Abstract
Smad proteins are key intracellular signaling effectors for the transforming growth factor-β superfamily of peptide growth factors. Following receptor-induced activation, Smads move into the nucleus to activate transcription of a select set of target genes. The activity of Smad proteins must be tightly regulated to exert the biological effects of different ligands in a timely manner. Here, we report the identification of Smurf2, a new member of the Hect family of E3 ubiquitin ligases. Smurf2 selectively interacts with receptor-regulated Smads and preferentially targets Smad1 for ubiquitination and proteasome-mediated degradation. At higher expression levels, Smurf2 also decreases the protein levels of Smad2, but not Smad3. In Xenopus embryos, ectopic Smurf2 expression specifically inhibits Smad1 responses and thereby affects embryonic patterning by bone morphogenetic protein signals. These findings suggest that Smurf2 may regulate the competence of a cell to respond to transforming growth factor-β/bone morphogenetic protein signaling through a distinct degradation pathway that is similar to, yet independent of, Smurf1.
Members of the transforming growth factor-β (TGF-β) family of peptide growth factors, which include TGF-β, bone morphogenetic proteins (BMPs), and activins, regulate a broad range of cellular processes from cell growth and differentiation to apoptosis (1–3). They also serve as inductive signals during development to direct cell fate determination and tissue patterning (4, 5). The signaling responses to TGF-β and other family members are mediated by a heteromeric complex of two types of transmembrane serine/threonine kinase receptors at the cell surface and their intracellular substrates, the Smad proteins (2, 3, 6, 7). Following ligand binding, the type II receptor kinases phosphorylate and thereby activate the type I receptor cytoplasmic domains. The Smads then act as type I receptor-activated signaling effectors, which, following receptor-induced phosphorylation, move into the nucleus to activate transcription of a select set of target genes (8, 9).
The structurally related Smad proteins can be divided into three classes, based on their sequences and functions. The first class is the receptor-regulated Smads. These Smads are phosphorylated by activated receptors at their C-terminal SSXS sequence and dictate the nature of the receptor-induced responses. Smad1, Smad5, and Smad8 are phosphorylated by the activated BMP receptors and mediate BMP responses, whereas Smad2 and Smad3 are activated by activin and TGF-β receptors. While these two groups of Smads each have distinct target genes, they can also antagonize each other's responses, and this may explain some mutually exclusive BMP and activin responses. Once activated, these receptor-regulated Smads associate with a second class of Smads, the “common mediator” Smad (i.e., Smad4 in vertebrates). Smad4 thus participates in the different Smad signaling pathways. A third class of Smads acts as antagonists of these signaling pathways. Among them, Smad6 preferentially inhibits BMP signaling, whereas Smad7 preferentially inhibits activin and TGF-β signaling. When overexpressed, Smad6 and Smad7 can interact with various type I receptors and nonselectively inhibit signaling by various members of the TGF-β superfamily (10–12).
The activities of Smad proteins are regulated at both the transcriptional and posttranslational levels, thereby allowing alterations in the biological effects of Smads (10, 11, 13). Several recent reports revealed that Smads undergo ubiquitin–proteasome-mediated degradation (14, 15). This process of regulated degradation has been implicated in a variety of cellular responses such as the heat shock response, cell cycle progression, DNA repair, signal transduction, and transcription (16). It is now understood that protein ubiquitination is carried out by a sequence of three enzymes, an E1 ubiquitin-activating enzyme, E2 ubiquitin-conjugating enzymes, and E3 ubiquitin ligases. Among these, E3 ubiquitin ligases play a crucial role in defining substrate specificity and subsequent protein degradation by the 26S proteasomes. Smurf1 (Smad ubiquitination regulatory factor 1), a member of the Hect family of E3 ubiquitin ligases, has been found to interact with the BMP-activated Smad1 and Smad5, thereby triggering their ubiquitination and degradation (14). Hect domain proteins represent a major subclass of E3 ligases and contain a conserved cysteine, located toward the carboxyl end of the Hect domain, which is capable of forming a thioester bond with ubiquitin (17). Ubiquitin is first transferred from an appropriate E2 enzyme to this cysteine residue of the E3 ligase. This E3-ubiquitin thioester is then the donor for amide bond formation with the protein substrate. Another motif often found in the Hect family of E3 ligases is the WW domain, which derives its name from the presence of two highly conserved tryptophans and a conserved proline in an approximately 30-amino acid region (18). The WW domains have a preference for binding to small proline-rich sequences, PPXY motifs, and different WW domains possess differential substrate specificity.
The WW domains of Smurf1 have been shown to interact with Smad1 and Smad5, but not with Smad2 and Smad3, through a PPXY motif in the linker region of the Smads (14). Here, we report the identification of Smurf2, a Smurf1-related member of Hect domain E3 ligases. We demonstrate that Smurf2 interacts with receptor-regulated Smads (i.e., Smad1, Smad2, and Smad3) but not with the common mediator Smad (Smad4). Smurf2 preferentially targets Smad1 for ubiquitination and degradation, has a much weaker effect on Smad2 protein levels, but does not affect Smad3 levels. Increased Smurf2 levels in Xenopus embryo profoundly affect ventral mesoderm formation by decreasing Smad1 signaling. These findings suggest that Smurf2 regulates the competence of a cell and/or developing organism to respond to TGF-β superfamily signals.
Methods
Cloning of Human Smurf2 cDNA and Construction of Expression Plasmids.
