Abstract
The REF family of evolutionarily conserved heterogeneous ribonucleoprotein (hnRNP)-like proteins consists of one central RNP-type RNA binding domain flanked by Arg–Gly-rich regions of variable length. Members of this protein family bind directly to RNA and the mRNA export factor TAP/Mex67p, and it has been suggested that they facilitate the recruitment of TAP/Mex67p to cellular mRNPs. We show that the variable regions are necessary for binding of REFs to RNA and to TAP. Antibodies specific to REFs prevent their interaction with RNA in vitro. After microinjection into Xenopus oocytes, these antibodies inhibit mRNA nuclear export. This inhibition of export is observed whether or not the mRNAs are generated by splicing. The antibodies do not interfere with pre-mRNA splicing or with the nuclear export of constitutive transport element (CTE)-containing RNAs (directly mediated by TAP), so REF proteins must play a critical role in mRNA nuclear export, acting downstream of splicing and upstream of TAP/Mex67p. We also show that recombinant REFs stimulate directly the export of mRNAs that are otherwise exported inefficiently. Together, our data indicate that REFs are directly implicated in the export of mRNAs from the nucleus. More generally, we show that spliced and unspliced mRNAs use common export factors to reach the cytoplasm.
The REF proteins belong to a superfamily of RNA binding proteins containing ribonucleoprotein (RNP)-type RNA binding domains (RBD, ref. 1). The distinguishing feature of the REF family is the presence of two highly conserved motifs in their N and C termini: REF-N and REF-C boxes. Between the conserved motifs and the RBD, REF proteins have regions of variable length (N-vr and C-vr), which are related to the RGG boxes present in many heterogeneous RNP (hnRNP) proteins (refs. 1 and 2; see also Fig. 2A).
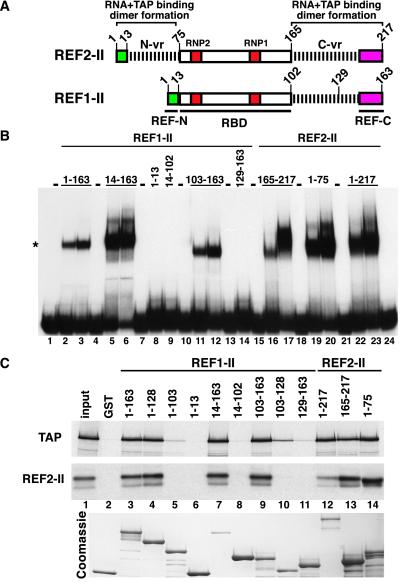
Figure 2.
REF variable regions are required for TAP and RNA binding. (A) Domain organization of REF proteins as described by Stutz et al. (2). RBD with the conserved RNP1 and RNP2 motifs; REF-N and REF-C, conserved N- and C-terminal motifs; N-vr and C-vr represent the N- and C-terminal variable regions specific to each member of the family. Numbers indicate the position in the amino acid sequence. (B) An electrophoretic mobility retardation assay was performed with the purified recombinant proteins indicated above the lanes. Proteins exhibiting RNA binding activity were tested at 10 and 25 ng/μl, whereas proteins without RNA binding activity were added at 100 ng/μl (lanes 8, 9, and 14). The position of the free RNA probe (lane 1) and of the REF/RNA complexes (*) is shown on the left. (C) 35S-methionine-labeled TAP and REF2-II were synthesized in vitro in rabbit reticulocyte lysates or E. coli lysates, respectively. Five-microliter aliquots were incubated with glutathione agarose beads precoated with the recombinant proteins indicated above the lanes. One-tenth of the input and one-quarter of the bound fractions were analyzed by SDS/PAGE followed by Coomassie stain and fluorography.
Yra1p, a Saccharomyces cerevisiae member of the REF family, is an essential nuclear protein first identified from its RNA-annealing activity (3). More recently, Yra1p was shown to be involved in the export of mRNA from the nucleus in yeast cells (2, 4). In mouse, REFs are encoded by at least three different genes (ref 1–3) and differ at multiple positions in the variable regions because of deletions and/or amino acid changes (2). In contrast, all murine REF proteins are 98% identical in the RBD and 100% in the conserved boxes (2). The complexity of the murine subfamily is further increased by the expression of multiple splice variants (2). Murine REF1-II is generated by alternative splicing of REF1-I (also named Aly; see ref. 5) and lacks the N-terminal variable region (see Fig. 2A), whereas murine REF2-I and REF2-II differ by one single amino acid insertion in REF2-I (Q198, ref. 2). REF1-I (Aly) was first identified as a protein interacting with LEF-1, a transcription factor that participates in the regulation of the T-cell receptor α enhancer (5). In this context, it was proposed that Aly facilitates the interaction of multiple proteins in the T-cell receptor α enhancer complex. Subsequently, human REF (also called BEF) was shown to increase transcriptional activation by proteins containing a basic region-leucine zipper DNA binding domain (bZIP, ref. 6). Thus, REFs may participate in multiple steps of mRNA biogenesis including transcription and transport.
