Abstract
A miniaturized approach was developed for quantitative permethylation of oligosaccharides, which involves packing of sodium hydroxide powder in microspin columns or fused-silica capillaries (500 μm i.d.), permitting effective derivatization in less than a minute at microscale. Prior to mass spectrometry, analytes are mixed with methyl iodide in dimethyl sulfoxide solution containing traces of water before infusing through the microreactors. This procedure minimizes oxidative degradation and peeling reactions and avoids the need of excessive clean-up. Picomole amounts of linear and branched, sialylated and neutral glycan samples were rapidly and efficiently permethylated by this approach and analyzed by matrix-assisted laser desorption/ionization mass spectrometry.
Whereas structural aspects of oligosaccharides have been studied by mass spectrometry (MS) for many years, the development of matrix-assisted laser desorption/ionization (MALDI) and electrospray ionization (ESI) MS for this class of compounds has accelerated substantially the acceptance of MS-based methodologies during the last decade. In the structural analysis of complex glycans originating from various isolated glycoproteins,1–12 MALDI-MS in conjunction with exoglycosidase digestion and a tandem (MS/MS) operation have become particularly popular.13
Although MALDI-MS structural analysis of most glycans can basically be performed in their native forms, there are several reasons for conversion of such compounds into their methylated derivatives. These include an easy determination of branching, interglycosidic linkages and the presence of configurational and conformational isomers. Permethylation also stabilizes the sialic acid residues in acidic oligosaccharides, yielding more predictable ion products when subjected to MS/MS experiments. Moreover, methylated sugars deem to ionize more efficiently than their native counterparts. In conjunction with ESI and collision-induced dissociation (CID), permethylated sugars also yield a most detailed structural information.14–20
Most permethylation procedures, employed over a number of years in carbohydrate analysis, are derived from two successful methodologies. The first, originally described by Hakomori,21 utilizes the dimethyl sulfoxide anion (DMSO−, commonly refereed to as ‘dimsyl anion’) to remove protons from the sample analyte molecules prior to their replacement with methyl groups. The second, and currently more widespread approach, introduced in 1984 by Ciucanu and Kerek,22 is based on the addition of methyl iodide to DMSO containing powdered sodium hydroxide (NaOH). The original procedure was modified more recently.23 The current popularity of the modified procedure stems from its rapidity, experimental simplicity, ‘cleaner’ reaction products,24 and the effectiveness for replacing protons at both oxygen and nitrogen sites in oligosaccharides. This methylation procedure has now been used for derivatization of carbohydrates,22,25,26 other polyols,27 and fatty acids, including their hydroxyl derivatives.28 However, it has gained a particular popularity with complex oligosaccharides and glycolipids.
Although the previously described procedures for methylation of sugars have now been used successfully in various MS structural studies, they appear less satisfactory when approaching the range of low picomole to femtomole quantities of glycoprotein samples. This is primarily due to oxidative degradation and peeling reactions associated with the high pH resulting from dissolving NaOH powder prior to liquid-liquid extractions. These side reactions are adversely prominent with small samples (low picomole to femtomole). In order to reach reproducibly the low glycoprotein amounts in modern glycoanalysis of biological fluids and tissues, we have thoroughly investigated the derivatization conditions leading to suitable MALDI-MS/MS of such derivatives in this work.
The principal aim of this study has been the development of an on-line permethylation procedure for neutral and acidic oligosaccharides, leading to the use of capillary reactors packed with powdered NaOH. Highly quantitative conversion of the minute quantities of glycoprotein-derived glycans is demonstrated under optimized operational parameters, including flow rate, concentration of methyl iodide, and the length of reactor capillary. This microscale permethylation procedure is compatible with the previously reported microscale glycan release methods11,29–31 in glycoprotein analysis. The new derivatization procedure has been quantitatively compared with the widely used method,22,23 as well as a version using a small spin column.
EXPERIMENTAL
Materials
Maltoheptaose, pancreatic bovine ribonuclease B, fetuin from fetal calf serum, human α1-acid glycoprotein, proteomicsgrade trypsin, PNGase F, tris-(hydroxymethyl)amino-methane hydrochloride (Tris-HCl), ethylenediaminetetraacetic acid (EDTA), and ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA) were obtained from Sigma (St. Louis, MO, USA). The MALDI matrix, 2,5-dihydroxybenzoic acid (DHB), and NaOH were purchased from Aldrich (Milwaukee, WI, USA). Chloroform, methyl iodide, and sodium chloride were received from EM Science (Gibbstown, NJ, USA). Dithiothreitol (DTT), iodoacetamide, and 3-([3-cholamidopropyl]dimethylammonio)-1-propane-sulfonate (CHAPS) were purchased from Bio-Rad Laboratories (Hercules, CA, USA). Ammonium bicarbonate was received from Mallinckrodt Chemical Company (Paris, KY, USA), and sodium pyrophosphate was obtained from J.T. Baker, Inc. (Phillipsburg, NJ, USA). Acetonitrile (ACN) and hydrochloric acid solution N/10 were purchased from Fisher Scientific (Fair Lawn, NJ, USA).