BLAST search of databases of human-expressed sequence tags was performed to identify human cDNA sequences that share high homology to Smurf1 sequence. cDNA clones 323430, 37867, 42163, and 360440, obtained from the I.M.A.G.E. Consortium, were sequenced and served as the basis to assemble the full-length coding sequence. Expression plasmids for N-terminal hemagglutinin (HA), Myc-tagged or untagged full-length human Smurf2 protein, or defined regions of Smurf2 were generated by PCR of coding sequences and inserted into EcoRI and SalI or HindIII sites of the cytomegalovirus promoter-driven mammalian expression plasmid pRK5 (19) or derivatives. PCR-based approaches were also used to generate a ubiquitin–ligase-inactive mutant of Smurf2, in which cysteine 716 was replaced by glycine.
Expression plasmids encoding N-terminally FLAG-tagged Smad1, Smad2, Smad3, and C-terminal FLAG-tagged Smad4 were described before (20, 21).
Yeast Two-Hybrid Interaction Assays.
The Lex-A-based yeast two-hybrid system was used as described (22). Bait plasmid containing the coding sequences for individual Smads in pEG202 were described before (23, 24). Smad3NLΔPY, which lacks the PPGY sequence (amino acids 181–184) in Smad3, in bait plasmid pEG202 and Smurf2 fragments in prey plasmid pJG4-5 were generated by PCR-mediated approaches and subcloning.
Transfection, Immunoprecipitation, and Pulse–Chase Analyses.
COS-1 and 293 cells were transiently transfected using LipofectAmine (GIBCO/BRL) in a 10-cm-diameter dish. Per transfection, 2 μg of each expression plasmid was used unless otherwise indicated in the figure legends, and “empty” vector DNA was added as needed to keep the total DNA amounts the same. After transfection (36–44 h), cells were lysed in 1 ml of whole cell extract buffer (10 mM Hepes, pH 7.9/300 mM NaCl/0.1 mM EGTA/20% glycerol/0.2% Nonidet P-40 with protease inhibitors) and frozen in liquid nitrogen and then thawed on ice. In experiments that used proteasome inhibitors, transfected cells were incubated overnight with 10–20 μM lactacystin before harvest. The lysates (250 μl) were then diluted 1:1 with water and subjected to immunoprecipitation with anti-FLAG antibody-conjugated beads (Sigma) or anti-HA (Covance, HA11) or anti-Myc (Santa Cruz Biotechnology, 9E10) monoclonal antibodies absorbed in protein G-Sepharose (Amersham Pharmacia). After washing the adsorbed beads in coimmunoprecipitation buffer (10 mM Tris-HCl, pH 8.0/150 mM NaCl/1% Nonidet P-40), the protein complexes were subjected to SDS/PAGE and immunoblotting.
For pulse–chase analysis, 293 cells were metabolically labeled for 1 h with 0.12 mCi/ml 35S-Pro-mix (NEN) in methionine/cysteine-free medium at 36 h after transfection. After washing and incubating in DMEM + 10% FBS for the indicated time, cells were lysed and immunoprecipitations with anti-FLAG antibody-conjugated beads were performed. The protein complexes were then resolved by SDS/PAGE and visualized by autoradiography and phosphorimager to quantify the amount of 35S-labeled Smad1 present at each time point.
Xenopus Animal Cap Assays.
Xenopus Smurf1, human Smad, and Smurf2 mRNA was synthesized by in vitro transcription using Message Machine kit (Ambion). The mRNA was injected into both animal poles of two-cell stage embryos. The animal caps were dissected from the embryos at blastula stage 9 and assayed at gastrula stage 11 or tadpole stage 32 by RT-PCR for the expression of marker genes. The PCR primers and conditions used in the RT-PCR assays have been previously described (25).
Whole-Mount in Situ Hybridization of Xenopus Smurf2 in Xenopus Embryo.
A partial Xenopus Smurf2 cDNA was isolated by screening a Xenopus gastrulation stage cDNA library (Research Genetics, Huntsville, AL) using human Smurf2 cDNA as hybridization probe and subcloned into pRK7 vector. A digoxigenin-UTP-labeled, antisense xSmurf2 hybridization probe was synthesized from the EcoRI-linearized pRK7-xSmurf2 plasmid as template using SP6 RNA polymerase. Whole-mount in situ hybridization was performed as described (26).
Results
Isolation of Human Smurf2.
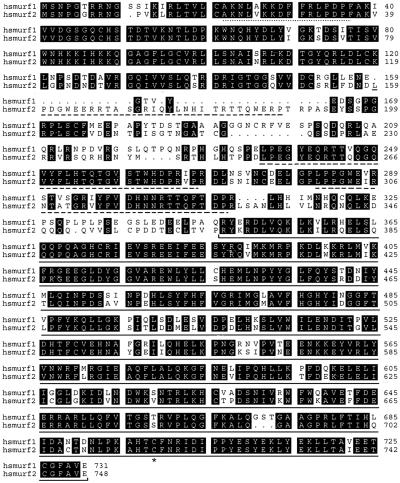
Using a combination of expressed sequence tag database searches and yeast two-hybrid interaction assays using Smad2 as bait, we isolated human cDNAs for a protein we named hSmurf2 (human Smad ubiquitination regulatory factor 2). The corresponding polypeptide encodes a protein of 748 amino acids, with 80% overall sequence identity with Smurf1 (Fig. 1). Like Smurf1, Smurf2 contains the distinctive structural features of the Hect subclass of E3 ubiquitin ligases. These features include an amino-terminal phospholipid/calcium-binding C2 domain, two WW domains, which are predicted to facilitate protein–protein interactions by binding to PPXY motifs on partner proteins, and a carboxyl-terminal Hect domain that catalyzes ubiquitination on the target proteins (17). The predicted Smurf2 polypeptide is slightly longer than the Smurf1 sequence, due to an insertion of 31 amino acids in its N-domain, which contains another WW domain-like sequence motif (Fig. 1). Human Smurf2 cDNAs were found in a variety of tissues, suggesting a widespread distribution similar to Smads (data not shown).