REF proteins bind RNA directly and form conserved interactions with members of the NXF family of mRNA export factors including human TAP (also named NXF1) and NXF2 and the yeast homolog of TAP, Mex67p (2, 4, 7). The domains of REFs involved in these interactions have not yet been defined. When splicing reactions are performed in vitro (in HeLa nuclear extracts), human REF associates with mRNAs in a splicing-dependent manner (8, 9), suggesting that REFs may interact with components of the splicing machinery. Several lines of evidence support this hypothesis. First, human REF copurifies with spliceosomes (10). Second, a fraction of human REF localizes to nuclear speckles (9, 11), sites of enrichment of splicing factors (12). Third, human REF is a component of a 335-kDa protein complex deposited by the spliceosome 20–24 nts upstream of a splice junction (8, 13).
In this study, we have defined the domains of REF interacting with RNA and TAP, and we have investigated the role of vertebrate REFs on the export of mRNAs from the nucleus. We show that the variable regions are required for binding of REFs to RNA and TAP. Antibodies specific to REFs, which prevent their binding to RNA in vitro, inhibit mRNA nuclear export when injected into Xenopus laevis oocytes. This export inhibition is observed whether or not the mRNAs have been generated by splicing. We show that microinjection of recombinant REFs stimulates directly the export of mRNAs that are otherwise exported inefficiently. Together, our data indicate that REF proteins play a direct role in the export of mRNAs, most likely by recruiting TAP to mRNP export complexes.
Materials and Methods
DNA Constructs.
Most TAP and REF constructs used in this study have been described (2). REF cDNA fragments were cloned by PCR using the Expand high-fidelity PCR system (Boehringer) with murine REF1-II and REF2-II cDNAs as templates. PCR fragments were cloned into the NcoI–BamHI sites of vector pGEXCS. For expression in HeLa cells, full-length REF1-II and REF2-II cDNAs were excised from the corresponding pGEXCS constructs as NarI–BamHI fragments and inserted into the AccI–BamHI sites of pEGFP-C1 vector (CLONTECH).
Antibodies and Immunoblotting.
Rabbit antibodies were raised against the recombinant RBD of REF1-II (residues 14–102) expressed in Escherichia coli as a glutathione S-transferase (GST) fusion. The antibodies were purified over an affinity column made by coupling the antigen to Affigel-10 (Bio-Rad). For Western blots, the polyclonal antibodies were diluted 1:2,000 in PBS with 5% fat-free milk and 0.1% Triton X-100. Bound primary antibody was observed by enhanced chemiluminescence detection.
GST Pull-Downs and In Vitro RNA Binding Assays.
Gel retardation and GST pull-down assays were performed as described (2). 35S-labeled proteins were synthesized in vitro by using the combined transcription/translation (TnT) kit from Promega. GST fusions were expressed in E. coli BL21(DE3) pLysS and purified as described (15). The amount of recombinant proteins used in each binding reaction is indicated in the figure legends.
X. laevis Oocyte Microinjections.
All DNA templates for in vitro synthesis of labeled RNAs have been described. These were pBSAd1 and Ad-CTE pre-mRNAs (17); dihydrofolate reductase (DHFR) mRNA; histone H4 mRNA; U1ΔSm, U5ΔSm, and U6Δss snRNAs; and human initiator methionyl tRNA (16, 17). AdHML81, Fushi tarazu (Ftz), and β-globin pre-mRNAs have been described (8, 14). Ftz-218 and β-globin-247 cDNAs were kindly provided by Hervé Le Hir (Brandeis University, Waltham, MA) [FluorImager (Fuji, FLA-2000)]. Oocyte injections and analysis of microinjected RNA by denaturing gel electrophoresis and autoradiography were performed as described (17). Quantitation was done by PhosphorImager. The concentrations of antibodies and recombinant proteins in the injected samples are indicated in the figure legends.
Immunofluorescence and Heterokaryon Assays.
HeLa cells were transfected with pEGFP-C1 constructs by using FuGENE6 (Roche). Approximately 20 h after transfection, cells were fixed with 3.7% formaldehyde for 10 min and subsequently permeabilized with 0.5% Triton X-100 for 15 min. Indirect immunofluorescence and heterokaryon assays were performed as described (18).
Results
Antibodies to the RBD Prevent REFs Binding to RNA But Not to TAP.
