of exon 9 to exon 10 (Fig. 2A, Ex9–10 substrates). In this context, only a small stimulation was observed upon deletion of the In100 element (Fig. 2 A and B). We then asked whether the intronic element could affect the splicing of the upstream exon 8 to 9 splicing unit (Fig. 2A, Ex8–9 substrate). In100 did not have any detectable effect on the efficiency of this splicing event (data not shown). Finally, we examined the splicing efficiency of exon 8 to 10 in the presence or absence of the In100 region (Fig. 2A, Ex8–10 substrates). In vitro splicing experiments again revealed only a slight stimulation of splicing upon deletion of the 100-nt element (Fig. 2 A and C), indicating that In100 did not stimulate splicing of exon 8 to 10. These results thus uncover the requirement for a competing splicing event to observe the full inhibitory activity of the In100 element. Furthermore, it suggests a mechanism for In100 function in which it mediates a negative effect on the inclusion of exon 9.
Figure 2.
In100 requires the context of competing splice sites. (A) Structure of splicing substrates. The thinner line and shaded rectangle represent β-globin sequences substituted in the C2/C3Gloi2 substrates. The gray box in parentheses depicts the presence or the absence of the In100 element. The table shows the splicing efficiency (%) as determined from three independent experiments. (B) Splicing of Ex9–10 substrates. Substrates were incubated in HeLa nuclear extracts for the time (in hours) indicated above each lanes. (C) Splicing of Ex8–10 substrates. (D) Splicing of 553 derivatives. Position of the splicing products generated from the use of the proximal (Prox.) or distal (Dist.) splice site is indicated by the white or black triangles, respectively. (E) Splicing of the (C2/C3)Gloi2 derivatives in transfected HeLa cells. Position of exon 9 inclusion and skipping mRNAs is shown.
The 3′ splice site of the alternative exon was inactivated by changing the conserved AG to a CU dinucleotide, thus creating a substrate in which proximal and distal 5′ splice sites were competing for pairing with a common downstream 3′ splice site (Fig. 2A, C2/C3–553 substrates containing or lacking In100, respectively). The presence of In100 in these substrates promoted distal splice site utilization, showing that In100 can function in the context of competing 5′ splice sites (Fig. 2D, compare lanes 2 and 3 with lanes 5 and 6). We next investigated if the effect of In100 required the presence of an intact upstream intron 8. Substrates were designed in which the upstream intron 8 and associated 3′ splice site were substituted with sequences from the human β-globin intron 2 (Fig. 2A, C2Gloi2/C3Gloi2). When incubated in nuclear extracts or transfected in HeLa cells, the C2Gloi2 substrate was spliced predominantly following the exon-skipping pathway to a level similar to that obtained with the C2 substrate (data not shown and Fig. 2E, compare lane 3 to lane 1). C3Gloi2, which was constructed by deleting the In100 element from C2Gloi2, yielded a significant shift toward the production of the exon-inclusion mRNA (Fig. 2E, lane 4). These results show that the native intron 8 sequence (including the 3′ splice site of exon 9) is not essential for In100 to retain its inhibitory activity on exon 9 inclusion. However, the In100 element did not have any effect when tested in a completely heterologous context, such as the previously described (28) DUP51 substrate (data not shown). Taken together, these results indicate that In100 is at least partially sensitive to context. These observations further point to the 5′ splice site of the alternative exon as an important requirement for In100 function.
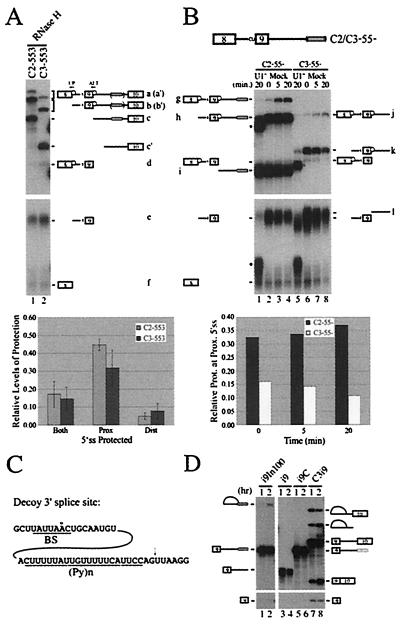
U1 snRNP Occupancy of the Competing 5′ Splice Sites Does Not Correlate with Splicing.
