Abstract
Several recent findings have indicated that the promyelocytic leukemia gene product (PML) oncogenic domains (PODs) are involved in proteasome-mediated degradation of ubiquitinated proteins. We wanted to examine the intracellular distribution of PML protein in the presence of a proteasome inhibitor. We used high-resolution microscopy to study the distribution of PML protein and other POD-associated proteins along with the proteasomes themselves under normal conditions and in cells treated with the proteasome inhibitor, MG132. Inhibition of the proteasomes in MCF-7, HeLa, and IB-4 cell lines resulted in a radical redistribution of the POD-associated proteins PML, Sp100, and SUMO-1. After 6–10 h of MG132 treatment, PML, Sp100, and SUMO-1 were no longer detectable in the PODs and accumulated mainly in the nucleolus. Moreover, MG132 treatment changed the cellular distribution of the proteasomes. Interestingly, this included the accumulation in euchromatin areas of the nucleus and within the nucleoli. Several non-POD-associated proteins did not change their cellular distribution under the same conditions. The accumulation of POD-associated proteins and proteasomes in the nucleoli of MG132-treated cells indicates that these proteins may target the nucleoli under normal conditions and that the nucleolus may have a function in the regulation of proteasomal protein degradation.
Promyelocytic leukemia gene product (PML) oncogenic domains [PODs, also termed nuclear domain 10 (ND10) or PML bodies] are nuclear structures that are specifically disrupted in human acute promyelocytic leukemia cells. The most extensively studied component, PML, is a RING-finger motif protein. The t(15;17) chromosomal translocation in acute promyelocytic leukemia fuses PML with the retinoic acid receptor α (RAR-α) gene to form the oncoprotein PML–RAR-α. The PODs are disrupted in acute promyelocytic leukemia cells, and the fusion protein is distributed throughout the nucleoplasm in a fine granular pattern. Retinoic acid or arsenic trioxide, used in the clinical treatment of acute promyelocytic leukemia, leads to the reconstitution of PODs, indicating that a tight relationship exists between nuclear organization and malignant phenotype. In addition to PML protein, PODs accumulate several other cellular proteins such as Sp100, SUMO-1, INT-6, CBP/p300, HAUSP, HSP70, and a fraction of RB (1). It has been suggested that the PODs are involved in many different cellular functions such as transcriptional regulation, growth suppression, and apoptosis (2).
Several recent studies indicate that the PODs are involved in the proteasomal degradation of ubiquitinated proteins. The ubiquitin–proteasome pathway is the major protein degradation system in eukaryotic cells. Proteins targeted for degradation are covalently modified by polyubiquitin followed by the proteasome-mediated degradation (3). The association of PODs with the ubiquitination/deubiquitination process is demonstrated by the presence of the Ub-dependent hydrolase, HAUSP, in the PODs (4). SUMO-1 covalently modifies a number of proteins including PML and Sp100 (5) and is proposed to play a role in modulating intracellular localization of the proteins rather than targeting them for degradation (6, 7). HAUSP removes the ubiquitin, but not the SUMO-1, from its substrate before proteasomal degradation (8). Proteasomal protein degradation generates peptides that can become associated with MHC class-I antigens, targeted by T cells. The PML protein can regulate MHC expression in untransformed fibroblasts and induce proteins involved in antigen processing, such as proteasomal LMP-2 and LMP-7, and antigen presentation (9). Interferons can increase the supply of antigenic peptides by inducing the expression of components involved in proteasomal degradation. The expression of PML and Sp100 is enhanced by interferons. Several recent studies showed that PML and PODs play a role in the cellular response to interferons (10, 11).
The involvement of proteasomes in the degradation of cellular and viral proteins can be studied by the use of proteasome inhibitors such as MG132 (Cbz-leu-leu-leucinal), that inhibit the chymotrypsin-like activity of the proteasome. MG132-treated cells accumulate polyubiquitinated proteins and subsequently die (12).
Several viral proteins such as ICP0, an immediate early gene product of HSV-1 and adenovirus-encoded E1A associate with and disrupt the PODs (13). ICP0 has been shown to interact with enzymes belonging to the ubiquitin-specific protease family (4) and to induce the proteasome-dependent degradation of PML and loss of its SUMO-1-modified isoforms (8). Degradation of the SUMO-1 modified Sp100 is also induced by HSV-1 infection (14).