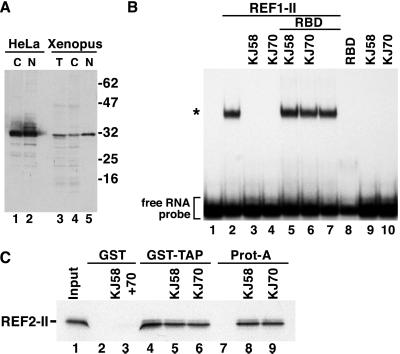
To determine whether vertebrate REFs play a role in mRNA nuclear export, we raised two distinct rabbit antisera (KJ58 and KJ70) against the recombinant RBD of murine REF1-II produced in E. coli. As expected from the conservation of the RBD (2), the antisera recognized both murine Aly and REF2-II obtained by in vitro translation (not shown). When used in Western blots, the antibodies labeled a major band with the expected mobility in HeLa cells and Xenopus oocyte lysates (Fig. 1A). After affinity purification, these antibodies prevented REF1-II binding to an RNA probe in vitro (Fig. 1B, lanes 3 and 4). Similar results were obtained with REF2-II (not shown). The inhibitory effect of the antibodies was relieved by the addition of excess recombinant RBD (Fig. 1B, lanes 5 and 6). The antibodies on their own and the recombinant RBD have no effect on RNA mobility (Fig. 1B, lanes 7–10).
Figure 1.
Anti-REF antibodies prevent REF binding to RNA. (A) Protein samples from HeLa cytoplasmic (C) or nuclear extracts (N), total Xenopus oocytes (T), or cytoplasmic (C) and nuclear (N) fractions were analyzed by Western blot using anti-REF antibodies. (B) An electrophoretic mobility retardation assay was performed with a labeled RNA probe and recombinant GST-REF1-II protein (20 ng/μl). Lane 1, free RNA; lane 2, REF/RNA complexes; lanes 3 and 4, when affinity-purified REF antibodies (KJ58 and KJ70) were included in the reaction, formation of REF/RNA complexes was prevented. The inhibitory effect was abolished if purified GST-RBD was added together with the antibodies to the binding reaction (lanes 5 and 6). Controls show that the RBD fusion and the antibodies on their own have no effect on RNA mobility (lanes 7–10). The concentration of the antibodies in the binding reactions was 0.5 mg/ml and that of GST-RBD 1 mg/ml. The position of the free RNA probe (lane 1) and of the REF/RNA complexes (*) is shown on the left. (C) 35S-methionine-labeled REF2-II was synthesized in vitro in rabbit reticulocyte lysates. Ten-microliter aliquots were incubated with affinity-purified antibodies. After a 20-min incubation period, samples were divided and assayed by binding to glutathione agarose beads precoated with GST-TAP or to protein-A Sepharose beads. Bound fractions were analyzed by SDS/PAGE and fluorography.
To test whether these antibodies also prevent REF binding to TAP, in vitro-translated REF2-II was preincubated with the antibodies. Samples then were halved and assayed for binding to immobilized GST-TAP on glutathione agarose beads or to protein-A Sepharose beads. Preincubation of REF2-II with the antibodies did not prevent its interaction with TAP (Fig. 1C, lanes 4–6), although REF2-II must have been bound to the antibodies because it could be quantitatively selected on protein-A Sepharose beads (Fig. 1C, lanes 8 and 9). Thus, antibodies raised to the RBD of REFs prevent binding to RNA but not to TAP.
The Variable Regions of REFs Are Required for Interaction with RNA and TAP.
To characterize further the effects of the antibodies, we mapped the regions of REFs interacting with RNA and TAP. To this end, we compared the RNA and TAP-binding properties of various protein fragments derived from REF1-II and REF2-II. These proteins exhibit 95% identity in their RBD and 100% in the conserved boxes (ref. 2, Fig. 2A). However, they differ at multiple positions within the C-vr region. Moreover, REF1-II lacks the N-vr region present in REF2-II.
As reported (2), both REF1-II and REF2-II bound to the RNA probe in vitro (Fig. 2B, lanes 2 and 3 and lanes 22 and 23, respectively), indicating that the N-terminal variable region is not strictly required for binding. Consistent with this, we found that all of the REF fragments containing at least one variable region bound RNA (Fig. 2B, lanes 5, 6, 11, 12, 16, 17, 19, and 20). Neither the RBD (fragment 14–102, Fig. 2B, lane 9) nor the conserved boxes (fragments 1–13 or 129–163, Fig. 2B, lanes 8 and 14) exhibited detectable RNA binding activity. This indicates that either of the variable regions, but not the RBD, is required for binding to RNA. Consequently, antibodies raised to the RBD may prevent the variable regions from binding to RNA indirectly, probably by steric hindrance.