Permethylation
For both the spin-column and capillary derivatization versions, the amount of NaOH used was the same, aside from the experiments involving the optimization of the capillary length. All permethylation conditions, including sample amount, amount of methyl iodide, and water/chloroform sample extraction, were maintained constant to ensure a valid quantitative comparison.
In-solution permethylation
Maltoheptaose and all N-glycans derived from glycoproteins were permethylated according to the procedure of Ciucanu and Costello.23 Briefly, methyl iodide, a trace of water, and NaOH powder were suspended in DMSO and mixed for 10 min at room temperature. Typically, 1–10 μg sample were suspended in 30 μL of DMSO, to which 3.6 mg of NaOH powder, 0.3 μL of water and 5.6 μL of methyl iodide were added.
Spin-column permethylation
Spin columns obtained from Harvard Apparatus (Holliston, MA, USA) were packed with NaOH mesh beads (Fig. 1(A)). First, NaOH beads were suspended in ACN, thus preventing atmospheric moisture absorption. Next, NaOH beads were packed in the spin column to about 3-cm depth. Prior to sample application, the spin column was spun down at 1000 rpm for 2 min. The NaOH-packed spin column was then washed several times with DMSO.
Figure 1.
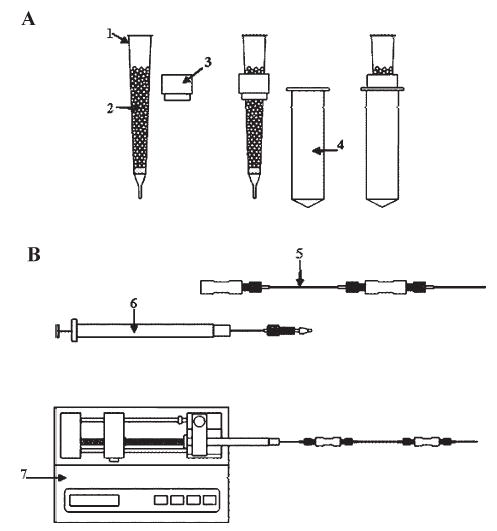
Schematic representation of the different permethylation techniques. (A) Spin-column setup: (1) spin column; (2) NaOH beads; (3) column holder; (4) micro-centrifuge tube; (B) 500 μm i.d. fused-silica capillary setup: (5) 500 μm i.d. fused-silica capillary; (6) syringe; (7) syringe pump.
The ratios of DMSO, methyl iodide, water, and the used sample were the same as in the in-solution permethylation. The sample, methyl iodide, and trace of water were mixed right before being applied to the spin column. Next, the sample was passed through, utilizing a low spin speed (1000 rpm) while it was collected.
Capillary permethylation
Fused-silica capillaries (500 μm i.d.) from Polymicro Technologies (Phoenix, AZ, USA) were used. Tubes, nuts and ferrules from Upchurch Scientific (Oak Harbor, WA, USA) were employed to assemble the capillary set. To protect the packing material from moisture, NaOH powder was suspended in ACN immediately after crushing the beads. The powdered NaOH in ACN was then packed inside 500 μm i.d. fused-silica capillaries by using pressure. A 100-μL Hamilton syringe and a syringe pump from KD Scientific, Inc. (Holliston, MA, USA) were employed to introduce a sample solution into the capillary. The capillary setup was assembled after packing the capillary with NaOH and ACN. Then, DMSO was infused into the packed capillaries with NaOH to replace ACN prior to analysis (Fig. 1(B)).
Capillary permethylation employed the same chemical ratios used in the case of in-solution permethylation. Samples were prepared in DMSO, containing methyl iodide and a trace of water. Next, the samples were infused through the packed capillary at an appropriate flow rate by means of a syringe pump, while they were collected at the capillary end. The removal of DMSO was accomplished by the use of chloroform as described for the conventional permethylation approach.
Extraction of permethylated samples in conventional procedure
Permethylated glycans and maltoheptaose were extracted with chloroform and washed repeatedly with water. For the in-solution permethylation, ice-cold water was added to the derivatization mixture and placed in an ice-bath prior to the addition of chloroform. The mixture was then vortexed for several minutes. The aqueous layer was then discarded and the chloroform layer washed repeatedly with water. The pH of the aqueous layer was continuously monitored with pH indicators, while 5-fold washing with water was deemed sufficient to eliminate residual NaOH, any side products and excess methyl iodide.
