Abstract
A convenient and precise mass spectrometric method for measurement of the deamidation rates of glutaminyl and asparaginyl residues in peptides and proteins has been developed; the rates of deamidation of 306 asparaginyl sequences in model peptides at pH 7.4, 37.0°C, 0.15 M Tris⋅HCl buffer have been determined; a library of 913 amide-containing peptides for use by other investigators in similar studies has been established; and, by means of simultaneous deamidation rate measurements of rabbit muscle aldolase and appropriate model peptides in the same solutions, the use of this method for quantitative measurement of the relative effects of primary, secondary, tertiary, and quaternary protein structure on deamidation rates has been demonstrated. The measured rates are discussed with respect to the hypothesis that glutaminyl and asparaginyl residues serve, through deamidation, as molecular timers of biological events.
Keywords: deamidation, biological clocks, peptides, mass spectrometry
The hypothesis that glutaminyl and asparaginyl residues in peptides and proteins serve, through deamidative transformation into glutamyl and aspartyl residues, as molecular timers of biological events such as protein turnover, development, and aging (1–4) was originally based on the suggestion and then experimental proof that the deamidation of cytochrome c occurs in vivo (5–6) and on reasoning that deamidation is seriously disruptive to biological systems unless it is being used for compensating beneficial biological purposes.
Subsequently, it was shown that the first-order deamidation half times in pH 7.4, 37°C ionic strength 0.15–0.20 phosphate buffer of glutaminyl and asparaginyl residues vary over a range of at least 1 day to 9 years as a function of primary sequence and likely over an even wider range as a function of secondary, tertiary, and quaternary structure (3, 7–11). The dependence of deamidation on pH, temperature, ionic strength, and other solution properties was also demonstrated (3, 12–13). It was additionally shown that the overall in vivo compositions and specific sequence distributions of amide residues in peptides and proteins are supportive of the amide molecular clock hypothesis (1–3, 14).
The first two specific amide clocks to be identified were those that control the in vivo turnover rates of cytochrome c (6, 9) and rabbit muscle aldolase (15–16). Measurements of in vivo turnover rates, in vivo steady-state concentrations, in vivo and in vitro deamidation rates of these proteins, and in vitro deamidation rates of appropriate model peptides demonstrated both sequence dependence and three-dimensional structure dependence of the deamidation of these two proteins. Sequence-controlled deamidation of the C-terminal sequence … Thr-Asn-Glu in cytochrome c apparently leads to a changed three-dimensional structure that accelerates a second deamidation of cytochrome c. The resulting deamidated forms of cytochrome c are rapidly catabolized in vivo with rates equivalent to the turnover rate. In the case of rabbit muscle aldolase, sequence-controlled deamidation of the C-terminal sequence … Ileu-Ser-Asn-His-Ala-Tyr and in vivo turnover proceed at the same rates, the former apparently timing the latter. In both proteins, comparisons to model peptides also showed general suppression of deamidation of other amides in the proteins by means of three-dimensional structure effects. Deamidation of histone peptides was also reported and discussed in terms of development and aging (10). Since this initial work, much progress has been made. See, for example, recent experiments on deamidation of phenylalanine hydroxylase (17), histone (18), crystallin (19), trisphosphate isomerase (20), and ribonuclease (21).
It has been proposed that the deamidation of asparaginyl peptides at neutral pHs proceeds by way of a cyclic five-membered imide formed from the asparaginyl α carbon and side chain and the peptide bond nitrogen of the next residue toward the carboxyl end of the peptide (11, 22). This mechanism explains the production of both aspartyl and isoaspartyl peptides during deamidation, which has been widely observed.
The general approach that was first taken toward understanding the effects of primary, secondary, tertiary, and quaternary peptide and protein structure on deamidation was that of measuring the effects of primary structure in model peptides and then inferring the effects of secondary, tertiary, and quaternary structure by comparison with the deamidation rates of peptides and proteins of interest (2–4, 7–10, 16). Therefore, the deamidation rates of about 60 model peptides and several proteins were determined by means of the labor-intensive techniques available in the 1970s. Although a substantial amount of experimental data, which is beyond the scope of this discussion, has subsequently accumulated concerning deamidation, and new techniques have been used, progress has been impeded by lack of knowledge of the complete library of sequence-controlled deamidation rates and by lack of a fast and accurate means of determining the rates of deamidation of each amide in a protein, with reference to the model peptide rates.
We have, therefore, synthesized, by means of Merrifield solid-phase peptide synthesis (23–24), the entire library of 800 possible nearest-neighbor amide sequences in pentapeptides, which involve the 20 ordinary amino acid residues, and an additional 113 peptides of interest to the effects of residues between 2 and 6 positions distant from the amides in up to 13 residue peptides.
We have devised a means of measuring the rates of deamidation of these peptides by direct loop injections into an ion-trap mass spectrometer. This method permits the determination of an 18-point deamidation curve with an inherent deamidation precision error of less than 1% by means of 18 loop injections requiring about 2 h total mass spectrometer time. Although we have also shown that measuring several peptides of differing molecular weights simultaneously can markedly accelerate these measurements, we prefer to measure the peptides individually except where simultaneous multiple measurements have special value as in work on proteins.