To map the TAP binding domain of REFs, 35S-methionine-labeled TAP was synthesized in vitro and assayed for binding to glutathione agarose beads coated with recombinant full-length REFs or various REF fragments fused to GST. TAP could be selected on beads coated with REF1-II, REF2-II (Fig. 2C, lanes 3 and 12), and with all of the REF fragments carrying at least one variable region and a conserved box. This was observed for fragment 103–163 from REF1-II and 1–75 or 165–217 from REF2-II (Fig. 2C, lanes 9, 13, and 14). Curiously, it was not necessary for the conserved motif and the variable region to be adjacent to each other as fragment 1–128 from REF1-II, having the N-REF box and the C-vr region separated by the RBD, bound TAP (Fig. 2C, lane 4). Neither the RBD (fragment 14–102) nor the conserved boxes (fragments 1–13 or 129–163) exhibited detectable TAP binding activity (Fig. 2D, lanes 6, 8, and 11), but the C-vr region of REF1-II, in the absence of the conserved boxes, bound TAP, albeit with a reduced affinity (fragment 103–128, Fig. 2C, lane 10). Thus, either of the variable regions with at least one conserved box is sufficient for TAP binding. These interactions are specific as the C-terminal domain of TAP (fragment 371–619) did not bind to the REF fragments tested (not shown). Because REFs form multimers in vitro (6) and all of the REF fragments interacting with TAP can form dimers with REF2-II (Fig. 2C Middle), it is unclear whether REF binding to TAP requires dimer formation. In summary, the variable regions are implicated in dimer formation as well as in binding to RNA and to TAP. Due to the lack of structural information, however, it is difficult to predict how these regions participate in these multiple, nonexclusive interactions and how antibodies raised to the RBD interfere with RNA but not with TAP binding.
REF Proteins Mediate Nuclear Export of Unspliced mRNAs.
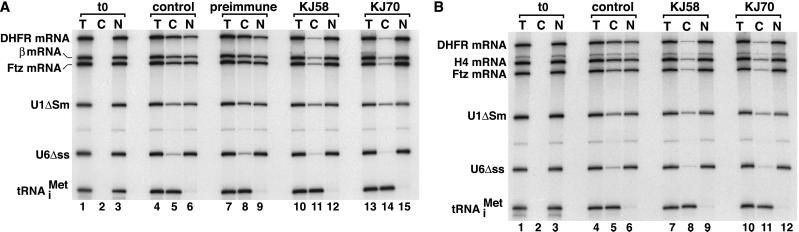
We tested the effect of anti-REF antibodies on the export of various mRNAs that do not naturally contain introns, such as histone H4 mRNA, or that were synthesized by using cDNA as template. Affinity-purified antibodies were injected into Xenopus oocyte nuclei together with a mixture of labeled RNAs. This mixture consisted of DHFR, β-globin, and fushi tarazu (Ftz) mRNAs, U1ΔSm and U6Δss snRNAs, and the human initiator methionyl tRNA (tRNAMet). U6Δss RNA is not exported from the nucleus and serves as an internal control for nuclear injection (19). U1ΔSm RNA is exported normally but, unlike the wild-type U1, is not subsequently reimported into the nucleus. Immediately after injection, all RNAs were nuclear (Fig. 3, lanes 1–3). After a 90-min incubation period, in control oocytes and oocytes injected with affinity-purified preimmune serum, about 50% of the DHFR, Ftz, and β-globin mRNA and 42% of the U1 snRNA were cytoplasmic (Fig. 3A, lanes 4–6 and 7–9). tRNA export was complete (Fig. 3A, lanes 4–9). Microinjection of anti-REF antibodies resulted in significant inhibition of DHFR, Ftz, and β-globin mRNA export, as only 10% of these mRNAs was detected in the cytoplasm (Fig. 3A, lanes 10–15). This effect was specific, as export of tRNA and U1ΔSm RNA was not affected. Fig. 3B shows that the anti-REF antibodies also inhibited the export of histone H4 mRNA. The observation that inhibition of mRNA export could be obtained reproducibly by using antibodies purified from the two different antisera and in several independent experiments (e.g., Fig. 3 and not shown) indicates that these effects are specific. These antibodies prevent formation of REF/RNA complexes in vitro, so we conclude that REF binding to mRNAs is required for their efficient export.
Figure 3.
Anti-REF antibodies inhibit mRNA nuclear export. (A and B) Xenopus oocyte nuclei were injected with affinity-purified anti-REF antibodies along with mixtures of 32P-labeled RNAs, as indicated. As controls, oocytes were injected either with PBS (lanes 4–6) or preimmune serum purified following the same procedure as for the immune serum. RNA samples from total oocytes (T) and cytoplasmic (C) and nuclear (N) fractions were collected immediately after injection (t0, lanes 1–3) or 90 min after injection and analyzed on acrylamide/urea-denaturing gels. One oocyte equivalent of RNA, from a pool of 10 oocytes, was loaded per lane. The concentration of antibodies in the injected samples was 5 mg/ml.
REFs Act Downstream of Splicing and Upstream of TAP.