The tight association of PODs with proteasome-mediated protein degradation prompted us to analyze the subcellular distribution of proteasomes, different POD components, and non-POD-associated proteins in cells with inhibited proteasomal activity.
Materials and Methods
Cells and Cell Culture Conditions.
The cell lines used in this study were MCF-7 (human breast carcinoma), HeLa (human cervical carcinoma), and IB-4 (EBV-immortalized LCL). The cells were grown at 37°C with 5% CO2 in Iscove's modified Dulbecco's cell culture medium containing 10% FBS/2 mM L-glutamine/100 units/ml penicillin/100 units/ml streptomycin. The absence of mycoplasma contamination was examined routinely by Hoechst 33258 staining.
Antibodies.
A rabbit polyclonal anti-PML serum (a gift from H. de The, Institut d'Hematologie de Universite Paris VII, Paris) or a mouse monoclonal anti-PML (PG-M3, Santa Cruz Biotechnology) were used for PML staining. FITC-conjugated swine–anti-rabbit (Dakopatts) and Texas Red streptavidin (Vector) were used as secondary antibodies. DNA was stained by Hoechst 33258. Nucleolar antigen B23/nucleophosmin was detected using a mouse mAb, NPM (a gift from P.K. Chan, Baylor College of Medicine, Houston). A mouse mAb against fibrillarin was used (a gift from E. Tan, Scripps Research Institute, La Jolla, CA). Sp100 was detected by a rabbit polyclonal anti-Sp100 (a gift from H. de The). Staining of the proteasomes was performed using a mouse mAb directed against 20S proteasome, clone HP810 (Affiniti Research Products, Mamhead, Exeter, U.K.). A rabbit polyclonal serum was used to detect SUMO-1 (Santa Cruz Biotechnology).
Proteasome Inhibition and Immunofluorescense Staining.
The cells were grown in 6-well plates and then incubated for 6 h (IB-4 cells) or overnight (MCF-7 or HeLa cells) in the presence of 5–20 μM carbobenzoxy-L-leucyl-L-leucyl-L-leucinal (MG132, Calbiochem) diluted in DMSO (Merck). Mock samples and DMSO-treated cells were cultured in parallel as controls. In addition to inhibition of proteasomal activity, 20 μg/ml cycloheximide (Sigma) was included to inhibit protein synthesis. Following proteasome inhibition, the cells were washed with PBS, trypsinized, and cytospinned onto glass slides. Cells were fixed with methanol-acetone (1:1) at −20°C for 15 min and then rehydrated in PBS for 30 min. Antibodies were diluted in blocking buffer (2% BSA/0.2% Tween-20/10% glycerol/0.05% Na3N in PBS). Cells were incubated with the primary antibody in a moist chamber for 60 min at room temperature and then washed three times with PBS. The secondary antibody was incubated for 30 min at room temperature followed by three PBS washes. The glass slides were mounted with 70% glycerol containing 2.5% DABCO anti-fading agent (Sigma). Images were collected using a Leitz DM RB microscope, equipped with Leica PL Fluotar 100×, 40×, and PL APO Ph 63× oil immersion objectives. Composite filter cubes were used for the FITC, Texas Red, and Hoechst 33258 fluorescence, respectively. The pictures were captured with a Hamamatsu dual mode cooled CCD camera (C4880), recorded and analyzed on a Pentium PC computer equipped with an AFG VISIONplus–AT frame grabber board using Hipic4.0.4 (Hamamatsu), Image-Pro Plus (Media Cybernetics). Digital images were assembled using Adobe photoshop software. Three-dimensional reconstituted images were collected using Zeiss Axiophot microscope equipped with 63× oil Plan-Apochromat NA 1.4, and 100× oil Plan-Neofluar NA 0.7–1.3 objectives, illuminated with Osram HBO 200W mercury short arc lamp. A program developed by us, stereotrooper, was used to produce images. This program also gives information about the depth distribution by creating a maximal-intensity projected image pair representing a three-dimensional reconstitution of the original object as single or stereoscopic image (15). To remove out-of-focus blur, the program uses the nearest-neighbor deconvolution deblurring algorithm. The following excitation filters were used: single band UV exciter for Hoechst (84360), single band blue exciter for FITC (84490), and single band green exciter for TRITC (84555). The images were captured with a PXL cooled camera (Photometrics, Munich, Germany) and analyzed on a Webforce Indy Silicon Graphics computer using the Isee 5.1 graphical programming system (Inovision, Raleigh, NC) or Pentium PC computer with linux os.