Moreover, we have demonstrated that this method can be used to measure the deamidation rates of specific amides within a protein, while the rates of deamidation of the peptides to be compared with that protein are also simultaneously being measured within the same solution. The model peptides are included with the protein during the rate experiment, and the reaction mixture is digested with an appropriate proteolytic enzyme before direct injection into the mass spectrometer. The mass regions of all of the amide-containing peptides are then simultaneously scanned.
We herein report the deamidation rates of 306 asparaginyl pentapeptides. We also report the deamidation rates of the first two asparaginyl residues near the C terminus of rabbit muscle aldolase along with the rates of simultaneously measured analogous model peptides, which allow the determination of the effects of primary and secondary structure on these aldolase deamidations. All of these rates of deamidation were determined at pH 7.4 in 0.15 M Tris⋅HCl buffer at 37.0°C with peptide and protein concentrations of 1.0 × 10−3 M.
Materials and Methods
Synthesis of Peptides.
The peptides were synthesized by means of Merrifield solid-phase peptide synthesis (23, 24) in an Advanced ChemTech Model 396 MBS synthesizer with a 96-well reaction block. Derivatives used were 9-fluorenylmethoxycarbonyl (Fmoc)-Ala, Fmoc-Arg(2,2,4,6,7-pentamethyldihydrobenzofuran-5-sulfonyl), Fmoc-Asp(O-t-butyl), Fmoc-Asn, Fmoc-Cys(acetamidomethyl), Fmoc-Glu(O-t-butyl), Fmoc-Gln, Fmoc-Gly, Fmoc-His(Trityl), Fmoc-Ile, Fmoc-Leu, Fmoc-Lys(t-butyloxycarbonyl), Fmoc-Met, Fmoc-Phe, Fmoc-Pro, Fmoc-Ser(t-butyl), Fmoc-Thr(t-butyl), Fmoc-Trp, Fmoc-Tyr(t-butyl), Fmoc-Val, and Wang resin substituted with 0.67 meq/g Fmoc-Gly. These were all purchased from Peptides International.
Each well of the synthesizer initially contained 0.1 g of Wang resin. Double couplings for 45 min each with a 3-min 1.5-ml wash of 50:50 N-methylpyrrolidone (NMP)/dimethylformamide (DMF) between couplings were used. Reagents for each coupling were 0.5 ml of 0.5 M derivative in NMP; 0.5 ml of coupling reagent that was 0.5 M 2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate and 0.5 M 1-hydroxybenzotriazole in DMF; 0.25 ml of neutralizer that was 2 M N,N-diisopropylethylamine in NMP; and 0.25 ml of DMF. Double deblocking with 1.5 ml of 20% piperidine in DMF was carried out once for 5 min, followed by a second 15-min deblocking. Resin and side-chain protecting groups were removed with 1.5 ml of scavenger-containing trifluoroacetic acid (TFA) solution at room temperature for 2 h. The TFA solution was TFA/anisole/ethylmethylsulfide/ethanedithiol in the proportions 93:3:3:1, respectively, by volume. The product was filtered and the resin washed once with 1 ml of TFA. Eleven separate syntheses were performed, with 96 peptides synthesized simultaneously in most cases.
The peptides were precipitated and washed three times with 15 ml of methyl-t-butyl ether, vacuum dried, dissolved in 18.2 MΩ distilled and purified H2O, divided into five parts in low-temperature vials, freeze dried, and stored at −80°C.
The acetamidomethyl-blocking groups of cystine were approximately 20% removed in TFA. Deamidation rates of both the blocked and unblocked peptides were measured by mass spectrometry. Deamidation rates for the blocked peptides will be reported elsewhere.
Yields were determined by hydrolysis with 6 N HCl in H2O at 95°C for 48 h followed by amino acid analysis in a Thermoquest HPLC with a P4000 pump, AS3000 autosampler, and UV6000LP detector combined with a Pickering PCX5200 derivatizer (Pickering Laboratories, Mountain View, CA), column, ninhydrin, and lithium buffer system. Yields averaged about 70% with the remaining peptide either not washed from the resin or solubilized during the ether washes. In most cases, impurities in the peptides were so minimal as to be indistinguishable from background in the mass spectrometer.
Deamidation of Peptides.
Peptide deamidation reactions were carried out in pH 7.4, 37.0°C, 0.15 M Tris⋅HCl buffer. Peptide concentrations were 1.0 × 10−3 M. These reactions were carried out in 1.5-ml polypropylene centrifuge vials with screw caps and rubber O-rings. The vials were also sealed in trays in a hydrated environment to prevent evaporation. Half of the vials in each experiment, alternating in reaction times, were fitted with pressed-in 0.002-inch-thick Teflon film liners and tops. Each vial contained 100 μl of solution.
The deamidation reactions took place in a specially constructed incubation chamber controlled at 37.00 ± 0.02°C throughout the 3-month period of these experiments. This chamber was continuously monitored by means of three thermisters and one glass thermometer; one of the thermisters and the thermometer were calibrated to ASTM standards.
Incubation times varied between 4 and 102 days. These were adjusted to provide measured points over a range of at least one-fourth and at most six deamidation half times for each individual peptide or protein. At timed intervals, vials were removed from the incubator and frozen at −80°C. All portions for an individual rate were measured in a single set of mass spectrometry runs with the analyses alternated to eliminate drift errors during the course of the mass spectrometry.