Figure 1.
Alignments of the predicted amino acid sequences of human Smurf1 and Smurf2. Identical amino acids are highlighted in black, the critical cysteine at position 716 is marked with an asterisk, and gaps are introduced to optimize the alignment. Conserved domains are underlined; C2 domain, dotted line; WW domains, broken line; Hect domain, solid line.
Interaction of Smurf2 with Receptor-Regulated Smads.
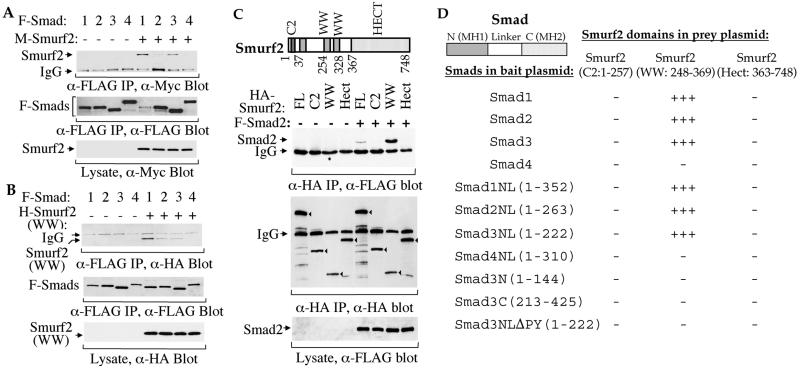
Smurf1 has been reported to selectively interact with Smad1 and Smad5, but not with Smad2 and Smad4, thereby specifically targeting the BMP pathway-specific Smads for degradation (14). To gain insight into the function of Smurf2, we first assessed its ability to interact with different Smads in transfected cells. Using epitope-tagged proteins in coimmunoprecipitation and Western analyses, we found that Smurf2 interacted efficiently with Smad1, associated weakly with Smad2 and Smad3, but did not interact with Smad4 (Fig. 2A). This observation is consistent with the presence of the PPXY motif in the linker region of most Smads but not in the common mediator Smad, Smad4. Similar to the receptor-independent interaction of Smurf1 with Smad1, coexpression of the activated BMP type I receptor ALK2 or ALK6 with Smad1, or of the activated TGF-β type I receptor TβRI with Smad2 or Smad3, had no effect on the associations (data not shown). We also assessed the association of the WW domains of Smurf2 (amino acids 248–369) with Smad1, Smad2, or Smad3. Similar to full-length Smurf2, the WW domains of Smurf2 had a higher affinity for Smad1 than for Smad2 or Smad3 (Fig. 2B). The N-terminal C2 (amino acids 1–257) and the C-terminal Hect (amino acids 363–748) domains of Smurf2 did not associate with Smad2 (Fig. 2C) or Smad1 (data not shown), even though the former segment contained a WW-like motif (amino acid 159–188). The WW domains of Smurf2 (amino acids 248–369) interacted more efficiently with Smad2 than full-length Smurf2 (Fig. 2C). It is difficult to compare the efficiencies of interaction of full-length Smurf2 and its WW domains with Smad1, since expression of full-length Smurf2 dramatically decreased the protein level of Smad1 (Fig. 2A).
Figure 2.
Physical interactions of Smurf2 with Smads. (A) Full-length Smurf2 interacts with Smad1, Smad2, and Smad3 in mammalian cells. COS-1 cells were transfected with expression plasmids for FLAG (F)-tagged full-size Smad1, Smad2, Smad3, or Smad4 or Myc (M)-tagged Smurf2, as marked. Cell lysates were subjected to anti-FLAG immunoprecipitation followed by anti-Myc immunoblotting. (Top) Coprecipitation of M-Smurf2 with F-Smad1, Smad2, or Smad3 is shown. The levels of F-Smads in the immunoprecipitates (Middle) and M-Smurf2 in total cell lysates (Bottom) are shown as indicated. (B) WW domains of Smurf2 (amino acids 248–369) interact with Smad1, Smad2, or Smad3. Lysates of COS-1 cells, transfected with F-Smad1, Smad2, Smad3, or Smad4 and/or HA-tagged WW domains of Smurf2 expression plasmids, were subjected to anti-FLAG immunoprecipitation followed by anti-HA immunoblotting to detect association of HA-Smurf2 (WW) with F-Smad1, Smad2, or Smad3 (Top). The levels of F-Smads in the immunoprecipitates (Middle) and H-Smurf2 (WW) in the total cell lysates (Bottom) are shown as indicated. (C) WW domains (amino acids 248–369) but not C2 or Hect domains of Smurf2 interact with Smad. COS-1 cells were transfected with HA-tagged Smurf2 (FL), HA-Smurf2 fragments (C2, WW, or Hect) and/or F-Smad2, as indicated. Immunoprecipitation with anti-HA antibodies was followed by anti-FLAG immunostaining to detect Smurf2-associated Smad2 (top). (Middle) The anti-HA immunoprecipitated Smurf2 or its fragments (indicated by arrowheads) is shown. (Bottom) The expression level of Smad2 is shown. (D) Yeast two-hybrid assays demonstrate the interaction of Smad1, Smad2, and Smad3, but not Smad4, with the WW domain segment, and the requirement of the linker (L) region and the PPXY sequence of Smads. Interactions were scored by measuring the β-galactosidase activity from − (negative) to +++ (strongly positive).