Results
Nucleolar Accumulation of PML Proteins in Proteasome Inhibitor-Treated Cells.
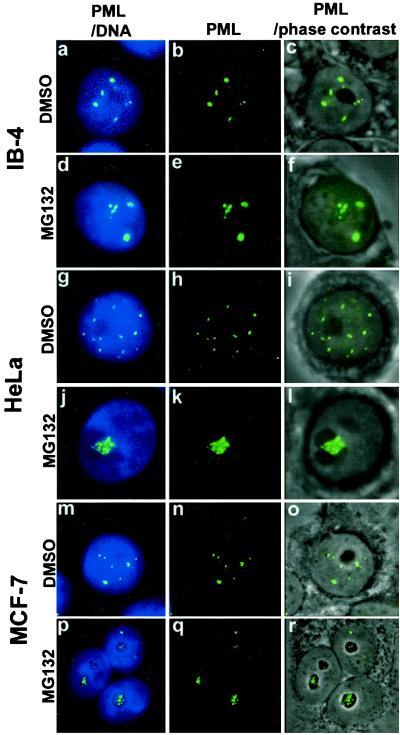
PML protein accumulates in distinct nuclear bodies, excluding the nucleolus, and associates with the nuclear matrix. To study the nuclear distribution of the protein in proteasome inhibitor-treated cells, a lymphoblastoid cell line IB-4 was cultured in the presence of 5 μM MG132 for 6 h. In the control DMSO-treated cells, PML was located in the PODs that were distributed throughout the nucleus, excluding the nucleoli in agreement with all previous reports (Fig. 1 a–c). In IB-4 cells treated with MG132, there was an accumulation of PML in the nucleoli and in a few extra nuclear dots (Fig. 1 d–e). The study was extended to two carcinoma lines, HeLa and MCF-7. After overnight treatment with 5 μM MG132, a similar nucleolar-staining pattern, as in IB-4 cells, was found (Fig. 1 g–r). PML accumulated in the form of large clumps throughout the nucleolus in both lines. There were small numbers of PML dots left in the nucleoplasm.
Figure 1.
Subnuclear distribution of PML after DMSO or MG132 treatment of MCF-7, HeLa, or LCL IB-4 cell lines. Combination of DNA (blue) and PML (green) is shown in the left panels (a, d, g, j, m, p), middle panels show PML alone (b, e, h, k, n, q), combination of phase-contrast and immunofluorescence of PML is shown in right panels (c, f, i, l, o, r). IB-4 cells cultured in the presence of DMSO (a–c) with PML distributed throughout the nucleoplasm excluding the nucleoli. Cells cultured with 5 μM MG132 for 6 h (d–f) show staining of PML inside the nucleoli (d, f) in addition to some nuclear dots. HeLa cells cultured in the presence of DMSO (g–i) or with 5 μM MG132 for 15 h (j–l). PML accumulation upon MG132 treatment is shown in one of three nucleoli (l). MCF-7 cells cultured in the presence of DMSO (m–o) or with 5 μM MG132 for 15 h (p–r). PML accumulates in the nucleoli of MG132-treated cells (r).
Removal of proteasome inhibitor from MCF-7 cells and replacement with normal cell culture media showed that the nucleolar accumulation of PML upon MG132 treatment was reversible. Twenty hours after the replacement, the PML staining was exclusively localized to nuclear bodies, excluding nucleoli (data not shown).
