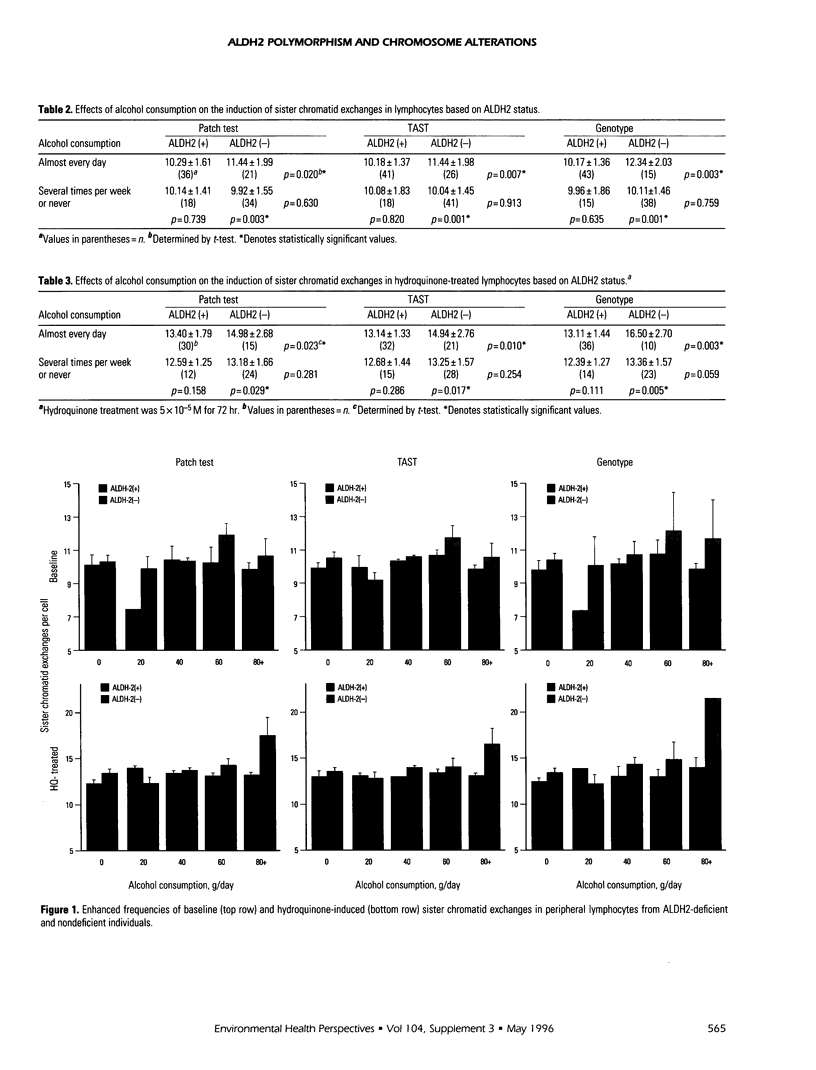
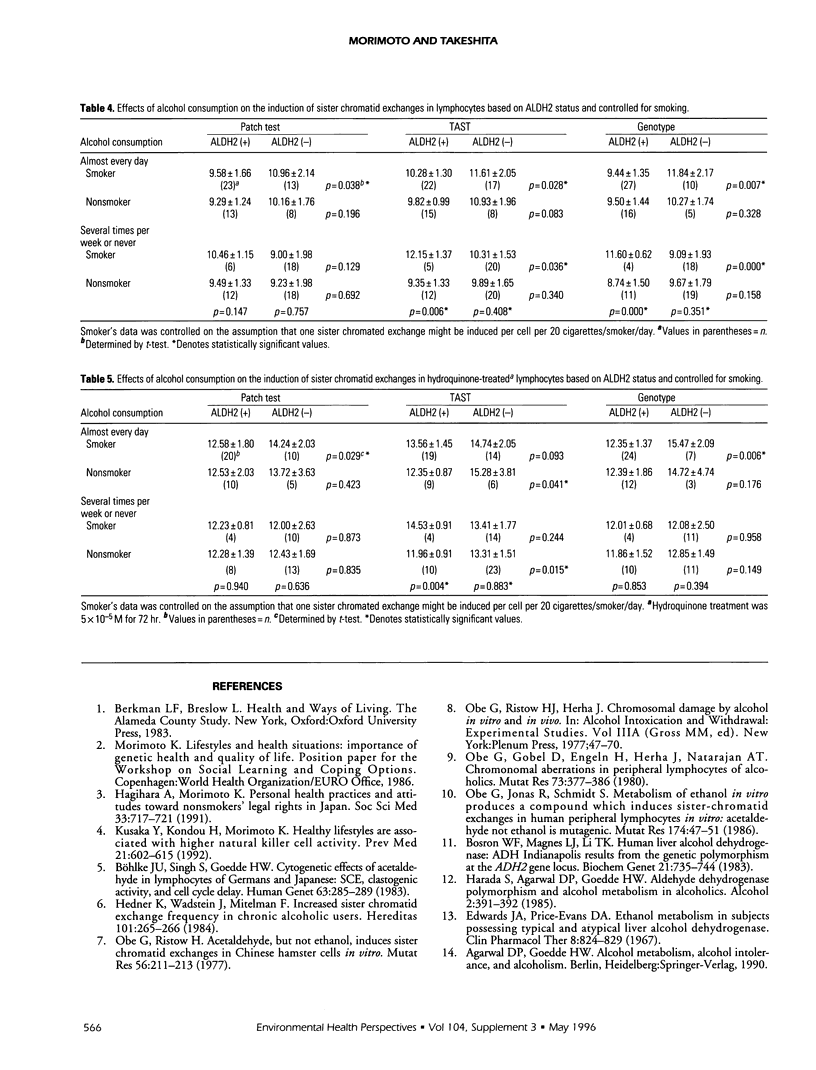
Abstract
Excessive drinking of alcohol is now widely known to be one of the major lifestyle choices that ca effect health. Among the various effects of alcohol drinking, cytogenetic and other genotoxic effects are of major concern from the viewpoint of prevention of alcohol-related diseases. Alcohol is first metabolized to acetaldehyde, which directly causes various types of chromosomal DNA lesions and alcohol-related diseases, and is then further detoxified to the much less toxic metabolite acetate. About 50% of Oriental people are deficient in the aldehyde-dehydrogenase 2 isozyme (ALDH2) that can most efficiently detoxify acetaldehyde. We have performed a series of experiments to investigate how the genetic deficiency in ALDH2 affects the behavioral pattern for alcohol drinking and the sensitivity of peripheral lymphocytes to the induction of chromosome alterations by exposure to alcohol and alcohol-related chemicals. We found great effects of the ALDH2 genotypes on alcohol sensitivity and alcohol-drinking behavior. We also show that lymphocytes from habitual drinkers with the deficient ALDH2 enzyme had significantly higher frequencies of sister chromatid exchanges than those from ALDH2-proficient individuals.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bosron W. F., Magnes L. J., Li T. K. Human liver alcohol dehydrogenase: ADH Indianapolis results from genetic polymorphism at the ADH2 gene locus. Biochem Genet. 1983 Aug;21(7-8):735–744. doi: 10.1007/BF00498920. [DOI] [PubMed] [Google Scholar]
- Böhlke J. U., Singh S., Goedde H. W. Cytogenetic effects of acetaldehyde in lymphocytes of Germans and Japanese: SCE, clastogenic activity, and cell cycle delay. Hum Genet. 1983;63(3):285–289. doi: 10.1007/BF00284666. [DOI] [PubMed] [Google Scholar]