In the case of aldolase, ICN rabbit muscle aldolase precipitated from 6 M ammonium sulfate was centrifuged, dissolved in H2O, and diluted to 1.0 × 10−3 M. The solution also contained the peptides Gly-Ser-Asn-His-Gly and Gly-Ala-Asn-Ser-Gly at 1.0 × 10−3 M and was 0.015 M in pH 7.4 Tris⋅HCl buffer. This solution was divided into 50-μl portions in polypropylene vials, placed at 37.0°C, removed from the incubator at 14 1-day intervals, and frozen at −80°C. Before mass spectrometric analysis, 3-μl portions of the reaction mixtures were each mixed with 16 μl of a 0.5 mg/ml solution of Sigma T-1426 TPCK treated trypsin or ICN 100478 α-chymotrypsin respectively. The trypsin mixtures were incubated at 37.0°C for 2 hours, and the chymotrypsin mixtures were incubated at 25°C for 10 min before dilution for mass spectrometry. The tryptic and chymotryptic digests were injected separately into the mass spectrometer, so 28 loop injections were performed. Trypsin produced the Ala-Leu-Ala-Asn-Ser-Leu-Cys-Gln-Gly-Lys peptide and chymotrypsin the Ileu-Ser-Asn-His-Ala-Tyr peptide.
Measurement of Deamidation by Mass Spectrometry.
Measurements were made in a Thermoquest LCQ mass spectrometer fitted with a Thermoquest electrospray source, a Thermoquest AS3500 autosampler, and a customized sample delivery system driven by a Harvard Apparatus PHD2000 syringe pump with 5- and 10-ml Hamilton glass syringes. The autosampler had a Teflon-coated needle assembly, a Tefzel valve rotor, and a PEEK valve body. The LCQ was powered at 240 V by four Exeltech MX1000 power modules connected to a battery bank serviced by two Lorain Flotrol A100F25 rectifiers (Lorain Corporation, Lorain, OH).
The 5-ml syringe contained 1.5% acetic acid in acetone, and the 10-ml syringe contained purified H2O. These were pumped to give a combined flow of 40 μl/minute of solution that was 0.5% acetic acid and 33% acetone into the electrospray source. The heated capillary was at 180°C. The solutions were combined, after sample introduction into the H2O stream, in a U.466 Upchurch PEEK static mixing T (Upchurch Scientific, Oak Harbor, WA). Each run consisted of a 50-μl loop injection of sample, which was monitored continuously for 5–7 min by a sequence of 4 high-resolution zoom scans centered on the expected charge/mass ratios and one 100–2,000 charge/mass low-resolution scan. Approximately two zoom scans per microliter of sample were recorded. Meticulous attention was paid to the materials in contact with the sample, and the hold-up volume and surface area of the sample delivery system were minimized. Teflon and Tefzel were used when available.
Before analysis, the samples were diluted 500-fold with H2O by pipetting of 1.5 μl into 749 μl in the injection vials. In the case of proteins and larger peptides, H2O was substituted for acetone in the injection system to avoid precipitation. All solutions throughout these experiments were handled in Teflon or polypropylene containers. No glass containers were used except for the syringe pump syringes. The aldolase samples were diluted only 50-fold before analysis; 75-μl injections were used; data were collected for 8 min per sample; and the LCQ automatic gain control was turned off to avoid suppression of detector sensitivity by the large amounts of proteolytic enzyme and aldolase fragments in the sample. The larger samples offset the lesser data collected per peptide, because each of four peptides was being scanned simultaneously.
A typical 4-h mass spectrometry run consisted of 36 samples, with 18 from each of 2 peptide rate experiments. These included peptides of different masses, and the samples were alternated to mutually wash the sample delivery system. We also carried out experiments wherein several peptides of differing mass were simultaneously measured. The throughput of these procedures can be increased by 10- to 20-fold by such combined measurements and/or by combined peptide syntheses of appropriately selected peptide sets. Precision, however, is compromised as the number of peptides rises. As we wished to keep precision error in these experiments below 1%, we did not combine peptides for the experiments reported here, except in the case of the protein experiments.
Calculation of Results.
Raw data from all of the sample-containing zoom scans from each LCQ run were summed, averaged, and recorded in Microsoft excel tables to give results as shown in Figs. 1 and 2. The point-by-point values of these curves were then corrected for baseline noise, adjusted for the naturally occurring isotopic ratios, and separated into the contributions from the undeamidated and deamidated peptides, which differ from each other in that deamidation increases the mass by 1 atomic mass unit. The rates of deamidation were then calculated and plotted as first-order rate curves as shown in Fig. 3. These calculations were performed in MATHCAD 8 PROFESSIONAL (Math Soft, Cambridge, MA) with overlays from MACRO EXPRESS 2000 (Insight Software Solutions, Bountiful, VT) that permit large numbers of calculations to be made sequentially and automatically. In the course of this work, we have measured and calculated about 500 deamidation rates involving 9,000 loop injections and requiring a total of about 1,000 h of mass spectrometer operation. With the aid of our computer programs, each 4 h of mass spectrometer operation requires about 30 min of manual computer manipulations to calculate the results, largely because our programs are not integrated into the LCQ software.