Since U1 snRNP is the initial spliceosomal component responsible for 5′ splice site recognition, we used a specific RNase H protection assay (23) to measure the U1 snRNP occupancy of the competing 5′ splice sites on pre-mRNAs containing or lacking In100. Standard splicing reactions were incubated for 20 min before the addition of oligonucleotides (ALT and UP oligos in Fig. 3A) specific for each 5′ splice site and RNase H. Following RNase H treatment, the resulting protected or fully digested fragments were separated on a polyacrylamide denaturing gel. Specifically, band a and a′ represent the doubly protected C2- or C3–553 pre-mRNAs, respectively. Bands b and b′ represent the C2- or C3–553 substrate with U1 snRNP bound at the proximal 5′ splice site. Finally, protection at the distal 5′ splice site is represented by fragment d for both substrates as well as the respective 3′ fragment c or c′ (containing or lacking In100, respectively). For both C2–553 and C3–553 transcripts, protection was almost completely abolished when the assay was performed in a HeLa nuclear extract depleted of functional U1 snRNP (data not shown). Intriguingly, although drastic differences in splice site utilization were observed between C2–553 and C3–553 (see Fig. 2D, lanes 2 and 3 and lanes 5 and 6), similar levels of U1 snRNP-dependent protection were detected on the competing 5′ splice sites for both substrates (Fig. 3A, lanes 1 and 2; compare band d with bands b or b′, respectively, and histogram below). In addition, the predominant 5′ splice site protected by U1 snRNP was the proximal 5′ splice site (i.e., the 5′ splice site of exon 9), in sharp contrast with the C2–553 splicing profile that indicated this alternative 5′ splice site was poorly used. These results suggested a more complex mechanism than a simple reduction of U1 snRNP-mediated recognition of the alternative 5′ splice site. One potential model to explain these results would be to propose a looping-out of exon 9 promoted by the interaction of factors bound to In100 and to sequences in the upstream intron, as was proposed for the hnRNP A1 pre-mRNA alternative splicing (29). Such exon 9 looping-out would bring the 3′ splice site of exon 10 in closer proximity to the distal 5′ splice site of exon 8, thus favoring its utilization without affecting the level of U1 snRNP binding to the 5′ splice site of the alternative exon (consistent with the RNase H protection assay results). However, this model is not supported by our observation that substitution of the upstream intron with a completely different sequence does not affect In100 activity (Fig. 2).
Figure 3.
U1 snRNP occupancy of casp-2 competing 5′ splice sites as determined by an RNase H protection assay. (A) Substrates containing (C2–553) or lacking (C3–553) In100 were tested in a U1 snRNP protection assay. The protected and cleaved RNA species were resolved on an 8% polyacrylamide denaturing gel as indicated. Data from five independent experiments are represented below in the histogram. U1 snRNP-dependent complexes responsible for the observed protection are depicted by a white sphere on each 5′ splice site. (B) The U1 snRNP RNase H protection assay was carried out as shown in A except that substrates lacking exon 10 and its associated 3′ splice site were used. As a control, the protection was also done with HeLa nuclear extracts depleted of functional U1 snRNP (U1−; lanes 1 and 5). (Histogram) Relative levels of protection at the proximal 5′ splice site were plotted over time for each substrate with the data from two independent experiments. (C) In100 contains a sequence resembling a 3′ acceptor site. Putative branch site (BS) and polypyrimidine track [(Py)n] are shown. The arrow above the sequence shows the potential cleavage site at the 3′ splice site AG. (D) The upstream portion of In100 can be used as a functional 3′ splice site. Standard splicing reactions were set up using substrates containing exon 9 and its 5′ donor site with downstream intron sequences terminated immediately downstream of In100 (i9In100) or control substrates as depicted in Fig. 5A.