We also characterized the association of Smurf2 with Smads in yeast two-hybrid interaction assays. Consistent with the coimmunoprecipitation results (Fig. 2 A–C), the WW domains of Smurf2 (amino acids 248–369), but not other regions of Smurf2 (e.g., C2 or Hect domains), interacted strongly with Smad1, Smad2, and Smad3 but not with Smad4. These assays also allowed us to localize the Smurf2 interaction to the linker region of the Smads, since Smurf2 interacted with Smad1NL, Smad2NL, and Smad3NL but not with the C- or N-domains of the Smads (Fig. 2D). Deletion of the PPGY sequence of the linker region in Smad3NLΔPY abolished the interaction with the WW domain segment of Smurf2 (Fig. 2D). This finding is consistent with the ability of WW domains to bind PPXY motifs and the similar requirement of this motif for binding of Smurf1 to Smad1 (14).
Expression of Smurf2 Down-Regulates Smad1 and Smad2 Protein Levels.
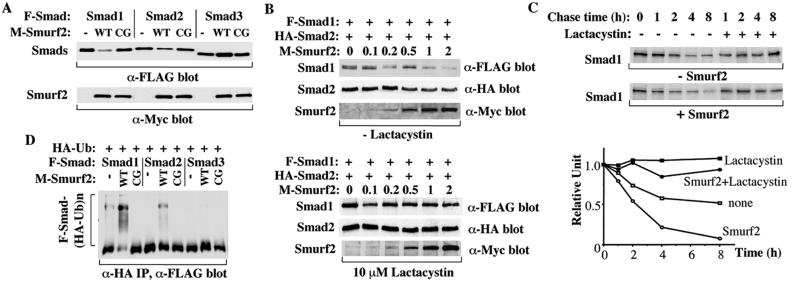
Since Smurf2 belongs to the Hect family of E3 ubiquitin ligases and can interact with Smads, we investigated whether Smurf2 expression affects the steady-state levels of Smad proteins in 293 cells. Coexpression of Smurf2 with Smad1 resulted in a considerable decrease in steady-state level of Smad1 protein and a smaller decrease of Smad2 level (Fig. 3A). However, Smurf2 expression did not decrease Smad3 level and sometimes conferred a slight increase in Smad3 levels (Fig. 3A). As control, we coexpressed similar levels of an inactive mutant of Smurf2(C716G), in which the cysteine that is believed to conjugate ubiquitin (17) was replaced by a glycine. In contrast to wild-type Smurf2, the Smurf2(C716G) mutant did not affect the Smad levels. The effect of Smurf2 on the Smad1 or Smad2 protein levels depended on the amount of Smurf2 proteins expressed in the cells (Fig. 3B, Upper). Thus, the protein levels of Smad1 or Smad2 decreased with increasing expression levels of Smurf2. This dose-dependent decrease was more obvious in the case of Smad1, and the required expression level of Smurf2 to visibly decrease the Smad1 level was much lower than for Smad2. These data suggested that Smurf2 preferentially decreased the Smad1 protein level. Smurf2 expression did not down-regulate the Smad1 or Smad2 levels in the presence of the proteasome inhibitor, lactacystin (Fig. 3B, Lower), indicating that Smurf2 induced Smad1 and Smad2 degradation through the proteasome.
Figure 3.
Effect of Smurf2 on Smad1 and Smad2 levels. Smurf2 expression results in a dramatic decrease of Smad1 protein level and a slight decrease of Smad2 protein level. COS-1 (A, D) or 293 (B, C) cells were transfected with the indicated mammalian expression plasmids. (A) Smurf2, but not the catalytic inactive mutant of Smurf2 (CG), decreases the Smad1 steady-state levels dramatically and the Smad2 steady-state levels slightly. Smurf2 does not decrease the Smad3 levels. Steady-state protein levels were determined by immunoblotting aliquots of the total cell lysates. (B) The decrease of Smad1 and Smad2 protein levels depends on the expression levels of Smurf2 and can be inhibited by lactacystin. Cells transfected with F-Smad1, HA-Smad2, and increasing amounts of Myc-Smurf2 expression plasmid were treated overnight without (Upper) or with (Lower) lactacystin before lysis of cells and immunostaining for steady-state protein levels. The amounts of Myc-tagged Smurf2 plasmid DNA used in transfections are shown in micrograms. (C) Smurf2 increases Smad1 turnover rate; 293 cells transfected with Smad1 and Smurf2 were pulse-labeled with [35S]methionine and then chased for the indicated times. 35S-labeled Smad1 in anti-Smad1 immunoprecipitates was detected by autoradiography of the gel and quantified by phosphorimaging and plotted relative to the amount present at time 0. (D) Ubiquitination of Smad1 and Smad2 in COS-1 cells in the presence of lactacystin. Cell lysates were subjected to anti-HA immunoprecipitation followed by immunoblotting to detect HA-ubiquitin-conjugated Smads. Multi-ubiquitinated species of Smads are indicated (F-Smad-(HA-Ub)n), whereas the lower band may represent an IgG band. Ubiquitination of Smad1 and Smad2 requires the activity of the Smurf2 Hect domain, as the Smurf2 C716G mutant (CG) does not induce ubiquitination of either Smad.
We also analyzed the effect of Smurf2 on the turnover of Smad1 using pulse–chase experiments in 293 cells. In the absence of Smurf2, Smad1 displayed a half-life of 6–8 h (Fig. 3C). However, its half-life was shortened to about 2 h in the presence of Smurf2. Again, lactacystin suppressed the decrease in Smad1 protein levels.