The effect on proteasome inhibitor treatment on non-POD-associated proteins was studied by immunofluorescence staining of the nuclear proteins p27, p21, cyclin D3, cyclin E, EBNA-2,and EBNA-6 in MG132-treated MCF-7 cells or LCL IB-4 (Table 1). p21, p27, cyclin D3, and cyclin E are known to be degraded by the proteasome pathway. In agreement with this, the staining intensity of cyclin D3, p21, and p27 was significantly increased after MG132 treatment. None of these proteins changed its nucleoplasmic distribution after MG132 treatment.
Table 1.
Effect of MG132 treatment on the distribution of different nuclear proteins in MCF-7 or IB-4 cells
| Nucleolar translocation | No effect on distribution |
|---|---|
| PML | p27 |
| Sp100 | p21 |
| EBNA-5 | Cyclin D3 |
| SUMO-1 | Cyclin E |
| 20S proteasome | EBNA-2 |
| EBNA-6 |
The Association of PML with Other Nucleolar Proteins.
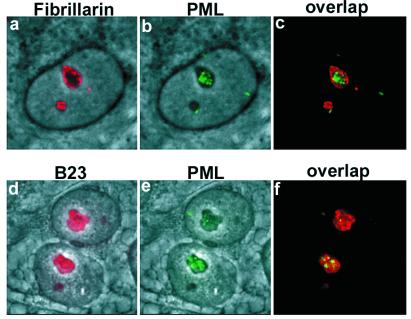
To verify that PML moved to the nucleolus and to define the subdomain targeted by PML, we double-stained MG132-treated MCF-7 cells for PML and fibrillarin or PML and B23 (nucleophosmin). Fibrillarin is a marker of the dense fibrillar centers (16), whereas B23 is localized to the granular center (17). Double staining of PML and fibrillarin showed that the PML protein accumulated mainly in areas lacking fibrillarin, although some colocalization was found at the border of fibrillarin regions (Fig. 2 a–c). Double staining for PML and B23 showed that the PML clumps were located both in areas with and without B23 (Fig. 2 d–f). Proteasome inhibitor treatment did not alter the staining pattern of B23 or fibrillarin (data not shown). These results suggest that PML can accumulate in both the fibrillar and granular regions but without preference for any of these two areas.
Figure 2.
Nucleolar localization of PML in MG132-treated MCF-7 cells. High magnification image of double staining for fibrillarin and PML or for B23 and PML. (a) Phase-contrast field combined with fibrillarin staining (red). (b) Phase-contrast and nucleolar PML (green) staining representing the same field as shown in a. (c) Overlap between fibrillarin and PML staining. (d) Phase-contrast field combined with nucleolar B23 staining (red). Panels e [phase-contrast and nucleolar PML staining (green)] and f overlap between B23 and PML.
Nucleolar Localization of PML Is Accompanied by Translocation of Other POD-Related Proteins.
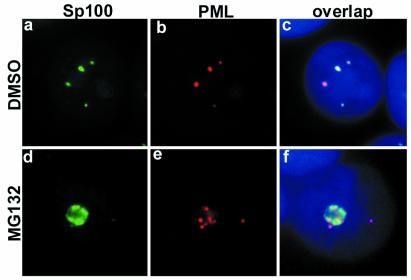
PML protein forms a shell of the PODs. Several distinct cellular proteins are localized to the PODs. Among them, Sp100 was the first protein to be described. To study whether the dislocation of PML from the PODs affected the subcellular localization of Sp100, we double-stained PML and Sp100 in MG132-treated MCF-7 cells. In untreated cells, PML and Sp100 were tightly colocalized, as expected (Fig. 3 a–c). In MG132-treated cells, Sp100 accumulated in the nucleoli, in addition to some single dots in the nucleoplasm (Fig. 3 d–f). Sp100 staining was clearly localized to the nucleoli without obvious colocalization with PML.
Figure 3.
Sp100 changes subcellular distribution and disassociates from PML upon MG132 treatment of MCF-7 cells. High magnification of Sp100 (green) and PML (red) double-staining of DMSO (a–c) or MG132-treated MCF-7 cells (d–f). (c and f) Overlap of Sp100 and PML. In DMSO-treated cells, Sp100 colocalized to a large extent with PML (c). After MG132 treatment, Sp100 completely changed its localization (d and f); it accumulated in the nucleoli along with PML but without any colocalization (f). DNA staining in blue.