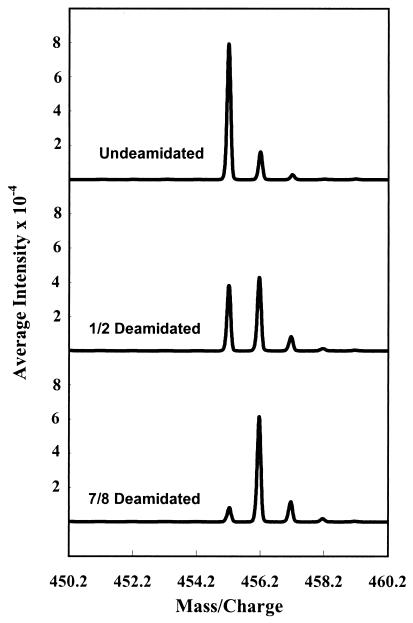
Figure 1.
Representative averaged zoom scans from direct 50-μl loop injection of the peptide Gly-Ala-Asn-His-Gly into the LCQ mass spectrometer. Scans are from the first, sixth, and fifteenth points of the deamidation experiment. Deamidation solutions were 1.0 × 10−3 M peptide, 0.15 M Tris⋅HCl, pH 7.4, and 37.0°C. Injected solution was 2 × 10−6 M peptide and 3 × 10−4 M Tris⋅HCl buffer. The graphs shown are actual and typical unsmoothed experimental data. The graphical base lines have been omitted to show the quality of these results.
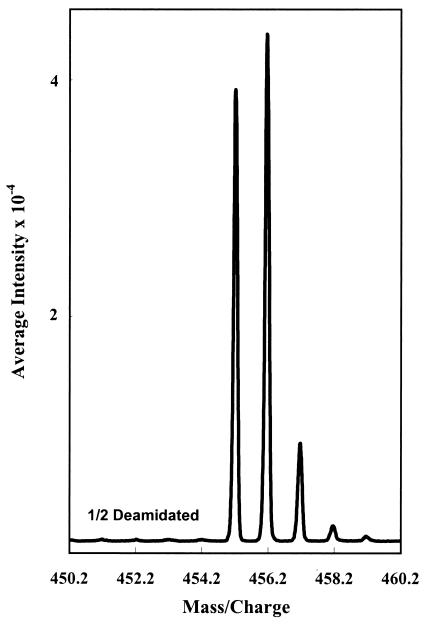
Figure 2.
Unsmoothed and uncorrected experimental data for averaged zoom scans of the sixth deamidation point in the deamidation of Gly-Ala-Asn-His-Gly. This graph is typical of actual experimental data obtained for the 306 peptides in these experiments.
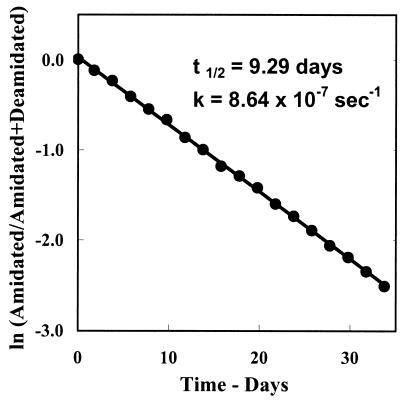
Figure 3.
First-order rate plot of the deamidation of the peptide Gly-Ala-Asn-His-Gly. Deamidation half time for the 1.0 × 10−3 M peptide in 0.15 M Tris⋅HCl, at pH 7.4, and 37.0°C was calculated to be 9.3 days, and the first-order rate constant k = 0.86 × 10−6⋅sec−1.
Results and Discussion.
The results are summarized in Tables 1 and 2. The first-order deamidation half times for the 306 asparaginyl peptides are between 1 day and 455 days with a distribution function as shown in Fig. 4. Clearly, the side chain of the amino acid residue on the carboxyl side of the asparaginyl residue has a larger effect on the deamidation rate than does the residue on the amino side. The residues on both sides affect the deamidation rate in an ordered way that is explainable from their structures and the proposed cyclic imide mechanism. Table 1 reports deamidation rates for amide-side Pro, but not for carboxyl-side Pro. For all 18 peptides with carboxyl-side Pro, our measurements showed deamidation half times of more than 1,000 days. Because Pro cannot participate in the five-membered imide ring, this result would be expected. The duration of our experiments did not, however, allow the careful and precise measurement of these rates, so the values will be reported later after they have been investigated more carefully.
Table 1.