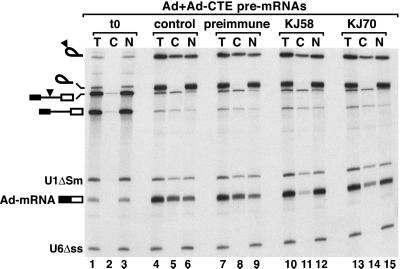
The experiments described above were performed with intronless mRNAs. Because REFs associate preferentially with spliced mRNAs (8, 9), the effects of the antibodies on splicing and export of spliced mRNAs were investigated. We also analyzed the effect of anti-REF antibodies on the export of RNAs bearing the constitutive transport element (CTE) of simian type D retroviruses, which is directly mediated by TAP (15) and thus, in principle, could be exported independently of REFs. Antibodies to REFs were injected into Xenopus oocyte nuclei together with four labeled RNAs. These were U1ΔSm and U6Δss snRNAs and two adenovirus-derived pre-mRNAs. One of these contained the SRV-1 CTE inserted in the intron (17). After a 90-min incubation period, both pre-mRNAs were quantitatively spliced (Fig. 4, lanes 4–15). About 65% of the resulting mRNA (Ad-mRNA) and 40% of the intron lariat bearing the CTE were found in the cytoplasm of both control oocytes and oocytes preinjected with preimmune serum (Fig. 4, lanes 4–9). Coinjection of anti-REF antibodies did not affect splicing efficiency but strongly inhibited the export of the spliced Ad-mRNA, as less than 10–15% of this mRNA reached the cytoplasm (Fig. 4, lanes 10–15). Export of the excised intron lariat bearing the CTE was only slightly reduced (Fig. 4, lanes 10–15). The CTE recruits TAP directly (15), so these results suggest that REFs participate in mRNA export upstream of TAP. The antibodies also inhibited the export of the spliced Ftz and β-globin mRNAs, but to a lesser extent (not shown). We therefore conclude that binding of REFs to spliced mRNAs is required for their efficient export to the cytoplasm as with mRNAs derived from cDNAs.
Figure 4.
REFs act downstream of splicing and upstream of TAP. Xenopus oocyte nuclei were injected with affinity-purified anti-REF antibodies and 32P-labeled pBS-Ad1 and pBSAd1-CTE pre-mRNAs (17) and U1 and U6 snRNAs. As controls, oocytes were injected either with PBS (lanes 4–6) or preimmune serum (lanes 7–9). RNA samples from total oocytes (T) and cytoplasmic (C) and nuclear (N) fractions were collected immediately after injection (t0, lanes 1–3) or 90 min after injection and analyzed as in Fig. 3. The mature products and intermediates of the splicing reaction are indicated diagrammatically on the left. The filled triangle represents the CTE.
REFs Stimulate mRNA Export Directly.
To investigate whether REFs can stimulate mRNA export directly, Xenopus oocyte nuclei were coinjected with purified recombinant REF2-II and a mixture of various cDNA-derived mRNAs that differ in export efficiency. U5ΔSm snRNA was used as a control because U1ΔSm snRNA comigrates with Ad-mRNA. After a 90-min incubation period, about 50% of the DHFR, Ftz, and β-globin mRNA moved to the cytoplasm (Fig. 5A, lanes 4–6). As reported (14), the unspliced Ad-mRNA was not efficiently exported (16% export, Fig. 5A, lanes 4–6). Coinjection of recombinant REF2-II, however, resulted in significant stimulation of Ad-mRNA export, as about 67% of this mRNA was detected in the cytoplasm (Fig. 5A, lanes 7–9). Nuclear exit of the efficiently exported mRNAs (e.g., DHFR, Ftz, and β-globin) was only slightly or not further stimulated, suggesting that REFs are not limiting for the export of these mRNAs. Export of tRNA and of U5ΔSm RNA was not affected (Fig. 5A, lanes 7–9). Only full-length REF2-II stimulates Ad-mRNA export, whereas its N-terminal, C-terminal, or RBD domains have no stimulatory effect when injected at the same concentration (Fig. 5A, lanes 10–18). REF1-II also stimulates Ad-mRNA export up to 50% (not shown).
Figure 5.
REF2-II stimulates mRNA export directly. (A and B) Xenopus oocyte nuclei were injected with mixtures of 32P-labeled RNAs and purified recombinant proteins as indicated. RNA samples from total oocytes (T) and nuclear (N) and cytoplasmic (C) fractions were collected immediately after injection (t0, lanes 1–3) or 90 min after injection and analyzed as in Fig. 3. The concentration of recombinant proteins in the injected was 14 μM. On the left of B, the numbers in brackets indicate the size of the transcripts. The stimulation of Ad-mRNA export obtained in three separate experiments was quantitated and expressed as fractional stimulation relative to the export activity in the absence of recombinant REF2-II. The mean value was (3.6 ± 0.4)-fold stimulation of export.
To find out whether REFs could stimulate export of other mRNAs, we decreased the export rate of β-globin and Ftz and mRNAs by reducing the length of the transcripts from 360 and 343 nts to 248 and 218 nts, respectively. Fig. 5B shows that REF2-II stimulates the export of the shortened mRNAs (lanes 7–9). The effect was more dramatic on the export of Ad-mRNA and Ftz-218 mRNA, which are exported least efficiently.
REFs Shuttle Independently of mRNA.