EBNA-5 also colocalizes with PML in the PODs of lymphoblastoid cell lines such as IB-4 (18). Recently, we showed that EBNA-5 translocated to the nucleoli after treatment with the proteasome inhibitor MG132 (19). To study the EBNA-5 and PML colocalization under these conditions, IB-4 cells were treated with 5 μM MG132 for 6 h. Both EBNA-5 and PML accumulated in the nucleoli in addition to a remaining number of nuclear dots, where the two proteins colocalized. The effect of EBNA-5 on PML translocation to the nucleoli was studied using transient transfection of MCF-7 cells with GFP-EBNA-5, followed by MG132 treatment for 6 h. The presence of EBNA-5 did not affect the nucleolar accumulation of PML in MG132-treated cells (data not shown).
Subcellular Distribution of PML in Relation to Proteasomes and SUMO-1.
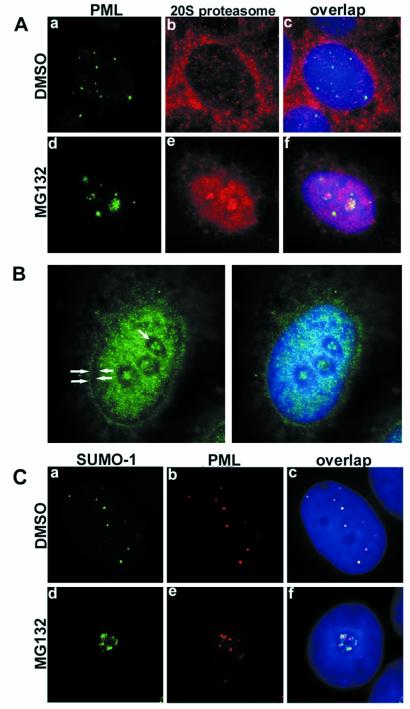
Proteasomes are distributed both in the nucleus and the cytoplasm of eukaryotic cells (20). We studied the subcellular distribution of proteasomes in relation to PML protein in untreated and MG132-treated MCF-7 cells. Immunostaining of proteasomes in untreated cells showed a preferentially cytoplasmic distribution in addition to weak nuclear staining (Fig. 4A, b). Double staining of PML and proteasomes did not indicate any colocalization between these proteins (Fig. 4A, c). In proteasome inhibitor-treated MCF-7 cells, the proteasomes accumulated in the nucleus and in the nucleolus (Fig. 4A, e). Proteasomes and PML were often found in the same nucleoli, showing distinct staining patterns (Fig. 4A, f). The extranucleolar, nucleoplasmic distribution of the proteasomes in the treated cells was not homogeneous, but clearly spared 0.3 to 0.5-μm wide perinucleolar areas along with a comparable wide rim at the nuclear border corresponding to the peripheral and perinucleolar heterochromatin (Figs. 4 A and B and 6).
Figure 4.
(A) Double staining of PML (green) and proteasomes (red) in DMSO- or MG132-treated MCF-7 cells. In DMSO-treated cells, proteasomes were homogeneously distributed throughout the nucleus excluding nucleoli (b). Double staining showed no obvious colocalization between the two proteins (c). MG132 treatment changed the nuclear distribution of both PML and proteasomes; they both accumulated in the nucleoli as shown in panel (d–f). (B) Subnuclear localization of proteasomes in MG132-treated MCF-7 cells. Proteasomes (green) accumulate in the euchromatin areas and nucleoli and avoid peripheral (solid arrowheads) and perinucleolar (concave arrowheads) heterochromatin. (C) a–c represent double staining for SUMO-1 (green) and PML (red) in DMSO-treated MCF-7 cells. Overlap image shows complete colocalization of the two proteins (c). Upon MG132 treatment, SUMO-1 and PML accumulated in the nucleoli (d and e) without colocalization. DNA staining in blue.
Figure 6.