We next tested whether Smurf2 enhanced Smad1 and Smad2 turnover through its ability to promote ubiquitination. We expressed HA-tagged ubiquitin, together with FLAG-tagged Smad1, Smad2, or Smad3, in the absence or presence of Smurf2 in COS-1 cells. In the absence of Smurf2, little ubiquitination was observed for Smad1, and no ubiquitination was detected for the other two Smads; however, in the presence of Smurf2, a strong ladder of high molecular weight, ubiquitin-conjugated Smad1 products was readily observed (Fig. 3D). Ubiquitin-conjugated Smad2 products of high molecular weight were also observed, although again the effect was less profound than the effect on Smad1. In contrast, ubiquitin was not detectably conjugated to Smad3 (Fig. 3D). We therefore conclude that Smurf2 preferentially promotes Smad1 degradation through polyubiquitination, with subsequent degradation through the proteasome. When expressed at a higher level, it can also promote degradation of Smad2 but not Smad3.
Smurf2 Specifically Inhibits the Biological Functions of Smad1.
To evaluate the biological effects of Smurf2, we used Xenopus animal cap assays. In this system, expression of a particular pathway-specific Smad in Xenopus embryos mimics the effects of the corresponding ligands or activated receptors (27–29). Thus, the Smads downstream from the BMP pathway induce expression of ventral/posterior mesoderm-specific genes, whereas Smad2, which transduces signals on behalf of activin, Vg1, and nodal, activates genes responsible for dorsal mesoderm differentiation. The coordinated action of these two types of Smads determines the expression of defined mesodermal marker genes and the type of mesodermal tissues that will form (30, 31).
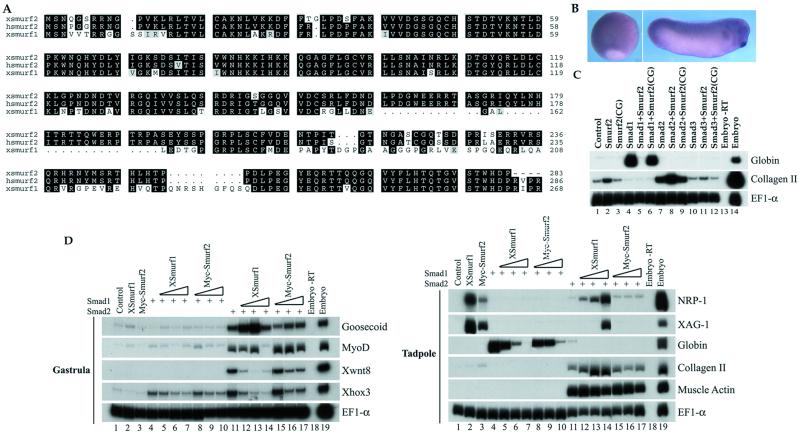
To study the potential involvement of Smurf2 in these pathways, we first verified that a Smurf2 homolog is expressed in the Xenopus embryos. We isolated a partial Xenopus cDNA that is 96% identical to the corresponding amino acid sequence in human Smurf2 and only 74% identical to Xenopus or human Smurf1 (Fig. 4A). We therefore conclude that this cDNA encodes a Xenopus homolog of human Smurf2 and refer to it as xSmurf2. Whole-mount in situ hybridization of staged embryos showed that xSmurf2 mRNA is ubiquitously expressed during gastrulation (Fig. 4B). At the tailbud stage (stage 32), xSmurf2 shows a low level of expression in the entire embryo with higher levels detected in the pharyngeal pouches, cement gland, brain, and eyes (Fig. 4B). These staining patterns resemble those of Xenopus Smad1 and Smurf1 (14, 29).
Figure 4.
Smurf2 inhibits the biological functions of Smad1 in Xenopus embryo assays. (A) Alignments of the N-terminal amino acid sequences of Xenopus Smurf1 with Xenopus and human Smurf2. Identical amino acids are highlighted in black, and gaps are introduced to optimize the alignment. (B) Whole-mount in situ hybridization of xSmurf2 in Xenopus embryo. (Left) Gastrula stage; (Right) tailbud stage. (C) Effect of Smurf2 on marker gene expression. Animal poles of two-cell stage embryos were injected with 2 ng of mRNA of Smurf2 or Smurf2C716G (CG), 1 ng of Smad1 mRNA, 0.5 ng of Smad2 mRNA, or combinations of these as indicated. Animal caps were dissected at stage 9 and assayed at tadpole stage 32 by RT-PCR for the expression of the marker genes shown. (D) Comparison of the activities of Smurf2 and Smurf1. Two-cell stage embryos were injected with 2 ng of Smurf1 or Myc-Smurf2 mRNA, 1 ng of Smad1 mRNA, 0.5 ng of Smad2 mRNA plus 0.5 ng, 1 ng, or 2 ng of Smurf1 or Myc-Smurf2 mRNA. The two lanes on the far right in all panels are control reactions of total embryonic RNA, with (+) or without (−) reverse transcription.