First-order deamidation half times of pentapeptides GlyXxxAsnYyyGly in days at pH 7.4, 37.0°C, 0.15 M Tris⋅HCl
| Yyy
|
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gly | His | Ser | Ala | Asp | Thr | Cys | Lys | Met | Glu | Arg | Phe | Tyr | Trp | Leu | Val | Ile | Mean | Median | |
| Xxx | |||||||||||||||||||
| Gly | 1.03 | 9.2 | 11.8 | 21.1 | 28.0 | 39.8 | 40.6 | 48.2 | 50.4 | 73.9 | 57.8 | 64.0 | 63.6 | 77.1 | 104 | 224 | 287 | 70.6 | 50.4 |
| Ser | 0.96 | 8.3 | 15.1 | 24.1 | 30.3 | 45.7 | 60.2 | 55.5 | 54.9 | 59.7 | 59.7 | 52.2 | 64.7 | 76.8 | 110 | 233 | 285 | 72.7 | 55.5 |
| Thr | 1.04 | 9.6 | 17.1 | 24.6 | 27.9 | 50.0 | 55.5 | 57.6 | 47.6 | 60.8 | 51.2 | 76.4 | 80.6 | 72.5 | 110 | 237 | 279 | 74.0 | 55.5 |
| Cys | 1.14 | 10.8 | 19.0 | 26.4 | 30.6 | 48.7 | 46.0 | 46.6 | 64.5 | 48.3 | 83.1 | 73.9 | 83.9 | 111 | 119 | 229 | 304 | 79.1 | 48.7 |
| Met | 1.04 | 10.2 | 15.2 | 22.1 | 26.4 | 43.6 | 49.6 | 60.4 | 56.9 | 72.4 | 58.8 | 61.9 | 74.0 | 92.7 | 113 | 211 | 275 | 73.2 | 58.8 |
| Phe | 1.15 | 10.2 | 18.1 | 24.2 | 27.4 | 39.0 | 46.5 | 58.2 | 58.6 | 62.4 | 61.2 | 69.5 | 75.1 | 102 | 118 | 203 | 287 | 74.2 | 58.6 |
| Tyr | 1.49 | 10.2 | 11.9 | 24.3 | 28.4 | 38.1 | 48.6 | 55.1 | 64.3 | 41.0 | 56.9 | 58.0 | 70.6 | 120 | 118 | 241 | 306 | 76.1 | 55.1 |
| Asp | 1.53 | 9.7 | 17.0 | 24.0 | 29.4 | 52.4 | 54.1 | 75.9 | 57.3 | 46.8 | 87.2 | 70.1 | 70.4 | 80.3 | 111 | 241 | 298 | 78.0 | 57.3 |
| Glu | 1.45 | 9.0 | 16.4 | 25.8 | 32.0 | 36.8 | 44.2 | 77.8 | 59.6 | 60.3 | 80.9 | 70.2 | 94.5 | 98.4 | 130 | 268 | 279 | 81.4 | 60.3 |
| His | 1.14 | 10.7 | 15.7 | 24.6 | 31.2 | 47.2 | 43.9 | 50.2 | 63.1 | 69.4 | 48.9 | 72.1 | 82.3 | 95.4 | 116 | 247 | 327 | 79.2 | 50.2 |
| Lys | 1.02 | 10.5 | 15.6 | 23.6 | 34.0 | 58.1 | 49.0 | 53.5 | 60.9 | 72.5 | 57.4 | 70.1 | 96.7 | 98.1 | 119 | 246 | 313 | 81.1 | 58.1 |
| Arg | 1.00 | 10.0 | 14.3 | 24.4 | 34.7 | 50.7 | 50.5 | 49.6 | 74.4 | 68.3 | 67.4 | 68.3 | 90.0 | 127 | 128 | 247 | 311 | 83.4 | 67.4 |
| Ala | 1.05 | 9.3 | 14.9 | 22.5 | 31.9 | 43.5 | 63.7 | 55.9 | 59.2 | 74.1 | 62.4 | 65.6 | 73.9 | 130 | 124 | 254 | 300 | 81.5 | 62.4 |
| Leu | 1.08 | 10.7 | 16.7 | 25.1 | 32.1 | 46.1 | 53.5 | 60.1 | 62.6 | 56.7 | 62.1 | 72.4 | 75.7 | 74.5 | 155 | 294 | 391 | 87.6 | 60.1 |
| Val | 1.23 | 10.2 | 18.2 | 27.5 | 33.5 | 49.9 | 63.2 | 63.8 | 65.7 | 64.8 | 67.4 | 66.6 | 79.2 | 88.9 | 154 | 291 | 366 | 88.9 | 64.8 |
| Ile | 1.26 | 11.5 | 14.5 | 25.9 | 33.8 | 46.3 | 52.7 | 64.4 | 58.8 | 58.6 | 66.4 | 61.5 | 79.3 | 86.7 | 154 | 295 | 384 | 87.9 | 58.8 |
| Trp | 1.75 | 11.3 | 15.5 | 30.7 | 43.6 | 38.9 | 83.1 | 59.4 | 64.2 | 75.7 | 73.9 | 71.1 | 92.6 | 135 | 133 | 226 | 286 | 84.8 | 71.1 |
| Pro | 1.18 | 12.8 | 18.9 | 31.8 | 48.6 | 63.1 | 60.0 | 67.8 | 78.4 | 92.0 | 72.9 | 100 | 114 | 122 | 181 | 364 | 455 | 111 | 72.9 |
| Mean | 1.20 | 10.2 | 15.9 | 25.1 | 32.4 | 46.5 | 53.6 | 58.9 | 61.2 | 64.3 | 65.3 | 69.1 | 81.2 | 99 | 128 | 253 | 318 | 81.4 | 61.2 |
| SD | 0.05 | 0.25 | 0.50 | 0.65 | 1.4 | 1.7 | 2.4 | 2.1 | 1.8 | 2.9 | 2.6 | 2.4 | 3.1 | 5.0 | 5.0 | 9.3 | 11.9 | 3.1 | 2.4 |
| %SD | 4.5 | 2.4 | 3.2 | 2.6 | 4.2 | 3.6 | 4.5 | 3.6 | 2.9 | 4.6 | 4.0 | 3.4 | 3.8 | 5.0 | 3.9 | 3.7 | 3.7 | 3.7 | 3.7 |
| Median | 1.14 | 10.2 | 15.7 | 24.5 | 31.6 | 46.2 | 51.6 | 57.9 | 60.3 | 63.6 | 62.3 | 69.8 | 79.3 | 97 | 119 | 243 | 302 | 78.5 | 60.3 |
Table 2.