Extraction of glycoproteins from tissue and release of their N-glycans
The N-glycans were enzymatically released from ribonuclease B, fetuin, and α1-acid glycoprotein using PNGase F. This enzymatic release was performed according to our previously published procedure.11 Briefly, individual glycoproteins or a mixture of the three model glycoproteins were suspended in 10 mM sodium phosphate buffer (pH 7.5) containing 0.1% mercaptoethanol. The sample was thermally denatured by incubation at 95°C for 5 min. Next, the sample was allowed to cool to room temperature prior to the addition of 5 mU of PNGase F. Finally, the reaction mixture was subsequently incubated for 3 h at 37°C.
Although there are many lysis buffers that are commercially available and commonly utilized in proteomics, none of them seems suitable for a glycomic analysis following protein and glycoprotein extractions. Accordingly, we have first focused on developing a lysis buffer that is efficient and suitable for the extraction of proteins and glycoproteins without interfering with the MALDI-MS analysis of released glycans. The total proteome sample was extracted by suspending a homogenized tissue in the lysis buffer composed of 20 mM Tris-HCl (pH 7.5), 150 mM sodium chloride, 1 mM disodium EDTA, 1 mM EGTA, 2.5 mM sodium pyrophosphate and 0.5% CHAPS. Next, the mixture was sonicated for 15 min and shaken for 1 h at 4°C. The extract was then centrifuged at 30 000 rpm for 1.5 h at 4°C. The supernatant layer containing a cytosolic part of the proteome was then reduced and alkylated prior to the addition of trypsin. Briefly, the extracted total proteome sample was suspended in 100 μL of 100 mM ammonium bicarbonate buffer solution, to which 40 μL of 10 mM DTT solution were added and incubated at 56°C for 45 min. After cooling, 40 μL of 55 mM iodoacetamide prepared in 100 mM ammonium bicarbonate buffer solution were added to the mixture and incubated at room temperature for 30 min in the dark. Next, trypsin was added to the reduced and alkylated mixture, continuing incubation at 37°C for 18 h. The action of trypsin was quenched through heating the reaction mixture at 95°C for 10 min. Then, 5 mU of PNGase F were added to the reaction mixture (to release N-glycans) and incubated at 37°C overnight. Peptides were eliminated from the mixture by passing the reaction mixture over a C18 cartridge, while collecting the eluent. Finally, the collected eluent containing released N-glycans was dried under vacuum and subsequently permethylated.
MALDI spotting
The dried permethylated sample was resuspended in 50:50 methanol/water solution, containing 2.5 mM sodium acetate, to promote complete sodium adduct formation in MALDI-MS. The sample was then spotted directly on the MALDI plate and mixed with an equal volume of the DHB matrix, which was, in turn, prepared by suspending 10 mg of DHB in 1 mL of 50:50 water/methanol solution to produce a 10 mg/mL matrix solution. The sample spot was then dried under vacuum to ensure uniform crystallization. Native maltoheptaose was utilized as an internal standard in the case of the optimization studies. The intensities of the permethylated oligosaccharides were reported as relative intensities to that of the internal standard.
Instrumentation
An Applied Biosystems 4700 proteomics analyzer (Applied Biosystems, Framingham, MA, USA) was utilized in this study. This instrument is equipped with an Nd:YAG laser with 355-nm wavelength. MALDI spectra were acquired in the positive-ion mode. MS data were further processed using DataExplorer 4.0 (Applied Biosystems).
RESULTS AND DISCUSSION
Optimization studies
An optimum chemical derivatization of analytes should modify quantitatively the required functional groups, yielding preferably a single, stable reaction product. Moreover, such an analyte modification should be fast, simple, and applicable to minute sample quantities. The last requirement is particularly crucial for the glycans derived from trace quantities of glycoproteins. Over the years, solid-phase derivatization has been particularly recognized as an effective approach at small sample scale.32 Two different versions of solid-phase treatment, a spin-column derivatization and a capillary reactor derivatization (Fig. 1), have been explored in this work and compared with the standard solution technique.22,23
In comparing different permethylation approaches, several variables affecting permethylation efficiency were evaluated: sample flow rate (residence time) through the reactor; length of the capillary; and amount of methyl iodide. The optimum conditions utilized for in-solution permethylation were used initially and optimized for the different approaches. Although each parameter was varied while the other parameters were kept constant, no major changes were observed for the studied parameter upon changing the values of the other parameters (data not shown). The spin-column technique involves a very simple procedure, in which the reaction can be completed through repeated passes of the sample over the packed NaOH. Therefore, the optimization experiments were primarily reserved for the capillary reactor technique.
Figure 2 illustrates the effect of sample flow rate on the relative MS intensity of a model permethylated glycan (maltopheptasoe), which is related to a residence time in the capillary or reaction time. As an important criterion for this solid-phase derivatization, the sample and the reagent should have sufficient interaction time to complete the derivatization. In the solid-phase approach, this needed time is considerably shorter than what is needed for the in-solution derivatization where both diffusion and convection are necessary.
Figure 2.