To investigate the function of Smurf2, RNAs encoding Smad1 or Smad2 were microinjected into the animal poles of two-cell stage embryos alone or together with RNA encoding full-size hSmurf2. Ectodermal explants (animal caps) were obtained from early blastulas and incubated to tailbud stages (stage 32), and gene expression was then assayed by RT-PCR. As shown in Fig. 4C, Smad1 induced expression of the ventral mesodermal marker globin. Coexpression of Smurf2 severely decreased Smad1-induced globin expression. In contrast, Smurf2 induced a low level of collagen II expression, a dorsal mesoderm marker, and moderately enhanced Smad2-mediated collagen II expression, although sometimes the effect was not seen (Fig. 4D). These effects of Smurf2 required the catalytic activity of the Hect domain, as the inactive mutant, Smurf2(C716G), did not affect Smad1-induced globin expression or Smad2-induced expression of collagen type II (Fig. 4C). Smad3 had little activity in these assays (Fig. 4C, ref. 31), and its activity was not affected by Smurf2 (Fig. 4C). These results suggest that Smurf2 primarily inhibits BMP/Smad1-mediated signals and may enhance activin/Smad2 signals as a consequence of decreased Smad1 levels. Similar effects on Smad2-mediated mesodermal induction have also been observed with Smurf1 (14) and are consistent with the hypothesis that, by down-regulating Smad1 signaling, the animal caps become sensitized to Smad2 induction, which leads to dorsalization of mesoderm (14, 31). This observation is also consistent with our findings in cultured cells that Smurf2 preferentially down-regulates the amount of Smad1 and only has a small effect on Smad2.
Because Smurf1 has been reported to inhibit Smad1 signaling (14), we compared Smurf2 and Smurf1 with each other in the Xenopus animal cap assays (Fig. 4D). At gastrula stages, Smad1 induced only the ventral mesoderm marker Xhox3, whose expression was reduced by the presence of either Smurf1 or Smurf2. Smad2, at this stage, induced a variety of genes expressed in both dorsal and ventral sides of the embryos. Coexpression of Smurf1 or Smurf2 with Smad2 reduced the ventral and ventral–lateral markers Xhox3 and Xwnt8, whereas it enhanced to some degree the induction of the dorsal markers goosecoid and MyoD (Fig. 4D, Upper). At tadpole stages, injection of either Smurf1 or Smurf2 RNA induced the cement gland marker XAG-1 and the neural marker NRP-1 in animal caps, and both Smurfs inhibited the induction of globin expression by Smad1. These results therefore suggest that Smurf1 and Smurf2 have similar activities in inhibiting BMP signals. Smurf2, however, acted as a milder antagonist of Smad1 activity than Smurf1. In the dose-response experiments, lower amounts of Smurf1 than Smurf2 blocked ventral marker induction by Smad1 and enhanced dorsal mesoderm induction by Smad2. Thus, Smurf2 exerted a milder decrease on globin expression than Smurf1 and had only a minimal effect on the expression of collagen II, XAG-1, and NRP-1, whereas Smurf1 induced or enhanced their expression (Fig. 4D). These data indicate that, although both Smurfs can inhibit Smad1 function, they are different in their capacity to do so, and this difference may influence the graded activity of BMPs.
Discussion
We have identified Smurf2, a new member of the Hect family of E3 ubiquitin ligases. Smurf2 selectively interacts with receptor-regulated Smads and preferentially targets Smad1 for ubiquitination and proteasome-mediated degradation. At high expression level, Smurf2 also slightly decreases the protein levels of Smad2 but not Smad3. In Xenopus embryos, ectopic Smurf2 expression specifically inhibits ventral mesoderm formation and Smad1 responses, thereby affecting embryonic patterning by BMP signals. Smurf2-mediated degradation represents a distinct degradation pathway that regulates BMP/activin signaling independent of the previously identified Smurf1 activity.
Smurf2 is a 748-aa-long enzyme, which, like Smurf1, contains two WW domains in the middle of the protein and a C-terminal Hect domain. WW domains mediate protein–protein interactions through their ability to interact with a PPXY motif (18). Consistent with the presence of this sequence in many Smads, the WW domains of Smurf2 (amino acids 248–369) interacted with Smad1, Smad2, and Smad3 in yeast two-hybrid and coimmunoprecipitation assays. Additionally, Smurf2 also interacted with Smad6 and Smad7 in coimmunoprecipitation assays (data not shown). The inability of Smurf2 to interact with Smad4 is consistent with the absence of a PPXY sequence in Smad4. The stronger affinity of Smurf2 for Smad1 than for Smad2 or Smad3, which is apparent in coimmunoprecipitation assays, is likely due to the participation of other Smad sequences in the interaction with Smurf2. The C-terminal Hect domain catalyzes ubiquitination of the target proteins and contains a highly conserved cysteine residue at position 716. Replacement of this cysteine by glycine inactivated the enzymatic activity of Smurf2 and, thus, its ability to induce degradation of Smad1 and Smad2.
Consistent with the physical interaction, expression of Smurf2 in cultured cells resulted in ubiquitination and degradation of Smad1, whereas it had only a small effect on Smad2 when it was overexpressed, and no effect on Smad3 or on Smad6 and Smad7 (data not shown). The preferential activity of Smurf2 on Smad1 was also apparent in Xenopus animal cap assays, in which ectopic expression of Smurf2 in embryos inhibited the developmental responses to Smad1/BMP signaling. Therefore, the ability of Smurf2 to enhance some responses of the Smad2 (activin/nodal) pathway is most likely a consequence of decreased Smad1 levels and may illustrate how the balance between BMP- and activin/nodal-activated Smads regulates gene expression responses.