It has been proposed that direct or indirect interactions in commitment complexes with factors bound at the 3′ splice site promote and stabilize complexes at the 5′ splice site (30, 31). The results shown above suggest that interactions which did not ultimately lead to productive splicing may contribute to the U1 snRNP-dependent protection observed on the 5′ splice site of exon 9. This prompted us to perform the same assay using substrates in which the native 3′ acceptor site (along with exon 10) had been deleted (Fig. 3B, C2–55- and C3–55-) to see if we could still detect a stable U1 snRNP protection at the alternative 5′ splice site. Strikingly, the C2–55- substrate, containing In100 but lacking the downstream native 3′ splice site, yielded a very strong U1 snRNP-dependent protection at the proximal 5′ splice site (Fig. 3B, lanes 2–4; band h and histogram below). However, upon deletion of In100 (C3–55-substrate), the level of protection was significantly reduced and actually decreased over time under splicing conditions (Fig. 3B, lanes 6–8; band k and histogram below). This result suggests that most of the protection observed at the proximal 5′ splice site in the C2–553 substrate is not derived from normal commitment complex interactions with the downstream 3′ splice site of exon 10. More importantly, these results show that this U1 snRNP-dependent protection is somehow mediated through sequences present in the In100 element.
In100 Contains a Potentially Functional 3′ Acceptor Site.
A close inspection of the In100 fragment revealed the presence of a stretch of sequence that highly resembles a bona fide splicing acceptor site (Fig. 3C): a branch site sequence (consensus: YNYYRÅY, where Y represents pyrimidines, R represents purine, and N represents any nucleotide; the small circle above the adenosine residue denotes the putative branch point nucleotide according to the branch site mammalian consensus sequence) followed by a long stretch of pyrimidine residues and (NY)AG (32, 33). Our splicing analysis using transfected cells or in vitro biochemical assays demonstrates that this site is not used in the wild-type casp-2 pre-mRNA context (see Fig. 1). To test if these sequences could function as a bona fide 3′ splice site in the absence of the native 3′ splice site of exon 10, we used a substrate containing exon 9 and its 5′ donor site with downstream intron sequences terminated after In100 (Fig. 3D, i9In100; left of gel panel). Splicing intermediates (lariat and 5′ exon) were detected for this minimal substrate after 2 h of incubation in nuclear extract, and final splicing products could be visualized upon a longer exposure of the autoradiogram (Fig. 3D, lanes 1 and 2 and data not shown). This splicing event was dependent on the presence of the In100 sequence because, under the same conditions, no splicing products were obtained for substrates in which the downstream intron was terminated just upstream of In100 or in which the In100 sequence was replaced with a filler sequence of the same size (i9 and i9C; Fig. 3D, lanes 3 and 4 and lanes 5 and 6, respectively). These results demonstrate that the 3′ splice site in In100 can support splicing, albeit with significantly lower efficiency than the corresponding C3i9 substrate containing the native 3′ splice site of exon 10 (Fig. 3D, lanes 7 and 8).
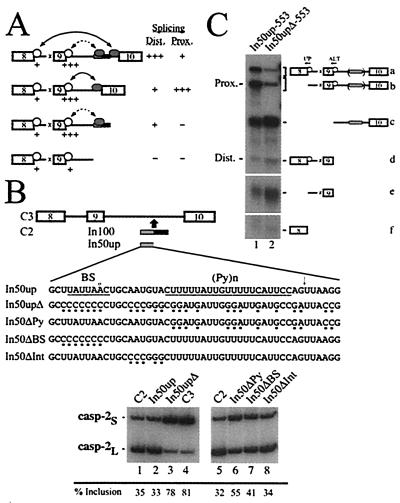
In100 Acts as a Decoy 3′ Splice Site.