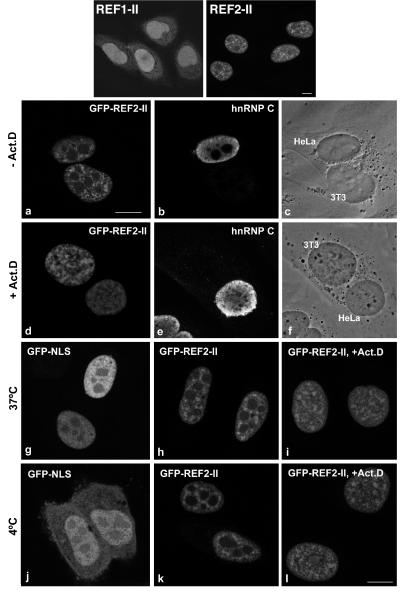
The results described above indicate that REFs are general export factors for mRNA. To investigate whether these proteins are exported in association with mRNAs, we monitored shuttling of REFs fused to green fluorescent protein (GFP) in human–mouse heterokaryons in the presence of the transcription inhibitor actinomycin D. First, the subcellular localization of REF1-II and REF2-II fused to GFP was investigated. GFP-tagged REF2-II exhibited a subcellular distribution similar to that of human REF (9, 11). The staining was widespread in the nucleoplasm with sites of highest concentration in speckled domains. REF1-II, which lacks the N-vr region, was widely distributed throughout the nucleus but did not concentrate in speckles (Fig. 6 Upper) and also was detected in the cytoplasm. Because of its cytoplasmic localization, REF1-II shuttling could not be analyzed.
Figure 6.
REFs shuttle independently of mRNA. (Upper) Subcellular localization of GFP-REF1-II or GFP-REF2-II in transfected HeLa cells. (a–f) HeLa cells expressing GFP-REF2-II were cocultured with mouse 3T3 cells and treated for 3 h with 20 μg/ml emetine in the absence (a–c) or presence (d–f) of actinomycin D (5 μg/ml), as indicated. After polyethylene glycol-induced fusion, the cells were incubated in medium containing the same inhibitors for 1 h. The resulting heterokaryons were observed by phase-contrast microscopy (c and f) and double-labeled with anti-hnRNP C antibodies (b and e). (g–l) HeLa cells were transfected with GFP-NLS or GFP-REF2-II. After 18–20 h, the cells were treated for 3 h with emetine in the presence or absence of actinomycin D. The cells then were incubated in media containing the same drugs for another 3 h at either 37°C or 4°C. Transcription inhibition in cells treated with actinomycin D can be seen by the absence of nucleolar exclusion of the hnRNP C and REF staining. (Bar, 10 μm.)
HeLa cells expressing GFP-REF2-II were cocultured with mouse 3T3 cells and treated with inhibitors of transcription and protein synthesis. After polyethylene glycol-induced fusion, the resulting heterokaryons were further incubated in medium containing both inhibitors. The non-shuttling hnRNP C protein was retained in the human nucleus (Fig. 6 b and e), whereas GFP-REF2-II was transported from the human into the mouse nucleus irrespective of the presence of actinomycin D (Fig. 6 a and d). This indicates that REF2-II does not need to associate with mRNA to transit to the cytoplasm.
To investigate whether REF2-II is exported by diffusion or requires an energy-driven mechanism, we performed a temperature-shift assay as described by Michael et al. (20). The rationale of this assay is that at 4°C, receptor-mediated nuclear transport is blocked, but diffusion is not affected. As a positive control for diffusion, we used GFP fused to the classical nuclear localization signal (NLS) from simian virus 40 large T antigen. GFP-NLS is exclusively nuclear at 37°C (Fig. 6g). At low temperature, GFP-NLS diffuses out of the nucleus, and, as NLS-mediated import is blocked, the protein progressively accumulates in the cytoplasm (Fig. 6j). In contrast, GFP-REF2-II is localized exclusively in the nucleus at both 37°C (Fig. 6h) and 4°C (Fig. 6k). Incubation of actinomycin D-treated cells at 4°C confirms that the protein does not diffuse passively to the cytoplasm when transcription is inhibited (Fig. 6 i and l). Thus, export of REF2-II is temperature sensitive and involves a carrier-mediated pathway. Moreover, its export is independent of mRNA traffic as shuttling of REF2-II occurs in the presence of transcription inhibitors such as actinomycin D.
Discussion
We investigated the role of REF proteins in nuclear export of mRNA and have presented evidence that binding of these proteins is required for efficient export irrespective of splicing. Because REF proteins are not important for CTE-dependent export (which is directly mediated by TAP), these proteins may function upstream of binding of TAP. REFs shuttle actively between the nucleus and cytoplasm (9), but this shuttling is independent of ongoing RNA synthesis.
Function of Conserved Domains Within REF Proteins.
Our investigation of the evolutionarily conserved domains within REF proteins has revealed that the RBD has no detectable RNA binding activity, nor is it important for TAP binding in vitro. The conservation of this domain and the observation that only full-length REFs can stimulate mRNA export efficiently (Fig. 5) indicate that the RBD must be important for function. Alternatively, the RBD may have a structural role critical for the proper folding and/or stabilization of interactions involving the flanking N- and C-terminal domains. These domains, each consisting of a conserved box and a variable region, are sufficient for REF binding to RNA and TAP and for dimer/multimer formation. Although these domains appear to be redundant for these activities, they are not equivalent, as only the N-vr region carries an NLS and targets vertebrate REFs to nuclear speckles (Fig. 6 and unpublished results).