- Edwards J. A., Evans D. A. Ethanol metabolism in subjects possessing typical and atypical liver alcohol dehydrogenase. Clin Pharmacol Ther. 1967 Nov-Dec;8(6):824–829. doi: 10.1002/cpt196786824. [DOI] [PubMed] [Google Scholar]
- Goedde H. W., Singh S., Agarwal D. P., Fritze G., Stapel K., Paik Y. K. Genotyping of mitochondrial aldehyde dehydrogenase in blood samples using allele-specific oligonucleotides: comparison with phenotyping in hair roots. Hum Genet. 1989 Mar;81(4):305–307. doi: 10.1007/BF00283679. [DOI] [PubMed] [Google Scholar]
- Hagihara A., Morimoto K. Personal health practices and attitudes toward nonsmokers' legal rights in Japan. Soc Sci Med. 1991;33(6):717–721. doi: 10.1016/0277-9536(91)90026-9. [DOI] [PubMed] [Google Scholar]
- Harada S., Agarwal D. P., Goedde H. W. Aldehyde dehydrogenase deficiency as cause of facial flushing reaction to alcohol in Japanese. Lancet. 1981 Oct 31;2(8253):982–982. doi: 10.1016/s0140-6736(81)91172-7. [DOI] [PubMed] [Google Scholar]
- Harada S., Agarwal D. P., Goedde H. W. Aldehyde dehydrogenase polymorphism and alcohol metabolism in alcoholics. Alcohol. 1985 May-Jun;2(3):391–392. doi: 10.1016/0741-8329(85)90100-4. [DOI] [PubMed] [Google Scholar]
- Hedner K., Wadstein J., Mitelman F. Increased sister chromatid exchange frequency in chronic alcoholic users. Hereditas. 1984;101(2):265–266. doi: 10.1111/j.1601-5223.1984.tb00926.x. [DOI] [PubMed] [Google Scholar]
- Kusaka Y., Kondou H., Morimoto K. Healthy lifestyles are associated with higher natural killer cell activity. Prev Med. 1992 Sep;21(5):602–615. doi: 10.1016/0091-7435(92)90068-s. [DOI] [PubMed] [Google Scholar]
- Morimoto K. Induction of sister chromatid exchanges and cell division delays in human lymphocytes by microsomal activation of benzene. Cancer Res. 1983 Mar;43(3):1330–1334. [PubMed] [Google Scholar]