Based on the above results, we propose a model in which the In100 regulatory element could act as a decoy 3′ splice acceptor site that would facilitate exon 9 skipping by engaging in the formation of nonproductive splicing complexes with the 5′ splice site of alternative exon 9 (Fig. 4A). To test this model, we first reintroduced into the C3 substrate a minimal fragment containing only the In100 decoy 3′ splice site sequence (Fig. 4B, In50up substrate). The expression of this construct in HeLa cells yielded predominantly products lacking exon 9, as judged by RT-PCR analysis (Fig. 4B, lane 2). This splicing profile was almost identical to the one observed with the C2 substrate containing the complete In100 element (Fig. 4B, lane 1). This observation indicates that the sequence containing the decoy 3′ splice site is sufficient to mediate an inhibitory effect on exon 9 inclusion. We then mutated this minimal sequence (In50upΔ substrate) to eliminate the splicing signals. Specifically, the branch site sequence (UAUUAAC) was replaced by a stretch of cytosines, the putative 3′ splice site AGs by GA or CC, and purines were interspersed throughout the polypyrimidine tract (Fig. 4B). Consistent with our model, the combination of these mutations promoted an increase in the level of exon 9 inclusion, similar to the effect observed upon complete deletion of the In100 element (Fig. 4B, compare lanes 1 and 2 with lanes 3 and 4, respectively). To further characterize the sequence requirement for In100 function, we tested the mutations described above in separate subsets. Mutating only the AG dinucleotides in the In50up sequence did not affect significantly the splicing profile (data not shown). Likewise, replacing the putative branch site sequence with a stretch of Cs only promoted a partial shift toward exon 9 inclusion (Fig. 4B; In50ΔBS, lane 7). It is possible that mutation of these sequences leads to the activation of cryptic branch points and/or AG-dinucleotide cleavage sites. However, we did not observe any difference in the size of RT-PCR products generated from mRNAs of mutant substrates. In contrast, introducing purines in the pyrimidine-rich sequence more strongly promoted exon 9 inclusion (Fig. 4B; In50ΔPy, lane 6). Finally, as a control, we changed nucleotides in a region between the splicing signals and observed no effect on the splicing profile of the transfected minigene (Fig. 4B; In50ΔInt, lane 8). These results show that a minimal fragment harboring the In100 decoy 3′ splice acceptor sequence is sufficient to mediate the inhibitory effect on exon 9 inclusion. Furthermore, this mutagenesis analysis demonstrates the requirement for an intact polypyrimidine tract and branch site sequence (although to a lesser extent) inside In50up for it to be fully active in suppressing exon 9 inclusion.
Figure 4.
In100 acts as a decoy 3′ splice site. (A) A model for function of In100 as a decoy 3′ splice site. White spheres stand for U1 snRNP-dependent 5′ splice site complexes, and gray spheres depict U2 snRNP-dependent complexes forming on In100 or on the native 3′ splice site of exon 10. Relative levels of U1 snRNP-dependent protection are represented by + signs below each 5′ splice site. Relative use of distal and proximal 5′ splice sites are indicated on the right. In this model, the 5′ splice site of alternative exon 9 engages in nonproductive splicing complexes with the decoy 3′ splice site in the In100 element, as shown by the dotted line between arrow heads. This in turn confers a competitive advantage to the exon 9-skipping splicing pathway, as shown by the long solid line between the arrow heads. (B) Minimal fragments containing the wild-type or the mutated 3′ acceptor site region of In100 were inserted back into the C3 construct (sequence of In50up fragments is indicated). Mutated nucleotides are indicated with small dots under the sequence. These constructs were transfected into HeLa cells, and the splicing profile was analyzed by RT-PCR as before. The numbers below the gels show the percentage of exon 9 inclusion as quantified using a PhosphorImager. (C) In50up and In50upΔ fragments were inserted in the C3–553 substrate (In50up- and In50upΔ-553 substrates, respectively), and the U1 snRNP RNase H protection assay was carried out as before. Positions of proximal and distal 5′ splice site protected fragments are as indicated.
Based on the RNase H protection results presented in Fig. 3, we hypothesized that the 3′ splice site-like sequence in the upstream portion of In100 could be promoting the U1 snRNP-dependent complexes detected on the 5′ splice site of exon 9. Since mutating the splicing signals in In50up was sufficient to abrogate inhibition by the element, the same mutations should result in a decrease of the U1 snRNP protection at the alternative exon 9 5′ splice site. Wild-type and mutant In50up fragments were inserted in the C3–553 substrate (In50up-553 and In50upΔ-553, respectively), and the U1 snRNP RNase H protection assay was carried out as before. Following incubation in HeLa nuclear extracts under splicing conditions, the In50up-553 substrate yielded a protection profile identical to that observe for the In100-containing C2–553 substrate (Fig. 4C, lane 1; see also Fig. 3A, lane 1). As predicted, introducing point mutations in the splicing signals present in the In50up sequence significantly reduced the level of protection at the alternative exon 5′ splice site (Fig. 4C, lane 2). These results lend strong support to our model that In100 acts as a decoy 3′ acceptor site that encumbers the use of exon 9 5′ splice site and thus competitively favors the splicing of exon 8 to exon 10.