Deamidation of rabbit muscle aldolase and model peptides in 1.0 × 10−3 M peptide or protein, 0.15 M Tris⋅HCl, pH 7.4, and 37.0°C in the same solution
| Peptide | t1/2 days | k × 106 sec−1 |
|---|---|---|
| Aldolase–IleuSerAsnHisAlaTyr | 9.4 | 0.85 |
| GlySerAsnHisGly | 8.3 | 0.97 |
| Aldolase–AlaLeuAlaAsnSerLeuCysGlnGlyLys | More than 150 days | |
| GlyAlaAsnSerGly | 11.4 | 0.70 |
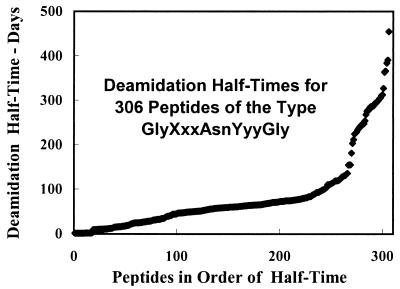
Figure 4.
Distribution function of deamidation half times for the 306 asparaginyl pentapeptides of the type Gly-Xxx-Asn-Yyy-Gly listed in Table 1. Half times are for 1.0 × 10−3 M peptide in 0.15 M Tris⋅HCl, pH 7.4, and 37.0°C.
The precision error of the values reported in Table 1 is generally less than 1%. Systematic errors, however, are also present. In some cases, the amidated and deamidated peptides were differentially retarded and slightly absorbed in the sample delivery system. Usually, the more positively charged peptides were absorbed more strongly, so the undeamidated peptides were absorbed more extensively than were the deamidated products. For this reason, we estimate that the absolute values of these deamidation rates have probable errors of about 5% or less.
At pH 7.4, these peptides are not homogeneous molecular species. For example, the pKs of the carboxyl, imidazole, and amino groups in the peptide Gly-Ser-Asn-His-Gly are 3.1, 6.4, and 7.8, respectively (25). Therefore, whereas the carboxyl and imidazole groups are mostly deprotonated at pH 7.4, the amino group, to a significant extent, is an equilibrium mixture of the protonated and deprotonated species, which have different but similar deamidation rates (25). Some of the variability seen in Table 1 is, therefore, likely to be the result of differences in pK of the peptides, but most of the variation arises from peptide sequence.
We have chosen Tris⋅HCl buffer for these experiments, because it is relatively benign in its direct effects on deamidation rate as compared, for example, to phosphate (3, 12). As these studies are expanded, the Tris⋅HCl rates will serve as a base to which the effects of other solute molecules can be added. Of central importance, however, is the comparison of peptide deamidation rates with those of proteins with corresponding sequences under identical solvent and environmental conditions. For this reason, the results reported in Table 2 are of special importance.
Table 2 shows that, as expected from previous work (15–16), the C-terminal Ser-Asn-His sequence in aldolase deamidates at essentially the same rate as does its corresponding model peptide—which was present in the same solution at the same time and was simultaneously measured in the mass spectrometer. The Tris⋅HCl value of 8.3 days for the deamidation of Gly-Ser-Asn-His-Gly, as compared with that for aldolase of 9.4 days, is in good agreement with that previously reported of 6.4 days for Gly-Ser-Asn-His-Gly in phosphate buffer (16) and aldolase of 8 days (15). Phosphate buffer is known to accelerate deamidation (12). It is also in good agreement with the reported in vivo turnover rate for rabbit muscle aldolase of 8 days (15).
Conversely, however, the second amide sequence from the C-terminal, Ala-Asn-Ser, deamidated with a half life of 11.4 days in the peptide but was not observed to deamidate at all in the protein during the 14-day duration of the experiment. In the protein, therefore, this sequence has a deamidation rate at least 10- to 20-fold slower than the model. This difference is likely because this amide is located in an α helix in the protein. The postulated cyclic imide intermediate involved in the deamidation of these asparaginyl peptides requires that the amide nitrogen of the residue toward the carboxyl end of the asparaginyl residue be available for imide formation. This would require disruption of the α helix in which this nitrogen is participating.
We suggest that strategically located asparaginyl residues may serve as useful quantitative detectors of helix formation in model peptide and other systems. This deamidation rate is very sensitive to helix and is easily measurable. Helix dependence of deamidation has also been reported in several model peptides (25).
We are convinced that this method for deamidation rate measurement of particular sequences in proteins, which we have demonstrated here with these two sequences in aldolase, will prove robust. Preliminary experiments that we have performed with several other proteins have shown that the majority of their amide-containing peptides are immediately measurable. Where disulfide bonds are present, it is helpful to include 1 μl of 0.1 M 1,4-dithiothreitol in the enzymatic digestion mixtures. Chromatographic and electrophoretic separations should be avoided entirely because they are time consuming and because they introduce the possibility of large systematic errors from differential losses of the amidated and deamidated peptides during measurement.