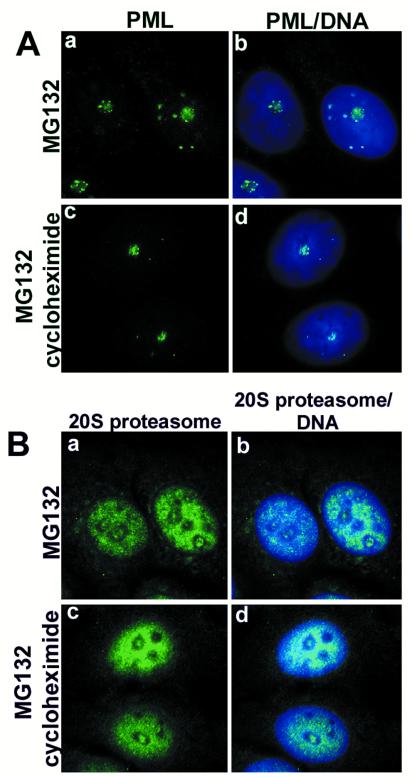
Effect of cycloheximide on the nucleolar accumulation of PML and proteasomes. (A) MCF-7 cells that were treated with MG132 alone (a and b) or with MG132 plus cycloheximide (c and d) over night did not change the nucleolar accumulation of PML (green). (B) The nucleolar accumulation of proteasomes (green) was not altered by the inhibition of protein synthesis in MCF-7 cells (a–d). DNA staining in blue.
The ubiquitin homologue SUMO-1 (small ubiquitin-like modifier protein) is another component of the PODs. SUMO-1 conjugation of PML is necessary for POD formation (21). In the untreated MCF-7 cells, SUMO-1 was distributed in nuclear dots that fully colocalized with PML (Fig. 4C, a–c). The distribution of SUMO-1 changed dramatically in MG132-treated cells. It accumulated in the nucleolus together with PML (Fig. 4C, d–e). However, the distribution pattern of the two proteins was distinct. The highly resolved image analysis showed that PML and SUMO-1 were not colocalizing in the nucleoli (Fig. 5).
Figure 5.
Three-dimensional stereoscopic reconstitution of PML (red) and SUMO-1 (green) double-stained cell (MCF-7) treated with MG132 showing that although both proteins accumulated in the nucleolus, they do not colocalize with each other any longer. The mathematically deblurred image was generated from a series of 11 optical sections, 0.3 μm apart. DNA staining in blue.
Cycloheximide Has No Effect on the Nucleolar Accumulation of PML and Proteasomes.
Next, we wanted to investigate whether the nucleolar accumulation of PML and proteasomes corresponded to newly synthesized proteins or was due to the translocation of nucleoplasmic proteins. Treatment of MCF-7 cells with MG132 alone or MG132 plus cycloheximide (to block protein synthesis) over night showed the same effect on the nucleolar accumulation of PML and proteasomes (Fig. 6 A and B). This indicates that the change of location was due to translocation of proteins already existing in the nucleoplasm.
Discussion
The nucleolus is the site of rRNA synthesis, and it is organized around the ribosomal RNA gene repeats. Ribosomal proteins are imported into the nucleolus where the assembly of ribosomal subunits occurs. Following assembly, the ribosome protein complexes are exported. The nucleolus contains a large number of proteins and is a highly dynamic structure with a constant flow of RNA and proteins. The nucleoli are also involved in the processing and export of certain mRNAs and in the processing and chemical modification of some tRNA precursors (22). The activity of such proteins as p19ARF, Mdm2, p53, and yeast Cdc14p is partly regulated by nucleolar sequestration. By preventing these proteins from reaching their targets in other regions of the cell, the nucleolus might function as a general sequestration site (23). Here, we show that PML, SUMO-1, Sp100, and proteasomes localized to the nucleoli in the proteasome inhibitor-treated cells, suggesting that nucleoli may be involved in the regulation of proteasome-dependent protein degradation. The nucleolar accumulation may also reflect a natural turnover of these proteins that involves trafficking through the nucleoli.