Decoy or pseudo splice sites have been implicated in pre-mRNA splicing regulation (11, 34, 35). In the P element third intron splicing, U1 snRNP is recruited to pseudo 5′ splice sites as part of a specific protein complex, thus preventing it from interacting with the normal 5′ splice site (34). Direct U1 snRNP binding to pseudosplice sites has also been reported in other pre-mRNAs, along with the presence of SR proteins in specific complexes (36–38). In the Rous sarcoma virus system, a negative regulator of splicing element can interact with a downstream 3′ splice site (39). In Caenorhabditis, an alternative exon in the U2AF65 pre-mRNA was found to contain multiple copies of the worm 3′ splice site consensus sequence (40). Similar to what we observed in this study, these sites, although seemingly normal, are not used as 3′ splice sites. However, mRNAs containing these sequences are retained in the nucleus (possibly by recruitment of U2AF itself to the alternative exon) where they might play a regulatory role (41). In our case, only one copy of a 3′ splice site consensus sequence is found, and this element is located in the intron downstream of the alternative exon. Nevertheless, it will be interesting to investigate the role of In100 at the cellular level.
Complexes Containing U2 snRNP Assemble on the Decoy 3′ Splice Site.
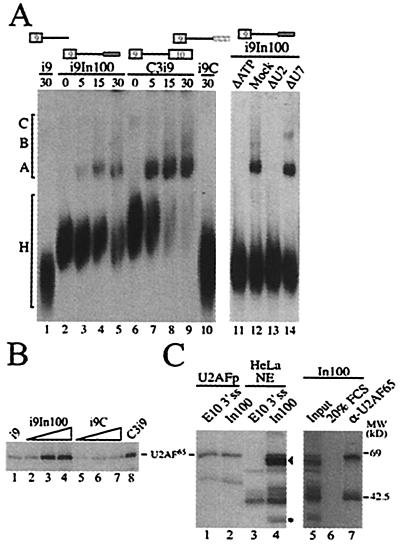
The formation of splicing complexes on In100 was assayed using aliquots of a splicing reaction with the i9In100 substrate. Complexes were detected that had a gel mobility similar to the pre-spliceosomal complexes A and B formed with the complete C3i9 splicing substrate (Fig. 5A, compare lanes 3–5 with lanes 7–9). After 30 min of incubation, the i9In100 substrate showed only a partial conversion of H to spliceosomal complexes, whereas the positive control C3i9 transcript was fully incorporated into spliceosomes at this time point (Fig. 5A, compare lanes 5 and 9, respectively). This suggested that the formation of splicing complexes on i9In100 was less efficient and/or that the complexes were less stable than those formed on a normal 3′ splice site. The formation of these splicing complexes with i9In100 was dependent on the presence of the In100 element because splicing reactions with i9 and i9C control substrates yielded no detectable complexes (Fig. 5A, lanes 1 and 10, respectively). We then tested if the formation of these complexes on i9In100 was dependent on the presence of ATP as well as intact U2 snRNA in the nuclear extract, which are both known to be required for the formation of spliceosomal A complex. As expected, no complexes were observed when i9In100 substrates were incubated in nuclear extracts depleted of ATP (Fig. 5A, lane 11) or depleted of functional U2 snRNP (Fig. 5A, lane 13). The formation of the complexes was not affected if extracts were preincubated in the presence of exogenous ATP or using a nonsplicing-related U7-specific 2′-O-methyl oligoribonucleotide (Fig. 5A; Mock and ΔU7, lanes 12 and 14, respectively). These results clearly indicate that the 3′ acceptor site in In100 can promote the formation of seemingly normal early splicing complexes that require ATP and intact U2 snRNP. In the IgM pre-mRNA, an exonic splicing inhibitor juxtaposed with a splicing enhancer was identified (42). Interestingly, an ATP-dependent complex containing U2 snRNP was also observed on this exonic inhibitory sequence, and the authors proposed that interactions between this complex and U1 snRNP-containing complexes at the 5′ splice site might induce formation of a “dead-end” pre-spliceosome, although biochemical evidence supporting this model has not been reported.
Figure 5.