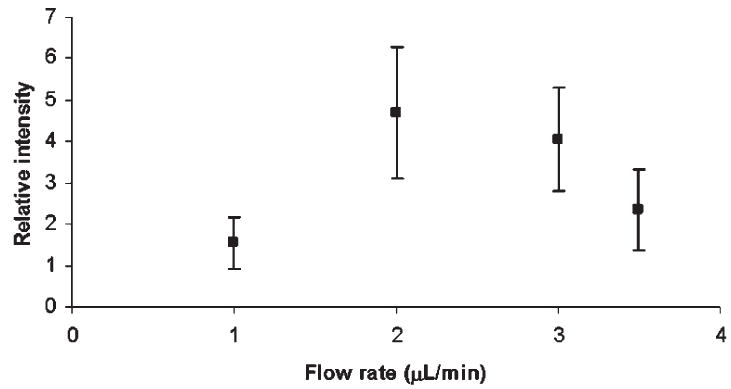
The effect of flow rate on the efficiency of capillary permethylation. Maltoheptaose was used as the analyte, while permethylation was performed with the 500 μm i.d. fused-silica capillary.
As seen in Fig. 2, the derivatization yield was low at 1 μL/min flow, and also at higher flow rates, but it reached optimum at 2–3 μL/min. Apparently, a fast flow through the reactor does not allow for a sufficient interaction at the surface of packed NaOH, while very low flow rates may result in a sample degradation. A prolonged interaction could prompt peeling and oxidative degradation.
In the in-solution technique (where the optimum reaction time was shown to be dependent on the NaOH/DMSO ratio23), a complete derivatization was only achieved at either low or high ratios and long reaction times. In comparison to our solid-phase approach, the low NaOH/DMSO ratio for in-solution permethylation resembles fast flow rates in the capillary permethylation (insufficient interaction time). Conversely, high NaOH/DMSO ratios in the in-solution approach are similar to a slow flow rate in the capillary permethylation, where the amount of NaOH and reaction time remain unchanged. Thus, the flow rate here ‘substitutes’ for the NaOH/DMSO ratio, as the permethylation efficiency depends decidedly on flow rate. The flow rate of 3 μL/min (near optimum) was chosen in further optimization studies described below.
The relative intensities of MALDI-MS peaks for heptaose were seen as relatively independent of the reactor capillary length at a constant residence time (Fig. 3). Obviously, the reaction time was sufficient with a length of 6–10 cm. The analyte residence time in a 12-cm long capillary is twice that in a 6-cm capillary, while the permethylation yield is similar. Figure 3 also seems to indicate smaller standard deviations at a shorter length. Accordingly, 8-cm long capillaries were utilized for the rest of this study.
Figure 3.
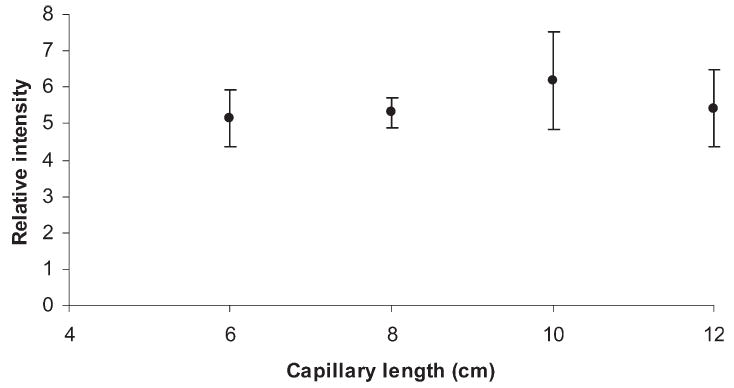
The effect of capillary length on the efficiency of capillary permethylation. Maltoheptaose was used as the analyte, while permethylation was performed using the 500 μm i.d. fused-silica capillary at 3 μL/min flow rate.
Finally, the effect of the amount of methyl iodide on permethylation efficiency was evaluated, as illustrated in Fig. 4. This evaluation was conducted for both linear oligosaccharides and branched N-glycans derived from glycoproteins. For the linear oligosaccharide permethylation (Fig. 4(A)), the efficiency slightly decreased as the amount of methyl iodide increased. However, this decrease was within the error of measurement variation, indicating no substantial effect for a linear oligosaccharide.
Figure 4.