It is becoming more apparent that the PODs are associated with proteasomal degradation of ubiquitinated proteins. The PODs and MTOC (microtubule organizing center) have been described as “proteolysis centers” or “aggresomes” to which misfolded proteins are recruited for degradation under normal conditions (24–26). Expression of a mutated form of influenza nucleoprotein that was misfolded and rapidly degraded by proteasomes led to the accumulation of the protein in the PODs and in the MTOC upon inhibition of proteasome activity (26). This was followed by the attraction of proteasomes polyUb and HSP70 to the same structures, suggesting that the PODs may function as a site for proteasomal degradation of ubiquitinated proteins. We have previously shown that a constitutively expressed form of HSP70 localized to the PODs (27). We have recently found that EBV-encoded EBNA-5 that accumulates in the PODs in untreated cells translocated to the nucleolus upon inhibition of proteasome activity (19).
The proteasomes are highly mobile structures. Bleaching experiments using fusion protein of the proteasome subunit LMP-2 and GFP showed that proteasomes move quickly within the nucleus and cytoplasm (28). The ability to move suggests that proteasomes are capable to search for their substrates and degrade them. Here, we show that blocking of proteasome activity led to the pronounced accumulation of proteasomes in the nuclei and nucleoli with a concomitant decrease in cytoplasmic localization. This may suggest a relative increase in the amount of ubiquitinated targets in the nucleus as compared with cytoplasm in proteasome inhibitor-treated cells.
In agreement with earlier studies, our experiments showed complete colocalization of PML and SUMO-1 in the PODs in untreated cells. Inhibition of proteasome activity separated them from each other, however. Although both accumulated in the nucleolus, the proteins did not show any obvious colocalization. This prompts us to suggest that nucleolus-localized PML is no longer SUMO-1-conjugated. According to our findings, Sp100 is not colocalized with PML in the nucleoli either. Taken together, our data suggest that the POD components of proteasome inhibitor-treated cells move to the nucleoli together, but the POD structure disassembled.
Treatment of cells with a low concentration of MG132 for 6 h (IB-4) or overnight (HeLa and MCF-7) kept the cells in good condition. Due to the inability of the MG132-treated cells to degrade proteins, we would expect an increase in levels of proteins assigned for degradation. Although the distribution of the POD-associated proteins changed dramatically in proteasome inhibitor-treated cells, the protein levels were not increased or were even decreased, as compared with untreated controls. In contrast, non-POD-associated proteins such as p21, p27, and cyclin D3, known to be degraded by the proteasome pathway, did not change their nuclear distribution but showed a marked increase in staining intensities. This suggests that POD components and proteasomes but not non-POD-associated polyubiquitinated proteasome substrates specifically target the nucleolus when proteasome-dependent protein degradation is blocked. This may reflect an alternative degradation pathway of POD components in the nucleoli.
Acknowledgments
We thank H. de The (Institut d'Hematologie de Universite Paris VII, Paris) for the rabbit serum against PML and Sp100, P. K. Chan (Baylor College of Medicine, Houston) for the B23/nucleophosmin antibody, and E. Tan (Scripps Research Institute, La Jolla, CA) for the fibrillarin antibody. This work was supported by The Swedish Cancer Society, Sweden, and the Cancer Research Institute/Concern Foundation.
Abbreviations
- PML
promyelocytic leukemia gene product
- POD
PML oncogenic domain
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.031566998.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.031566998
References