Analysis of complexes assembled on In100. (A) Gel electrophoresis analysis of splicing complexes formed on the substrates shown above the gels. Heterogeneous (H) and spliceosomal complexes (A–C) are indicated on the left. (B) U2AF65 interacts with the In100 element. UV cross-linking was performed following incubation of labeled RNA substrates with 30, 60, or 120 ng of purified U2AF65; 120 ng of U2AF65 protein was used for i9 and C3i9. Cross-linked proteins were resolved on 12.5% SDS/PAGE. (C) Nuclear factors specifically interact with In100. Labeled RNA substrates containing the In100 or the native 3′ splice site of exon 10 were subjected to UV cross-linking following incubation with purified U2AF65 or HeLa nuclear extracts (lanes 1 and 2 or lanes 3 and 4, respectively). In100 cross-linking reactions were subjected to immunoprecipitation with a monoclonal antibody directed against U2AF65 (lane 7) or FCS (lane 6). The black arrowhead and asterisk indicate positions of specific 65- and 35-kDa cross-linking proteins, as described in the text.
U2AF65 Interacts with the In100 Element.
Initial recognition of the 3′ splice site is mediated through U2AF, which binds tightly to the polypyrimidine track to promote recruitment and stabilization of U2 snRNP to the branch site sequence. We thus tested whether U2AF65 was interacting with the In100 element. 32P-labeled RNA substrates were incubated with purified U2AF65 proteins, and the samples were subjected to UV irradiation. Purified U2AF65 cross-linked to the i9In100 substrate at a level comparable to the one observed on the C3i9 splicing control (Fig. 5B, lanes 2–4 and 8, respectively). Direct immunoprecipitation experiments with α-U2AF65 antibodies also confirmed the interaction in nuclear extracts (data not shown). Only low basal levels of U2AF65 cross-linking were detected for the i9 and i9C substrates, even at the highest protein concentration used (Fig. 5B, lanes 1 and 5–7, respectively). This provides further evidence that In100 has features of a functional 3′ acceptor site even though this site is not used in the presence of the native downstream 3′ splice site.
The In100 sequence (harboring the decoy 3′ splice site) is located 140 nt downstream from the 5′ splice site of the alternative exon, and our experiments show that this distance is sufficient to support splicing. This suggests that the nonproductive nature of the interaction between In100 and the 5′ splice site of exon 9 is not likely due to a mere steric hindrance; instead, this effect may be mediated by trans-acting factors. In this sense, we observe a strong stimulation of exon 9 inclusion when molar excesses of In100 RNA are added to a HeLa splicing reaction with the C2 substrate (data not shown). As a first step toward the identification of nuclear factors interacting with In100, we carried out UV cross-linking with HeLa nuclear extracts. We also compared the cross-linking profile of In100 with the natural 3′ splice site of exon 10 (E10 3′ss substrate). As was observed for the corresponding splicing-competent substrates, purified U2AF65 cross-linked with similar efficiency to the isolated In100 and E10 3′ss RNAs (Fig. 5C, lanes 1 and 2). The UV cross-linking profile obtained when the E10 3′ss was incubated with nuclear extracts consisted of four major cross-linked proteins, all of which were also detectable with the In100 element (Fig. 5C, lanes 3 and 4). One of these products comigrated with the purified U2AF65 band (compare lanes 3 and 4 with lane 1) and was immunoprecipitated with a monoclonal antibody directed against U2AF65 (Fig. 5C, lane 7), confirming its identity. Interestingly, two products of estimated molecular mass of 60 and 35 kDa specifically cross-linked to the In100 sequence (Fig. 5C, compare lanes 3 and 4; indicated by a black rectangle and asterisk, respectively). We have previously reported the involvement of SC35, ASF/SF2, and hnRNP A1 in the regulation of casp-2 alternative splicing (17). However, we have observed that these splicing factors influenced casp-2 pre-mRNA splicing in an In100-independent fashion (data not shown). Several antibodies recognizing known splicing factors in the 30 to 35-kDa range have been tested in the cross-linking/immunoprecipitation assay. However, we have not yet found an antibody that would recognize the 35-kDa protein. Finally, we confirmed the identity of the 60-kDa doublet to be polypyrimidine tract-binding protein (PTB), and detailed characterization of the PTB involvement will be described elsewhere (43).