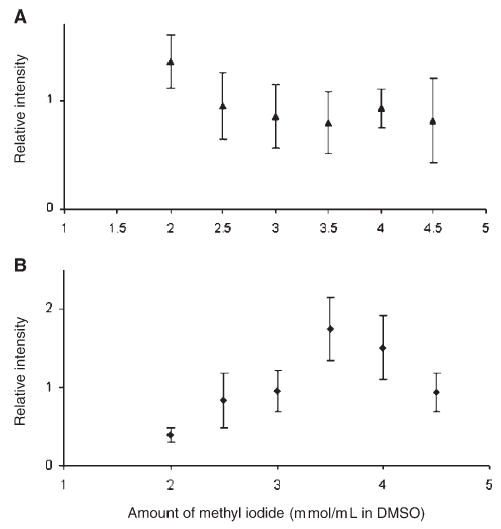
The amount of methyl iodide vs. efficiency of capillary permethylation for (A) maltoheptaose and (B) an N-glycan (m/z 2792) derived from α1-acid glycoprotein. Permethylation was performed using the 500 μm i.d. fused-silica capillary at 3 μL/min flow rate.
The situation was totally different for the more complex N-glycans, as illustrated in Fig. 4(B) with a branched glycan. Here, the optimum permethylation efficiency was attained at 3.5 mmol/mL of methyl iodide. Lower permethylation efficiencies were observed at lower and higher methyl iodide concentrations. Apparently, complex N-glycans require more methyl iodide than their linear counterparts. This is in agreement with the results of in-solution permethylation, since the optimum amount of methyl iodide for the solution technique is a function of reaction time and the NaOH/DMSO ratio.23 A faster in-solution permethylation is achieved using more methyl iodide. In the case of capillary permethylation, the reaction time is kept constant, so the amount of methyl iodide influences permethylation efficiency. Complex glycans require more methyl iodide than linear oligosaccharides owing to the steric hindrance due to branching.
The durability of the NaOH-packed capillaries for multiple and extended use was also investigated. Permethylation of maltoheptaose was performed at the end of every day during which the capillary was utilized extensively to permethylate different samples. The capillary was continuously flushed with DMSO when not in use. No noticeable loss in permethylation efficiency was observed after 7 days of continuous use. Although the study was not extended beyond this period, the possibility exists for using the same capillary for an even longer time.
Next, the efficiency of capillary permethylation was then compared directly to the spin-column technique and the standard procedure.23 We used N-glycans derived from fetuin for this comparison, as depicted in Fig. 5. Permethylation using NaOH powder packed in 8-cm (500 μm i.d.) capillaries clearly provided the best permethylation results, while the use of NaOH beads packed in a spin column was more efficient than in-solution permethylation method, but not nearly as efficient as the capillary method. This approach is clearly superior to the in-solution permethylation method, since it eliminates excessive sample handling or its degradation, resulting from the use of extremely basic aqueous solution, which may induce peeling reactions or oxidative degradation of glycans. This extreme condition seems completely avoided when NaOH is packed in capillaries or a spin column. The spin-column technique showed high permethylation efficiency, while the reaction was complete in less than 1 min. However, the capillary permethylation procedure is even better, being fast and highly effective for small amounts of sample. For every permethylated glycan case investigated in our laboratory, the capillary reactor technique was far superior to the in-solution procedure. While a solution permethylation of the N-glycan mixture derived from 0.1 μg of glycoprotein amounts was not feasible, the very same sample showed meaningful and reproducible results through capillary permethylation. A satellite peak preceding each peak in Fig. 5 is less than 5% intensity relative to the main peak and corresponds to underpermethylated glycans. The satellite peaks observed after the main peaks are due to adduct formation and impurities.
Figure 5.
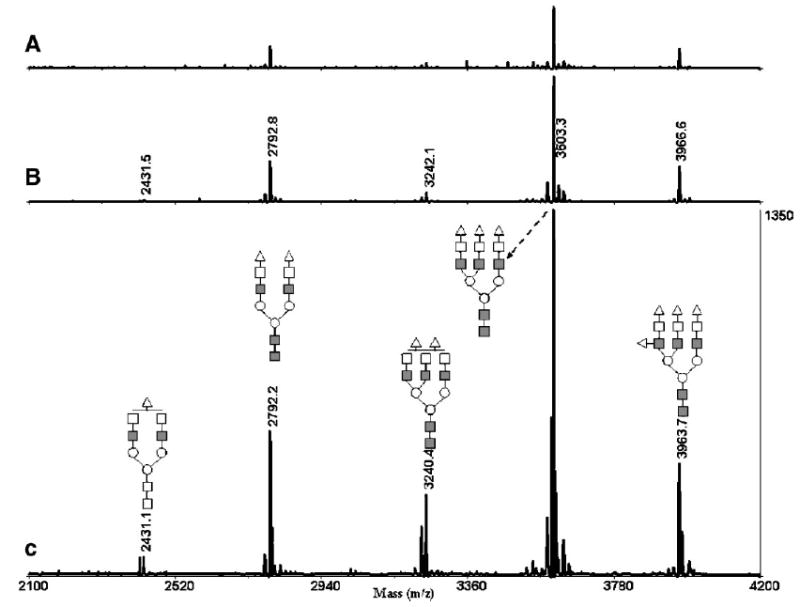
MALDI-TOFMS profile of permethylated N-glycans derived from 0.5 μg of fetuin using (A) the conventional in-solution method; (B) the microspin column; and (C) the 500 μm i.d. fused-silica capillary. Symbols: ■, N-acetylglucosamine; ○, mannose; □, galactose; ▵, N-acetylneuraminic acid.