REFs belong to a family of proteins having at least two members in different eukaryotic species, including S. cerevisiae. The variable regions differ between family members within each species, so it is possible that different REFs have different protein and/or RNA-binding specificities and specific functions. In yeast, only Yra1p is essential, but overexpression of Yra2p complements a YRA1 deletion (D. Zenklusen and F. Stutz, personal communication), so these proteins have overlapping functions. In vertebrates, alternative splicing also may alter the properties and function of specific REFs. Indeed, REF1-II, generated by alternative splicing of REF1-I (Aly), lacks the N-vr region. As a consequence, this protein is not targeted to nuclear speckles and is also detected in the cytoplasm (Fig. 6).
REFs Shuttle Independently of mRNA Export.
The observation that REF shuttling is independent of mRNA export does not exclude the possibility that REFs escort cellular mRNAs to the cytoplasm and have a cytoplasmic function, as do many hnRNP proteins (21). Shuttling of REFs is as efficient as shuttling of hnRNP A1, as 50% of the GFP–REF2-II signal was detected within the mouse nucleus of the heterokaryon within the 1-h postfusion incubation period. As with REFs, other RNA binding proteins such as hnRNP A1 (21), poly(A) binding protein II (18), and TAP (unpublished observations) shuttle independently of mRNA trafficking. One explanation for the shuttling of the RNA-free proteins is that binding to RNA may be required for their efficient release from import receptors in the nucleus. In the absence of RNA binding, these proteins may not dissociate efficiently from the import receptor and may engage in futile import/export cycles. This may provide a mechanism for regulating the levels and/or timing of the availability of export factors in the nucleus (21, 22).
Export of Spliced Versus cDNA-Derived mRNAs.
Previously, Luo and Reed (14) reported that in Xenopus oocytes, mRNAs generated by splicing of pre-mRNAs are efficiently exported, whereas mRNAs produced by transcription of the corresponding cDNAs are poor substrates for export. We have confirmed these observations for the mRNA derived from the major late region of adenovirus, which is not exported efficiently unless it is produced by splicing (ref. 14 and Figs. 4 and 5). In contrast, the Ftz and β-globin mRNAs used in this study are exported irrespective of prior splicing in vivo. Our export kinetics for DHFR mRNA are also more rapid than those reported by Lou and Reed (14), but are in agreement with those reported by several other laboratories (16, 23–26). These results indicate that some cDNA-derived mRNAs can be recognized as export substrates in Xenopus oocytes and can be exported efficiently. The export rate of the unspliced Ftz and β-globin mRNAs can be decreased by reducing the length of these transcripts, suggesting that the export efficiency of intronless mRNAs depends primarily on their ability to recruit export factors, in turn likely to be determined by the sequence and/or length of the mRNA. Inefficient export may reflect a reduced affinity for export factors, i.e., export factors are limiting for these particular substrates, so export can be stimulated by microinjection of excess of REFs (Fig. 5) or TAP (unpublished results). Consistently, excess of REFs does not stimulate the nuclear exit of efficiently exported mRNAs.
REFs are components of a multiprotein complex that binds upstream of splice junctions in a splicing-dependent manner (8). Thus, splicing guarantees binding of REFs on the spliced mRNA independently of its sequence (8, 9). This may provide a rationale for the observation that the expression of specific genes in mammalian cells is strongly stimulated by the presence of an intron, whereas the mRNA transcribed from the corresponding cDNA is expressed poorly (ref. 14 and references therein).
In conclusion, in vivo, REF proteins associate with spliced mRNAs in a sequence-independent manner (8, 9), whereas their association with unspliced mRNAs may be influenced by the sequence of the mRNA and/or the availability of the proteins. Based on the observation that anti-REF antibodies and excess CTE RNA (17, 25) inhibit the export of mRNAs, regardless of splicing, we propose that nuclear export of spliced or cDNA-derived mRNAs involves common factors including REFs and most likely TAP.
Acknowledgments
The technical assistance of Qiyu Li is gratefully acknowledged. We thank Maarten Fornerod, Hervé Le Hir, Iain W. Mattaj, and Melissa Moore for critical reading of the manuscript. We are grateful to Gideon Dreyfuss, Hervé Le Hir, Lynne Maquat, Melissa Moore, and Françoise Stutz for communicating results before publication. This study was supported by the Junta Nacional de Investigação Científica e Técnologica (Program PRAXIS XXI) Portugal and the European Molecular Biology Organization. B.J.B. was supported by grants from the Medical Research Council of Canada and the U.S. Breast Cancer Research Program.