This study reports on the characterization of cis-elements involved in alternative splicing of the casp-2 pre-mRNA. Our results provide biochemical evidence that cis-elements interacting with U2 snRNP may play a role in the regulation of alternative splicing. Our study also suggests that by mediating nonproductive interactions with a 5′ splice site, a sequence with features of a 3′ splice acceptor site may serve as a splicing repressor. The precise mechanism by which interactions with this decoy 3′ splice site are rendered inefficient for splicing remains unclear and will require further experimentation. Our results demonstrate that casp-2 exon 9 alternative splicing is distinct from previously characterized alternative splicing model systems. It is clear that complex and integrated mechanisms are involved in preventing the constitutive inclusion of exon 9 and thus maintain the balance of anti-apoptotic vs. pro-apoptotic isoforms of casp-2. It will be our future concern to study whether In100 plays a role in developmental and tissue-specific regulation of the casp-2 pre-mRNA alternative splicing.
Acknowledgments
We acknowledge Smitha Rajasekhar and Michael Nolan for their excellent technical assistance. We thank Drs. Marc T. McNally and Woan-Yuh Tarn for their generous gifts of 2′-O-methyl oligos, Dr. Rui-Ming Xu for providing purified U2AF65, and Drs. Gideon Dreyfuss, Maria Carmo-Fonseca, and Adrian Krainer for providing antibodies. We are grateful to Drs. Benoit Chabot, Marco Blanchette, and Douglas L. Black for providing various reagents, helpful discussions, and critical reading of the manuscript. We apologize to the colleagues whose work could not be included in the references because of space limitations. This work was supported by National Institutes of Health grants (to J.Y.W.). J.C. was supported by a postdoctoral fellowship from the Natural Sciences and Engineering Research Council of Canada. J.Y.W. is supported by a Scholarship from the Leukemia Society of America.
Abbreviation
- RT
reverse transcription
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.031564098.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.031564098
References
- 1.Wang J, Manley J L. Curr Opin Genet Dev. 1997;7:205–211. doi: 10.1016/s0959-437x(97)80130-x. [DOI] [PubMed] [Google Scholar]
- 2.Lopez A J. Annu Rev Genet. 1998;32:279–305. doi: 10.1146/annurev.genet.32.1.279. [DOI] [PubMed] [Google Scholar]
- 3.Lavigueur A, La Branche H, Kornblihtt A R, Chabot B. Genes Dev. 1993;7:2405–2417. doi: 10.1101/gad.7.12a.2405. [DOI] [PubMed] [Google Scholar]
- 4.Watakabe A, Tanaka K, Shimura Y. Genes Dev. 1993;7:407–418. doi: 10.1101/gad.7.3.407. [DOI] [PubMed] [Google Scholar]
- 5.Xu R, Teng J, Cooper T A. Mol Cell Biol. 1993;13:3660–3674. doi: 10.1128/mcb.13.6.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Black D L. Cell. 1992;69:795–807. doi: 10.1016/0092-8674(92)90291-j. [DOI] [PubMed] [Google Scholar]
- 7.Huh G S, Hynes R O. Genes Dev. 1994;8:1561–1574. doi: 10.1101/gad.8.13.1561. [DOI] [PubMed] [Google Scholar]
- 8.Ryan K J, Cooper T A. Mol Cell Biol. 1996;16:4014–4023. doi: 10.1128/mcb.16.8.4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Markovtsov V, Nikolic J M, Goldman J A, Turck C W, Chou M Y, Black D L. Mol Cell Biol. 2000;20:7463–7479. doi: 10.1128/mcb.20.20.7463-7479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chew S L, Baginsky L, Eperon I C. Nucleic Acids Res. 2000;28:402–410. doi: 10.1093/nar/28.2.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gontarek R R, McNally M T, Beemon K. Genes Dev. 1993;7:1926–1936. doi: 10.1101/gad.7.10.1926. [DOI] [PubMed] [Google Scholar]
- 12.Chan R C, Black D L. Mol Cell Biol. 1995;15:6377–6385. doi: 10.1128/mcb.15.11.6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang L, Ashiya M, Sherman T G, Grabowski P J. RNA. 1996;2:682–698. [PMC free article] [PubMed] [Google Scholar]