Applications to glycomic analysis
After securing that various model glycans (neutral, branched, and sialylated structures) were rapidly and effectively permethylated using the capillary reactor technique at picomole levels, we moved to the high-mannose N-glycans derived from ribonuclease B. The glycoprofile of Man5 through Man9 structures is shown in Fig. 6. The relative peak intensities of all structures in this profile are reflective of the typical abundance,33 now seen in many laboratories, suggesting that efficient permethylation occurred with all glycans in this sample type.
Figure 6.
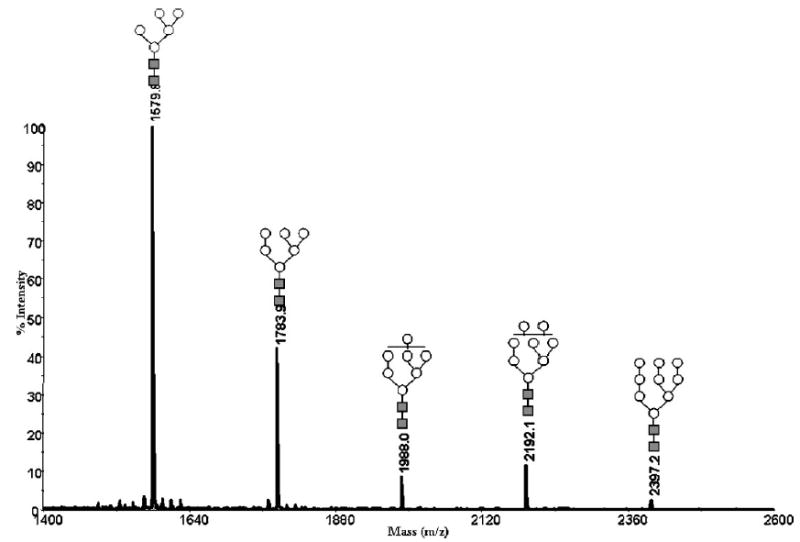
MALDI-TOFMS profile of permethylated N-glycans derived from 0.5 μg of ribonuclease B using the 500 μm i.d. fused-silica capillary. Symbols: ■, N-acetylglucosamine; ○, mannose.
The capillary permethylation procedure was also effective for the derivatization of sialylated glycans, as illustrated in Figs. 5 and 7 for the sialylated N-glycans derived from bovine fetuin and human α1-acid glycoprotein, respectively. Once again, the relative intensities of the permethylated sialylated glycans derived from both glycoproteins reflect their expected content. A mixture of N-glycans derived from 0.5 μg of a mixture of ribonuclease B, fetuin, and α1-acid glycoprotein was also derivatized successfully (Fig. 7). Thus, the ability to use the capillary permethylation approach is clearly effective for treatment of a heterogeneous mixture of glycans derived from different glycoproteins. Moreover, this form of permethylation is also very effective for derivatizing trace amounts of glycans, as deduced from the ability to permethylate glycans derived from submicrogram amounts of glycoproteins.
Figure 7.
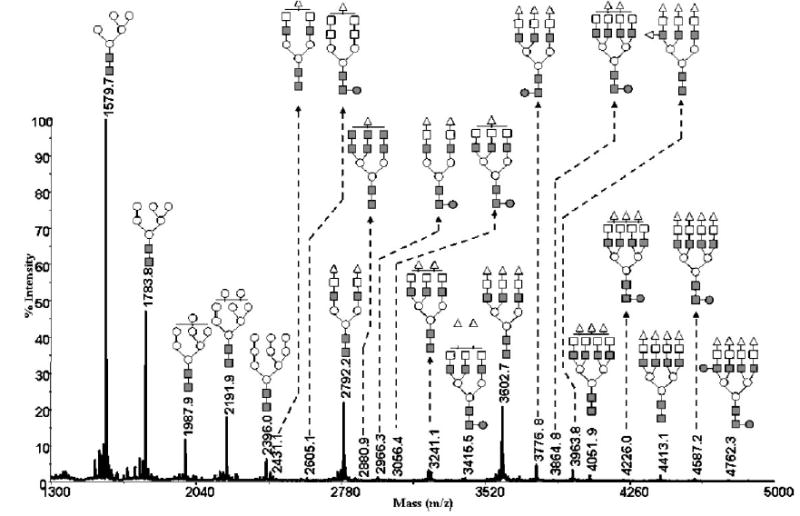
MALDI-TOFMS profile of permethylated N-glycans derived from a 0.5 μg mixture of ribonuclease B, fetuin, and α1-acid glycoprotein (same amount) using the 500 μm i.d. fused-silica capillary. Symbols: ■, N-acetylglucosamine; ○, mannose; □, galactose; ●, fucose; ▵, N-acetylneuraminic acid.