Studies of a wide variety of model peptides have shown that pentapeptide models of the sort summarized in Table 1 are good surrogates for longer peptides and proteins of similar sequences, with respect to primary structure effects on deamidation (25). It is evident that the deamidation rates of these asparaginyl pentapeptides at 37.0°C in pH 7.4, 0.15 M Tris⋅HCl buffer depend primarily on the adjacent carboxyl-side residue, as would be expected from the cyclic imide model.
For carboxyl-side residues without specific functionality, the order of deamidation rates is Gly, Ala, Leu, Val, and Ileu, entirely as expected from steric hindrance. Phe, Tyr, and Trp are between Ala and Leu and likely also contribute primarily steric effects. The series Ser, Thr, Cys, and Met follows the order of the dipole moments of the functional groups, which are geometrically available to stabilize imide formation as is the imidazole group of His. The Asp side chain is short enough for it, too, to contribute significantly to deamidation, but the longer Glu, Lys, and Arg side chains are inhibited by their length.
The amino side residues are also listed in Table 1 in order of their deamidation-enhancing effect. Although a smaller effect, this is easily distinguishable. This listing is approximately in the order of number of peptide rates above and below the medians, with allowances made for special structures. The order found is qualitatively similar to that of the carboxyl side.
Some special effects are notable. For example, peptides with paired nearest-neighboring basic and acidic residues, Glu-Asn-Lys, Glu-Asn-Arg, Asp-Asn-Lys, and Asp-Asn-Arg, clearly stand out as having deamidation half times about 50% higher than the similar singular Glu, Asp, Lys, and Arg analogues. In contrast, this same effect can be distinguished for Lys-Asn-Glu, Arg-Asn-Glu, Lys-Asn-Asp, and Arg-Asn-Asp, but it raises deamidation half times in these peptides only by about 10%. Detailed discussion and mechanistic considerations of the deamidation rates reported in Table 1 is beyond the scope of this report and will be discussed elsewhere.
The deamidation rates in Table 1 serve as a basis for the design of model peptides appropriate for the separation of primary, secondary, tertiary, and quaternary structure effects in experiments on proteins. Used in combination with the protein deamidation measurement technique exemplified in Table 2, they are suitable for the evaluation of nearest-neighbor sequence effects. Effects have also been observed from functional groups farther removed from Asn and from chain elongation (25). We are now maintaining a supply of the 800 possible Gly-Xxx-Asn-Yyy-Gly and Gly-Xxx-Gln-Yyy-Gly peptides and 113 other amide-containing peptides in our laboratory and are interested in making them available without cost to investigators wishing to make use of these peptide models in the comparative evaluation of deamidation of specific proteins.
Conclusions
The values reported in Table 1 strengthen the hypothesis that deamidations of peptide and protein amides serve as ubiquitous and versatile clocks for the timing of biological events. A necessary condition of this hypothesis is that amide clocks be available that are suitable for the timing of events throughout the useful range of biological processes. Although the evidence for this was good (3) on the basis of the approximately 30 asparaginyl peptides and 30 glutaminyl peptides previously synthesized and measured, it is far more robust now that a complete set of nearest-neighbor rates is becoming available. Fig. 4 shows that an abundance of nearest-neighbor asparaginyl sequences are available between deamidation half times of 1 day and 1 year. Glutaminyl sequences extend this range to more than 10 years (3).
Therefore, near-neighbor variations in combination with sequence effects of residues farther away in the peptide chain (25) and three-dimensional effects from secondary, tertiary, and quaternary structure surely provide a versatile set of deamidation rates suitable to the durations of most biological processes.
There is an additional possibility regarding protein alteration and turnover. Amides may serve as molecular indicators that the integrity of each protein molecule has been maintained. Because deamidation rates are suppressed by three-dimensional structures that impede the deamidation reaction, amides contained in these structures may serve as indicators that the structures are still intact. If an individual molecule is altered, deamidation within the altered region may then set off catabolism of that molecule or some other corrective process, or the altered region may deamidate to form a peptide or protein with a new function.
It is likely that the lifetime of a set of protein molecules is determined at the time of synthesis, perhaps through deamidation, as a useful compromise between the time it is expected to remain intact and useful to the organism and the effort that will be required to resynthesize it. For some proteins, of course, the time will be adjusted to remove the protein after it has completed its work because its continued existence would be harmful. Molecular clocks are needed, rather than molecule-by-molecule evaluation of proteins within the living system, because the wide variety of forms arising as the molecules age would be so great as to overwhelm even the versatility of a well-developed immune system. In addition to preprogrammed obsolescence, however, the possibility that amide evaluators in specific regions of each molecule may be keeping watch over the integrity of their regions should be considered.
Moreover, in some cases, deamidated peptides and proteins may have special biological functions wherein timed deamidation controls their delivery to the biological system. In these instances, which could control ordinary homeostatic functions or those related to timed processes such as development or aging, deamidation would serve as a functional molecular clock rather than a terminal marking clock.
Only a few biological events have been definitively shown to be regulated by amide molecular clocks. This is not surprising, because such demonstrations have, so far, required laborious and sophisticated experimentation. The original reasoning from which this hypothesis arose about 30 years ago (1–4) remains intact and is as follows.