- 14.Carstens R P, McKeehan W L, Garcia-Blanco M A. Mol Cell Biol. 1998;18:2205–2217. doi: 10.1128/mcb.18.4.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L, Miura M, Bergeron L, Zhu H, Yuan J. Cell. 1994;78:739–750. doi: 10.1016/s0092-8674(94)90422-7. [DOI] [PubMed] [Google Scholar]
- 16.Bergeron L, Perez G I, Macdonald G, Shi L, Sun Y, Jurisicova A, Varmuza S, Latham K E, Flaws J A, Salter J C, et al. Genes Dev. 1998;12:1304–1314. doi: 10.1101/gad.12.9.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang Z, Zhang W, Rao Y, Wu J Y. Proc Natl Acad Sci USA. 1998;95:9155–9160. doi: 10.1073/pnas.95.16.9155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar S, Kinoshita M, Dorstyn L, Noda M. Cell Death Differ. 1997;4:378–387. doi: 10.1038/sj.cdd.4400281. [DOI] [PubMed] [Google Scholar]
- 19.Chabot B. In: RNA Processing. Hames D, Higgins S, editors. Vol. 1. Oxford: Oxford Univ. Press; 1994. pp. 1–29. [Google Scholar]
- 20.Dignam J D, Lebovitz R M, Roeder R G. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krainer A R, Maniatis T. Cell. 1985;42:725–736. doi: 10.1016/0092-8674(85)90269-7. [DOI] [PubMed] [Google Scholar]
- 22.Black D L, Chabot B, Steitz J A. Cell. 1985;42:737–750. doi: 10.1016/0092-8674(85)90270-3. [DOI] [PubMed] [Google Scholar]
- 23.Eperon I C, Ireland D C, Smith R A, Mayeda A, Krainer A R. EMBO J. 1993;12:3607–3617. doi: 10.1002/j.1460-2075.1993.tb06034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konarska M M, Sharp P A. Cell. 1986;46:845–855. doi: 10.1016/0092-8674(86)90066-8. [DOI] [PubMed] [Google Scholar]
- 25.Chabot B, Frappier D, La Branche H. Nucleic Acids Res. 1992;20:5197–5204. doi: 10.1093/nar/20.19.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michaud S, Reed R. Genes Dev. 1991;5:2534–2546. doi: 10.1101/gad.5.12b.2534. [DOI] [PubMed] [Google Scholar]
- 27.Reed R, Maniatis T. Cell. 1986;46:681–690. doi: 10.1016/0092-8674(86)90343-0. [DOI] [PubMed] [Google Scholar]
- 28.Dominski Z, Kole R. Mol Cell Biol. 1991;11:6075–6083. doi: 10.1128/mcb.11.12.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blanchette M, Chabot B. EMBO J. 1999;18:1939–1952. doi: 10.1093/emboj/18.7.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu J Y, Maniatis T. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 31.Michaud S, Reed R. Genes Dev. 1993;7:1008–1020. doi: 10.1101/gad.7.6.1008. [DOI] [PubMed] [Google Scholar]
- 32.Mount S M. Nucleic Acids Res. 1982;10:459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Senapathy P, Shapiro M B, Harris N L. Methods Enzymol. 1990;183:252–278. doi: 10.1016/0076-6879(90)83018-5. [DOI] [PubMed] [Google Scholar]
- 34.Siebel C W, Fresco L D, Rio D C. Genes Dev. 1992;6:1386–1401. doi: 10.1101/gad.6.8.1386. [DOI] [PubMed] [Google Scholar]
- 35.Lou H, Yang Y, Cote G J, Berget S M, Gagel R F. Mol Cell Biol. 1995;15:7135–7142. doi: 10.1128/mcb.15.12.7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cook C R, McNally M T. Virology. 1998;242:211–220. doi: 10.1006/viro.1997.8983. [DOI] [PubMed] [Google Scholar]
- 37.Lou H, Neugebauer K M, Gagel R F, Berget S M. Mol Cell Biol. 1998;18:4977–4985. doi: 10.1128/mcb.18.9.4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McNally L M, McNally M T. J Virol. 1999;73:2385–2393. doi: 10.1128/jvi.73.3.2385-2393.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cook C R, McNally M T. J Virol. 1999;73:2394–2400. doi: 10.1128/jvi.73.3.2394-2400.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zorio D A, Lea K, Blumenthal T. Mol Cell Biol. 1997;17:946–953. doi: 10.1128/mcb.17.2.946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.MacMorris M A, Zorio D A, Blumenthal T. Proc Natl Acad Sci USA. 1999;96:3813–3818. doi: 10.1073/pnas.96.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kan J L, Green M R. Genes Dev. 1999;13:462–471. doi: 10.1101/gad.13.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Côté, J., Dupuis, S. & Wu, J. Y. (2001) J. Biol. Chem., in press. [DOI] [PMC free article] [PubMed]