The preferential targeting of Smad1 for ubiquitin-mediated degradation by Smurf2 resembles the activity of Smurf1. Smurf1 has been shown to selectively interact with the BMP pathway-specific Smad1 and Smad5, and not with Smad2 or Smad3 (14). Similarly to Smurf2, the interaction of Smurf1 with Smad1 also leads to ubiquitination and degradation of Smad1 and to inhibition of BMP/Smad1 signaling in Xenopus embryos and consequent dorsalization of the induced ventral mesoderm. On the other hand, Smurf1 and Smurf2 also display different activities. Thus, whereas Smurf1 is selective for Smad1 and Smad5 and has no affinity for Smad2, Smurf2 has a weak affinity for Smad2 and can cause a low level of ubiquitination and degradation of Smad2. In addition, differential effects of Smurf1 and Smurf2 were apparent in Xenopus assays. Whereas Smurf1 strongly inhibited Smad1 signaling, Smurf2 had milder effects. These differences of the Smurfs in inhibiting BMP signaling may be required for the establishment of a functional BMP signaling gradient in vivo. In early Xenopus embryos, a complete inhibition of BMP signaling leads to neural and dorsal mesoderm development, whereas a partial interference with BMP signaling results in the formation of cement gland, neural crest, and lateral mesoderm. When BMP signaling is not blocked and Smad1 is fully activated, epidermis and ventral mesoderm form. The complete and partial inhibition of Smad1 activity by Smurf1 and Smurf2 may differentially participate in the regulation of the levels of BMP signaling, thus influencing the patterning of early embryonic tissues.
The existence of two Smurfs with similar, yet distinct properties in vertebrates revealed another level of complexity of the BMP signaling mechanisms. It is already known that three Smads (Smad1, Smad5, and Smad8) mediate BMP signaling by at least four BMP type I receptors, and, although they mediate similar effects in vitro, the results from targeted gene inactivation in mice demonstrate distinct functions in vivo (32). Conceptually similarly, the two inhibitory Smads, Smad6 and Smad7, have similar activities in vitro, yet they display preferential inhibition activities in the TGF-β/activin and BMP signaling pathways (33–36). Therefore, it should not be surprising that two distinct Smurfs regulate independently from each other the BMP signaling pathways. A characterization of the temporal and spatial expression patterns during development, and of the developmental consequences of targeted gene inactivation, will most likely reveal the differential functions of Smurf1 and Smurf2.
Recently, it has been reported that the receptor-activated Smad2 is subject to constant degradation by the ubiquitin/proteasome pathway in the nucleus (15). However, Smurf-mediated ubiquitination of Smads is distinct from the nuclear ubiquitination of activated Smad2, as a Smad2 mutant lacking the PPXY motif still undergoes nuclear degradation. The N-terminal sequences of Smurf1 and Smurf2 contain a lipid/Ca2+-binding (C2) domain of 15 amino acid residues, which may determine the cytoplasmic location of the Smurfs (37). Accordingly, Smurf1 and Smurf2 are primarily located in the cytoplasm (data not shown). In addition, Smad degradation by Smurf2 occurs, similarly to Smurf1, independently of receptor activation. Thus, Smurf1 and Smurf2 regulate the available levels of Smads and may control the competence of a cell to respond to TGF-β superfamily signals. In the nucleus, Smad2 ubiquitination may involve E2 UbcH5 ubiquitin-conjugating enzymes (15), whereas the E3 enzyme involved in this process is not known. Since UbcH5-related enzymes can function in concert with Hect-domain-containing E3 proteins (38–40), it is likely that another member of the Hect family of E3 ubiquitin ligase may be involved in the degradation of nuclear Smad2 through a distinct mechanism from that of Smurfs. Further studies will be required to characterize the mechanisms that control cytoplasmic and nuclear degradation of Smads and their control in signaling by TGF-β family members.
Acknowledgments
We thank Y. Cheng for the ubiquitin expression plasmid and G. Thomsen (State University of New York, Stony Brook) for Xenopus Smurf1 plasmid; we also thank Dr. L. Samelson for critically reading the manuscript and preparing the PDF file of the manuscript. This research was supported by National Institutes of Health Grants CA63101 (to R.D.) and HD32105 (to A.H.-B.) and by intramural National Cancer Institute funding (to Y.Z.). A.H.-B. is a McKnight and Merck scholar.
Abbreviations
- TGF-β
transforming growth factor-β
- BMP
bone morphogenetic protein
- RT
reverse transcription
Footnotes
References
- 1.Derynck R, Choy L. In: The Cytokine Handbook. 3rd Ed. Thompson A, editor. Boston: Academic; 1998. pp. 593–636. [Google Scholar]
- 2.Massagué J. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 3.Whitman M. Genes Dev. 1998;12:2445–2462. doi: 10.1101/gad.12.16.2445. [DOI] [PubMed] [Google Scholar]
- 4.Hogan B L M. Genes Dev. 1996;10:1580–1594. doi: 10.1101/gad.10.13.1580. [DOI] [PubMed] [Google Scholar]