- 1.Hodges M, Tissot C, Howe K, Grimwade D, Freemont P S. Am J Hum Genet. 1998;63:297–304. doi: 10.1086/301991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seeler J S, Dejean A. Curr Opin Genet Dev. 1999;9:362–367. doi: 10.1016/s0959-437x(99)80054-9. [DOI] [PubMed] [Google Scholar]
- 3.Ciechanover A. EMBO J. 1998;17:7151–7160. doi: 10.1093/emboj/17.24.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Everett R D, Meredith M, Orr A, Cross A, Kathoria M, Parkinson J. EMBO J. 1997;16:566–577. doi: 10.1093/emboj/16.3.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sternsdorf T, Jensen K, Will H. J Cell Biol. 1997;139:1621–1634. doi: 10.1083/jcb.139.7.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muller S, Matunis M J, Dejean A. EMBO J. 1998;17:61–70. doi: 10.1093/emboj/17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duprez E, Saurin A J, Desterro J M, Lallemand-Breitenbach V, Howe K, Boddy M N, Solomon E, de The H, Hay R T, Freemont P S. J Cell Sci. 1999;112:381–393. doi: 10.1242/jcs.112.3.381. [DOI] [PubMed] [Google Scholar]
- 8.Everett R D, Freemont P, Saitoh H, Dasso M, Orr A, Kathoria M, Parkinson J. J Virol. 1998;72:6581–6591. doi: 10.1128/jvi.72.8.6581-6591.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng P, Guo Y, Niu Q, Levy D E, Dyck J A, Lu S, Sheiman L A, Liu Y. Nature (London) 1998;396:373–376. doi: 10.1038/24628. [DOI] [PubMed] [Google Scholar]
- 10.Chelbi-Alix M K, Quignon F, Pelicano L, Koken M H, de The H. J Virol. 1998;72:1043–1051. doi: 10.1128/jvi.72.2.1043-1051.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quignon F, De Bels F, Koken M, Feunteun J, Ameisen J C, de The H. Nat Genet. 1998;20:259–265. doi: 10.1038/3068. [DOI] [PubMed] [Google Scholar]
- 12.Lee D H, Goldberg A L. Trends Cell Biol. 1998;8:397–403. doi: 10.1016/s0962-8924(98)01346-4. [DOI] [PubMed] [Google Scholar]
- 13.Lamond A I, Earnshaw W C. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- 14.Chelbi-Alix M K, de The H. Oncogene. 1999;18:935–941. doi: 10.1038/sj.onc.1202366. [DOI] [PubMed] [Google Scholar]
- 15.Holmvall P, Szekely L. Appl Immunohistochem Mol Morphol. 1999;7:226–236. [Google Scholar]
- 16.Ochs R L, Lischwe M A, Spohn W H, Busch H. Biol Cell. 1985;54:123–133. doi: 10.1111/j.1768-322x.1985.tb00387.x. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt-Zachmann M S, Hugle-Dorr B, Franke W W. EMBO J. 1987;6:1881–1890. doi: 10.1002/j.1460-2075.1987.tb02447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szekely L, Pokrovskaja K, Jiang W Q, de The H, Ringertz N, Klein G. J Virol. 1996;70:2562–2568. doi: 10.1128/jvi.70.4.2562-2568.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pokrovskaja, K., Mattsson, K., Kashuba, E., Klein, G. & Szekely, L. (2000), www.sgm.ac.uk/JGVDirect/17322/17322ft.htm (DOI 10.1099/vir.0.17322-0). J. Gen. Virol. Direct. [DOI] [PubMed]
- 20.Brooks P, Fuertes G, Murray R Z, Bose S, Knecht E, Rechsteiner M C, Hendil K B, Tanaka K, Dyson J, Rivett J. Biochem J. 2000;346:155–161. [PMC free article] [PubMed] [Google Scholar]
- 21.Zhong S, Muller S, Ronchetti S, Freemont P S, Dejean A, Pandolfi P P. Blood. 2000;95:2748–2752. [PubMed] [Google Scholar]
- 22.Pederson T. Nucleic Acids Res. 1998;26:3871–3876. doi: 10.1093/nar/26.17.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olson M O, Dundr M, Szebeni A. Trends Cell Biol. 2000;10:189–196. doi: 10.1016/s0962-8924(00)01738-4. [DOI] [PubMed] [Google Scholar]
- 24.Wojcik C, Schroeter D, Wilk S, Lamprecht J, Paweletz N. Eur J Cell Biol. 1996;71:311–318. [PubMed] [Google Scholar]
- 25.Johnston J A, Ward C L, Kopito R R. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anton L C, Schubert U, Bacik I, Princiotta M F, Wearsch P A, Gibbs J, Day P M, Realini C, Rechsteiner M C, Bennink J R, Yewdell J W. J Cell Biol. 1999;146:113–124. doi: 10.1083/jcb.146.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szekely L, Jiang W Q, Pokrovskaja K, Wiman K G, Klein G, Ringertz N. J Gen Virol. 1995;76:2423–2432. doi: 10.1099/0022-1317-76-10-2423. [DOI] [PubMed] [Google Scholar]
- 28.Reits E A J, Benham A M, Plougastel B, Neefjes J, Trowsdale J. EMBO J. 1997;16:6087–6094. doi: 10.1093/emboj/16.20.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]