- Morimoto K., Wolff S. Cell cycle kinetics in human lymphocyte cultures. Nature. 1980 Dec 11;288(5791):604–606. doi: 10.1038/288604a0. [DOI] [PubMed] [Google Scholar]
- Morimoto K., Wolff S. Increase of sister chromatid exchanges and perturbations of cell division kinetics in human lymphocytes by benzene metabolites. Cancer Res. 1980 Apr;40(4):1189–1193. [PubMed] [Google Scholar]
- Muramatsu T., Higuchi S., Shigemori K., Saito M., Sasao M., Harada S., Shigeta Y., Yamada K., Muraoka H., Takagi S. Ethanol patch test--a simple and sensitive method for identifying ALDH phenotype. Alcohol Clin Exp Res. 1989 Apr;13(2):229–231. doi: 10.1111/j.1530-0277.1989.tb00317.x. [DOI] [PubMed] [Google Scholar]
- Obe G., Göbel D., Engeln H., Herha J., Natarajan A. T. Chromosomal aberrations in peripheral lymphocytes of alcoholics. Mutat Res. 1980 Dec;73(2):377–386. doi: 10.1016/0027-5107(80)90202-x. [DOI] [PubMed] [Google Scholar]
- Obe G., Jonas R., Schmidt S. Metabolism of ethanol in vitro produces a compound which induces sister-chromatid exchanges in human peripheral lymphocytes in vitro: acetaldehyde not ethanol is mutagenic. Mutat Res. 1986 May;174(1):47–51. doi: 10.1016/0165-7992(86)90075-8. [DOI] [PubMed] [Google Scholar]
- Obe G., Ristow H. J., Herha J. Chromosomal damage by alcohol in vitro and in vivo. Adv Exp Med Biol. 1977;85A:47–70. doi: 10.1007/978-1-4899-5181-6_4. [DOI] [PubMed] [Google Scholar]
- Takeshita T., Morimoto K., Mao X. Q., Hashimoto T., Furuyama J., Furyuama J. Phenotypic differences in low Km aldehyde dehydrogenase in Japanese workers. Lancet. 1993 Mar 27;341(8848):837–838. [PubMed] [Google Scholar]
- Takeshita T., Morimoto K., Mao X., Hashimoto T., Furuyama J. Characterization of the three genotypes of low Km aldehyde dehydrogenase in a Japanese population. Hum Genet. 1994 Sep;94(3):217–223. doi: 10.1007/BF00208273. [DOI] [PubMed] [Google Scholar]