- 5.Harland R, Gerhart J. Annu Rev Cell Biol. 1997;13:611–667. doi: 10.1146/annurev.cellbio.13.1.611. [DOI] [PubMed] [Google Scholar]
- 6.Derynck R, Feng X-H. Biochim Biophys Acta. 1997;1333:F105–F150. doi: 10.1016/s0304-419x(97)00017-6. [DOI] [PubMed] [Google Scholar]
- 7.Heldin C H, Miyazono K, ten Dijke P. Nature (London) 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 8.Derynck R, Zhang Y, Feng X-H. Cell. 1998;95:737–740. doi: 10.1016/s0092-8674(00)81696-7. [DOI] [PubMed] [Google Scholar]
- 9.Massagué J, Wotton D. EMBO J. 2000;19:1745–1754. doi: 10.1093/emboj/19.8.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, Derynck R. Trends Cell Biol. 1999;9:274–279. doi: 10.1016/s0962-8924(99)01579-2. [DOI] [PubMed] [Google Scholar]
- 11.Massagué J, Chen Y-G. Genes Dev. 2000;14:627–644. [PubMed] [Google Scholar]
- 12.ten Dijke P, Miyazono K, Heldin C H. Trends Biochem Sci. 2000;25:64–70. doi: 10.1016/s0968-0004(99)01519-4. [DOI] [PubMed] [Google Scholar]
- 13.Wrana J L. Cell. 2000;100:189–192. doi: 10.1016/s0092-8674(00)81556-1. [DOI] [PubMed] [Google Scholar]
- 14.Zhu H, Kavsak P, Abdollah S, Wrana J L, Thomsen G H. Nature (London) 1999;400:687–693. doi: 10.1038/23293. [DOI] [PubMed] [Google Scholar]
- 15.Lo R S, Massagué J. Nat Cell Biol. 1999;1:472–478. doi: 10.1038/70258. [DOI] [PubMed] [Google Scholar]
- 16.Hershko A, Ciechanover A. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 17.Huibregtse J M, Scheffner M, Beaudenon S, Howley P M. Proc Natl Acad Sci USA. 1995;92:2563–2567. doi: 10.1073/pnas.92.7.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rotin D. Curr Top Microbiol Immunol. 1998;228:115–133. doi: 10.1007/978-3-642-80481-6_5. [DOI] [PubMed] [Google Scholar]
- 19.Graycar J L, Miller D A, Arrick B A, Lyons R M, Moses H L, Derynck R. Mol Endocrinol. 1989;3:1977–1986. doi: 10.1210/mend-3-12-1977. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Feng X-H, Wu R-Y, Derynck R. Nature (London) 1996;383:168–172. doi: 10.1038/383168a0. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Feng X-H, Derynck R. Nature (London) 1998;394:909–913. doi: 10.1038/29814. [DOI] [PubMed] [Google Scholar]
- 22.Gyuris J, Golemis E, Chertkov H, Brent R. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 23.Wu R Y, Zhang Y, Feng X-H, Derynck R. Mol Cell Biol. 1997;17:2521–2528. doi: 10.1128/mcb.17.5.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng X-H, Zhang Y, Wu R Y, Derynck R. Genes Dev. 1998;12:2153–2163. doi: 10.1101/gad.12.14.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang C, Wilson P A, Mathews L S, Hemmati-Brivanlou A. Development (Cambridge, UK) 1997;124:827–837. doi: 10.1242/dev.124.4.827. [DOI] [PubMed] [Google Scholar]
- 26.Harland R M. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- 27.Baker J C, Harland R M. Genes Dev. 1996;10:1880–1889. doi: 10.1101/gad.10.15.1880. [DOI] [PubMed] [Google Scholar]
- 28.Graff J M, Bansal A, Melton D A. Cell. 1996;85:479–487. doi: 10.1016/s0092-8674(00)81249-0. [DOI] [PubMed] [Google Scholar]
- 29.Thomsen G H. Development (Cambridge, UK) 1996;122:2359–2366. doi: 10.1242/dev.122.8.2359. [DOI] [PubMed] [Google Scholar]
- 30.Lagna G, Hata A, Hemmati-Brivanlou A, Massagué J. Nature (London) 1996;383:832–836. doi: 10.1038/383832a0. [DOI] [PubMed] [Google Scholar]
- 31.Candia A F, Watabe T, Hawley S H, Onichtchouk D, Zhang Y, Derynck R, Niehrs C, Cho K W. Development (Cambridge, UK) 1997;124:4467–4480. doi: 10.1242/dev.124.22.4467. [DOI] [PubMed] [Google Scholar]
- 32.Weinstein M, Yang X, Deng C. Cytokine Growth Factor Rev. 2000;11:49–58. doi: 10.1016/s1359-6101(99)00028-3. [DOI] [PubMed] [Google Scholar]
- 33.Hayashi H, Abdollah S, Qiu Y, Cai J, Xu Y Y, Grinnell B W, Richardson M A, Topper J N, Gimbrone M A, Jr, Wrana J L, Falb D. Cell. 1997;89:1165–1173. doi: 10.1016/s0092-8674(00)80303-7. [DOI] [PubMed] [Google Scholar]
- 34.Imamura T, Takase M, Nishihara A, Oeda E, Hanai J, Kawabata M, Miyazono K. Nature (London) 1997;389:622–626. doi: 10.1038/39355. [DOI] [PubMed] [Google Scholar]
- 35.Nakao A, Afrakhte M, Moren M, Nakayama T, Christian J L, Heuchel R, Itoh S, Kawabata M, Heldin N E, Heldin C H, ten Dijke P. Nature (London) 1997;389:631–635. doi: 10.1038/39369. [DOI] [PubMed] [Google Scholar]
- 36.Hata A, Lagna G, Massagué J, Hemmati-Brivanlou A. Genes Dev. 1998;12:186–197. doi: 10.1101/gad.12.2.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plant P J, Yeger H, Staub O, Howard P, Rotin D. J Biol Chem. 1997;272:32329–32336. doi: 10.1074/jbc.272.51.32329. [DOI] [PubMed] [Google Scholar]
- 38.Scheffner M, Huibregtse J M, Howley P M. Proc Natl Acad Sci USA. 1994;91:8797–8801. doi: 10.1073/pnas.91.19.8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jensen J P, Bates P W, Yang M, Vierstra R D, Weissman A M. J Biol Chem. 1995;270:30408–30414. doi: 10.1074/jbc.270.51.30408. [DOI] [PubMed] [Google Scholar]
- 40.Hatakeyama S, Jensen J P, Weissman A M. J Biol Chem. 1997;272:15085–15092. doi: 10.1074/jbc.272.24.15085. [DOI] [PubMed] [Google Scholar]