The ability to use the capillary permethylation for a highly heterogeneous mixture is further illustrated for the total glycome derived from rat liver tissue. This will be important in the future creation of glycomic maps to compare the content of different biological samples. Permethylation of the total glycome derived from rat liver tissue was achieved using the capillary permethylation procedure, as demonstrated in Fig. 8. In conjunction with the accurate representation of glycans derived from model glycoproteins (Figs. 5–7), this preliminary result with a complex biological mixture suggests utility of the described permethylation procedure for future studies in functional glycomics.
Figure 8.
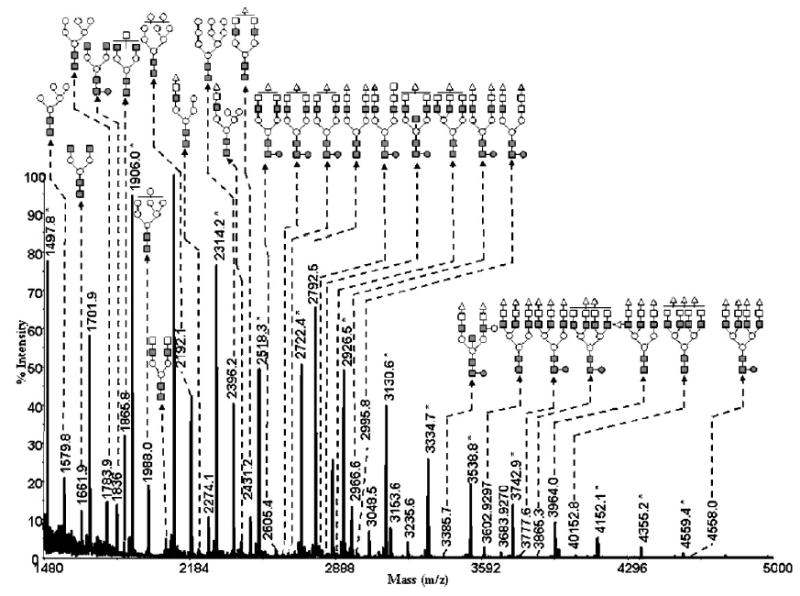
MALDI-TOFMS glycomic profile of permethylated N-glycans derived from 20 mg of rat liver tissue by using the 500 μm i.d. fused-silica capillary. Symbols: ■, N-acetylglucosamine; ○, mannose; □, galactose; ●, fucose; ▵, N-acetylneuraminic acid; ▴, N-glycolylneuraminic acid. Signals marked with asterisks originate from glucose ladders that are commonly present in tissue samples.
CONCLUSIONS
Permethylation of neutral and acidic glycans for MS investigations has been substantially improved through the utilization of NaOH-packed capillaries as reactors. This permethylation technique is facile, rapid and reproducible across the range of different glycan structures. The procedure has been optimized to work with submicrogram amounts of glycoproteins. The results presented here suggest its utility in the structural investigations of glycoproteins isolated through chromatographic fractionation and electrophoresis. It may also prove useful in developing quantitative glycomic maps from unfractionated biological materials for functional glycomic studies pertaining to different diseases.
Acknowledgments
This work was supported by Grant No. GM24349 from the National Institute of General Medical Sciences, US Department of Health and Human Services, and a center grant from the Indiana 21st Century Research and Technology Fund. The mass spectrometer used in this study was acquired as a result of support from Indiana Genomics Initiative (INGEN), which is funded in part by the Lilly Endowment, Inc.