The static properties of asparaginyl and glutaminyl residues are not unique and can be easily duplicated by some of the other 18 commonly occurring amino acid residues or others that might be used. The disruptive effect on peptide and protein structure from deamidation reactions, which introduce postsynthetic charged residues, is so great that, were they not especially useful, these amide residues would not be widely present in peptides and proteins. Therefore, it is likely that the instability of asparaginyl and glutaminyl residues is their primary biological function, and that they serve as easily programmable molecular clocks for the timing of biological processes (1–4).
We thank Professor and Mrs. R. B. Merrifield, in whose laboratory and with whose help we synthesized the peptides, and Professor Brian Chait for his advice and counsel concerning the mass spectrometry. We also thank the John Kinsman Foundation and other donors to the Oregon Institute of Science and Medicine for financial support.
Abbreviations
- DMF
dimethylformamide
- TFA
trifluoroacetic acid
- Fmoc
9-fluorenylmethoxycarbonyl
References
- 1.Robinson A B, McKerrow J H, Cary P. Proc Natl Acad Sci USA. 1970;66:753–757. doi: 10.1073/pnas.66.3.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robinson A B. Proc Natl Acad Sci USA. 1974;71:885–888. doi: 10.1073/pnas.71.3.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robinson A B, Rudd C. Curr Top Cell Regul. 1974;8:247–295. doi: 10.1016/b978-0-12-152808-9.50013-4. [DOI] [PubMed] [Google Scholar]
- 4.Robinson A B. Mech Ageing Dev. 1978;9:225–236. doi: 10.1016/0047-6374(79)90101-5. [DOI] [PubMed] [Google Scholar]
- 5.Flatmark T. Acta Chem Scand. 1964;18:1656–1666. [Google Scholar]
- 6.Flatmark T, Sletten K. J Biol Chem. 1968;243:1623–1629. [PubMed] [Google Scholar]
- 7.Robinson A B, Scotchler J W, McKerrow J H. J Am Chem Soc. 1973;95:8156–8159. doi: 10.1021/ja00805a032. [DOI] [PubMed] [Google Scholar]
- 8.Robinson A B, Tedro S. Int J Pept Protein Res. 1973;5:275–278. doi: 10.1111/j.1399-3011.1973.tb03461.x. [DOI] [PubMed] [Google Scholar]
- 9.Robinson A B, McKerrow J H, Legaz M. Int J Pept Protein Res. 1974;6:31–35. doi: 10.1111/j.1399-3011.1974.tb02355.x. [DOI] [PubMed] [Google Scholar]
- 10.Robinson A B, Scotchler J W. Int J Pept Protein Res. 1974;6:279–282. doi: 10.1111/j.1399-3011.1974.tb02385.x. [DOI] [PubMed] [Google Scholar]
- 11.Geiger T, Clarke S. J Biol Chem. 1987;262:785–794. [PubMed] [Google Scholar]
- 12.Scotchler J W, Robinson A B. Anal Biochem. 1974;59:319–322. doi: 10.1016/0003-2697(74)90040-2. [DOI] [PubMed] [Google Scholar]
- 13.Robinson A B, Scotchler J W. J Int Res Commun. 1973;1–8:8. [Google Scholar]
- 14.Robinson A B, Robinson L R. Proc Natl Acad Sci USA. 1991;88:8880–8884. doi: 10.1073/pnas.88.20.8880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Midelfort C F, Mehler A H. Proc Natl Acad Sci USA. 1972;69:1816–1819. doi: 10.1073/pnas.69.7.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKerrow J H, Robinson A B. Science. 1974;183:85. doi: 10.1126/science.183.4120.85. [DOI] [PubMed] [Google Scholar]
- 17.Solstad T, Flatmark T. Eur J Biochem. 2000;267:6302–6310. doi: 10.1046/j.1432-1327.2000.01715.x. [DOI] [PubMed] [Google Scholar]
- 18.Lindner H, Sarg B, Hoertnagl B, Helliger W. J Biol Chem. 1998;273:13324–13330. doi: 10.1074/jbc.273.21.13324. [DOI] [PubMed] [Google Scholar]
- 19.Takemoto L, Boyle D. J Biol Chem. 2000;275:26109–26112. doi: 10.1074/jbc.M002809200. [DOI] [PubMed] [Google Scholar]
- 20.Sun A, Yuksel K U, Gracy R W. Arch Biochem Biophys. 1995;322:361–368. doi: 10.1006/abbi.1995.1476. [DOI] [PubMed] [Google Scholar]
- 21.Capasso S, Salvadori S. J Peptide Res. 1999;54:377–382. doi: 10.1034/j.1399-3011.1999.00111.x. [DOI] [PubMed] [Google Scholar]
- 22.Clarke S. Int J Pept Protein Res. 1987;30:808–821. doi: 10.1111/j.1399-3011.1987.tb03390.x. [DOI] [PubMed] [Google Scholar]
- 23.Merrifield R B. J Am Chem Soc. 1963;85:2149–2154. [Google Scholar]
- 24.Merrifield R B. In: Peptides: Synthesis, Structures, and Applications. Gutte B, editor. New York: Academic; 1995. pp. 94–169. [Google Scholar]
- 25.Robinson, N. E., Robinson, A. B. & Merrifield, R. B. (2001) J. Pept. Protein Res., in press.