Abbreviations
- RNP
ribonucleoprotein
- hnRNP
heterogeneous RNP
- GST
glutathione S-transferase
- GFP
green fluorescent protein
- CTE
constitutive transport element
- RBD
RNA binding domain
- DHFR
dihydrofolate reductase
- NLS
nuclear localization signal
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.031586198.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.031586198
References
- 1.Burd C G, Dreyfuss G. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 2.Stutz F, Bachi A, Doerks T, Braun I C, Séraphin B, Wilm M, Bork P, Izaurralde E. RNA. 2000;6:638–650. doi: 10.1017/s1355838200000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Portman D S, O'Connor P, Dreyfuss G. RNA. 1997;3:527–537. [PMC free article] [PubMed] [Google Scholar]
- 4.Sträβer K, Hurt E. EMBO J. 2000;19:410–420. doi: 10.1093/emboj/19.3.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruhn L, Munnerleyn A, Grosschedl R. Genes Dev. 1997;11:640–653. doi: 10.1101/gad.11.5.640. [DOI] [PubMed] [Google Scholar]
- 6.Ching-Man A, Wagner S, Green M R. Mol Cell. 1999;4:219–228. doi: 10.1016/s1097-2765(00)80369-x. [DOI] [PubMed] [Google Scholar]
- 7.Herold A, Suyama M, Rodrigues J-P, Braun I C, Kutay U, Carmo-Fonseca M, Bork P, Izaurralde E. Mol Cell Biol. 2000;20:8996–9008. doi: 10.1128/mcb.20.23.8996-9008.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le Hir H, Izaurralde E, Maquat L E, Moore M J. EMBO J. 2000;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou Z, Luo M-J, Strasser K, Katahira J, Hurt E, Reed R. Nature (London) 2000;407:401–405. doi: 10.1038/35030160. [DOI] [PubMed] [Google Scholar]
- 10.Neubauer G, King A, Rappsilber J, Calvio C, Watson M, Ajuh P, Sleeman J, Lamond A, Mann M. Nat Genet. 1998;20:46–50. doi: 10.1038/1700. [DOI] [PubMed] [Google Scholar]
- 11.Wichmann I, Garcia-Lozano J R, Respaldiza N, Gonzalez-Escribano M F, Nuñez-Roldan A. Hum Immunol. 1999;60:57–62. doi: 10.1016/s0198-8859(98)00085-8. [DOI] [PubMed] [Google Scholar]
- 12.Misteli T, Spector D L. Curr Opin Cell Biol. 1998;10:323–331. doi: 10.1016/s0955-0674(98)80007-0. [DOI] [PubMed] [Google Scholar]
- 13.Le Hir H, Maquat L E, Moore M J. Genes Dev. 2000;14:1098–1108. [PMC free article] [PubMed] [Google Scholar]
- 14.Luo M-J, Reed R. Proc Natl Acad Sci USA. 1999;96:14937–14942. doi: 10.1073/pnas.96.26.14937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grüter P, Tabernero C, von Kobbe C, Schmitt C, Saavedra C, Bachi A, Wilm M, Felber B K, Izaurralde E. Mol Cell. 1998;1:649–659. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- 16.Jarmolowski A, Boelens W, Izaurralde E, Mattaj I W. J Cell Biol. 1994;124:627–635. doi: 10.1083/jcb.124.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saavedra C A, Felber B K, Izaurralde E. Curr Biol. 1997;7:619–628. doi: 10.1016/s0960-9822(06)00288-0. [DOI] [PubMed] [Google Scholar]
- 18.Calado A, Kutay U, Kühn U, Wahle E, Carmo-Fonseca M. RNA. 2000;6:245–256. doi: 10.1017/s1355838200991908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vankan P, McGuigan C, Mattaj I W. EMBO J. 1992;11:335–342. doi: 10.1002/j.1460-2075.1992.tb05056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michael M W, Choi M, Dreyfuss G. Cell. 1995;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- 21.Michael M W. Trends Cell Biol. 2000;10:46–50. doi: 10.1016/s0962-8924(99)01695-5. [DOI] [PubMed] [Google Scholar]
- 22.van der Houven van Oordt W, Diaz-Meco M T, Lozano J, Krainer A R, Moscat J, Caceres J F. J Cell Biol. 2000;149:307–316. doi: 10.1083/jcb.149.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kataoka N, Yong J, Kim V N, Velazquez F, Perkinson R A, Wang F, Dreyfuss G. Mol Cell. 2000;6:673–682. doi: 10.1016/s1097-2765(00)00065-4. [DOI] [PubMed] [Google Scholar]
- 24.Fischer U, Huber J, Boelens W C, Mattaj I W, Lührmann R. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 25.Pasquinelli A E, Ernst R K, Lund E, Grimm C, Zapp M L, Rekosh D, Hammarskjold M L, Dahlberg J E. EMBO J. 1997;16:7500–7510. doi: 10.1093/emboj/16.24.7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ullman K S, Shah S, Powers M A, Forbes D J. Mol Biol Cell. 1999;10:649–664. doi: 10.1091/mbc.10.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]