References
- 1.Harvey DJ. Methods Mol Biol. 1996;61:243. doi: 10.1385/0-89603-345-7:243. [DOI] [PubMed] [Google Scholar]
- 2.Harvey DJ, Bateman RH, Green MR. J Mass Spectrom. 1997;32:167. doi: 10.1002/(SICI)1096-9888(199702)32:2<167::AID-JMS472>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 3.Harvey DJ, Rudd PM, Bateman RH, Bordoli RS, Howes K, Hoyes JB, Vikers RG. Org Mass Spectrom. 1994;29:753. [Google Scholar]
- 4.Huang Y, Mechref Y, Novotny MV. Carbohydr Res. 2000;323:111. doi: 10.1016/s0008-6215(99)00254-2. [DOI] [PubMed] [Google Scholar]
- 5.Huang Y, Mechref Y, Tian J, Gong H, Lennarz WJ, Novotny MV. Rapid Commun Mass Spectrom. 2000;14:1233. doi: 10.1002/1097-0231(20000730)14:14<1233::AID-RCM16>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 6.Kuster B, Naven TJ, Harvey DJ. J Mass Spectrom. 1996;31:1131. doi: 10.1002/(SICI)1096-9888(199610)31:10<1131::AID-JMS401>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 7.Landberg E, Huang Y, Stromqvist M, Mechref Y, Hansson L, Lundblad A, Novotny MV, Pahlsson P. Arch Biochem Biophys. 2000;377:246. doi: 10.1006/abbi.2000.1778. [DOI] [PubMed] [Google Scholar]
- 8.Mechref Y, Baker A, Novotny MV. Carbohydr Res. 1999;313:145. doi: 10.1016/s0008-6215(98)00264-x. [DOI] [PubMed] [Google Scholar]
- 9.Mechref Y, Chen P, Novotny MV. Glycobiology. 1999;9:227. doi: 10.1093/glycob/9.3.227. [DOI] [PubMed] [Google Scholar]
- 10.Mechref Y, Ma W, Hao G, Novotny MV. Biochem Biophys Res Commun. 1999;255:451. doi: 10.1006/bbrc.1999.0176. [DOI] [PubMed] [Google Scholar]
- 11.Mechref Y, Novotny MV. Anal Chem. 1998;70:455. doi: 10.1021/ac970947s. [DOI] [PubMed] [Google Scholar]
- 12.Mechref Y, Zidek L, Ma W, Novotny MV. Glycobiology. 2000;10:231. doi: 10.1093/glycob/10.3.231. [DOI] [PubMed] [Google Scholar]
- 13.Mechref Y, Novotny MV. Chem Rev. 2002;102:2693. doi: 10.1021/cr0103017. [DOI] [PubMed] [Google Scholar]
- 14.Muhlecker W, Gulati S, McQuillen DP, Ram S, Rice PA, Reinhold VN. Glycobiology. 1999;9:157. doi: 10.1093/glycob/9.2.157. [DOI] [PubMed] [Google Scholar]
- 15.Reinhold VN, Sheeley DM. Anal Biochem. 1998;259:28. doi: 10.1006/abio.1998.2619. [DOI] [PubMed] [Google Scholar]
- 16.Sheeley DM, Reinhold VN. Anal Chem. 1998;70:3053. doi: 10.1021/ac9713058. [DOI] [PubMed] [Google Scholar]
- 17.Viseux N, de Hoffmann E, Domon B. Anal Chem. 1997;69:3193. doi: 10.1021/ac961285u. [DOI] [PubMed] [Google Scholar]
- 18.Viseux N, de Hoffmann E, Domon B. Anal Chem. 1998;70:4951. doi: 10.1021/ac980443+. [DOI] [PubMed] [Google Scholar]
- 19.Weiskopf AS, Vouros P, Harvey DJ. Rapid Commun Mass Spectrom. 1997;11:1493. doi: 10.1002/(SICI)1097-0231(199709)11:14<1493::AID-RCM40>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 20.Weiskopf AS, Vouros P, Harvey DJ. Anal Chem. 1998;70:4441. doi: 10.1021/ac980289r. [DOI] [PubMed] [Google Scholar]
- 21.Hakomori SI. J Biochem (Tokyo) 1964;55:205. [PubMed] [Google Scholar]
- 22.Ciucanu I, Kerek F. Carbohydr Res. 1984;131:209. [Google Scholar]
- 23.Ciucanu I, Costello CE. J Am Chem Soc. 2003;125:16213. doi: 10.1021/ja035660t. [DOI] [PubMed] [Google Scholar]
- 24.Gunnarson ALF. Glyconjugate J. 1987;4:239. [Google Scholar]
- 25.Ciucanu I, Konig W. J Chromatogr A. 1994;685:166. [Google Scholar]
- 26.Ciucanu I, Luca C. Carbohydr Res. 1990;206:731. [Google Scholar]
- 27.Ciucanu I, Gabris P. Chromatographia. 1987;23:574. [Google Scholar]
- 28.Ciucanu I, Kerek F. J Chromatogr. 1984;286:179. [Google Scholar]
- 29.Huang Y, Konse T, Mechref Y, Novotny MV. Rapid Commun Mass Spectrom. 2002;16:1199. doi: 10.1002/rcm.701. [DOI] [PubMed] [Google Scholar]
- 30.Huang Y, Mechref Y, Novotny MV. Anal Chem. 2001;73:6063. doi: 10.1021/ac015534c. [DOI] [PubMed] [Google Scholar]
- 31.Palm A, Novotny MV. Rapid Commun Mass Spectrom. 2005;19:1730. doi: 10.1002/rcm.1979. [DOI] [PubMed] [Google Scholar]
- 32.Rosenfeld JM. J Chromatogr A. 1999;843:19. doi: 10.1016/s0021-9673(99)00056-4. [DOI] [PubMed] [Google Scholar]
- 33.Fu D, Chen L, O’Neill RA. Carbohydr Res. 1994;261:173. doi: 10.1016/0008-6215(94)84015-6. [DOI] [PubMed] [Google Scholar]


