Abstract
The RET gene, encoding a receptor tyrosine kinase, is unusual among human protooncogenes in that its mutant alleles are implicated in a developmental defect involving enteric neurons as well as in tumorigenesis. The cells affected in both types of disorders are derived from the neural crest. Targeted disruption of mouse ret has revealed an additional role in kidney development. Here we report the analysis of a ret homolog in Drosophila melanogaster, an arthropod with no neural crest. Drosophila ret (D-ret) encodes a protein of 1,235 amino acids that has all of the domains identified in the vertebrate ret, including a cadherin motif. During embryogenesis, D-ret mRNA is first detected in the yolk sac at the late gastrula stage. In the postgastrula, D-ret is expressed in the foregut neurons, excretory system, peripheral ganglia, and the central nervous system. Thus, despite the wide divergence of early embryonic fate maps between vertebrates and invertebrates, D-ret is expressed in cells that are presumed to be the functional equivalents of the ret-expressing cells in vertebrates. Unexpectedly, D-ret is also expressed in the imaginal islands of the endodermal gut. These cells are proliferation-competent precursors for adult midgut that are diffusely embedded in the growth-arrested juvenile gut. These ret-expressing nonneuronal cells are strikingly analogous to vertebrate enteric neurons in their topography, but not in their cell fate. Our finding suggests a previously unrecognized phylogenetic relationship between the ret-expressing cells in vertebrates and the precursor reserves of metamorphosing insects.
Genes that have been recognized to play instructive roles in embryonic development in animals are often found mutated or deregulated in human tumors (1). The potential relationship between morphogenesis and oncogenesis has been suggested particularly strongly by the case of the ret protooncogene.
Loss-of-function mutations of human RET have been implicated in Hirschsprung's disease, a developmental disorder characterized by the partial absence of enteric neurons in the gut (2, 3). On the other hand, excess-of-function RET mutations are associated with multiple endocrine neoplasia type 2 (MEN2), an inherited cancer syndrome characterized by carcinomas of the thyroid C cells as well as occasional pheochromocytomas of the adrenal and neuromas of the enteric neurons (4–6). The allelic series of RET mutations found in these diseases suggests that the RET signal, required for normal morphogenesis, can be accidentally converted to a signal that leads to tumor outgrowth.
Targeted disruption of ret in mice results in perinatal lethality and has revealed both an essential role in innervating the entire length of the gastrointestinal track and an additional role in kidney development (7). These are features not immediately suggested by the human studies, because the null mutation of ret is homozygous lethal and the phenotypes of the Hirschsprung patients, who are heterozygotes, likely reflect a high degree of dosage sensitivity to human RET (8). The cells affected in MEN2 tumors and Hirschsprung's disease are derived from the neural crest, which is a population of cells diffusely migrating during embryogenesis.
Molecular probes used to study mammalian embryos detected ret expression in the sympatho-enteric subgroup of the neural crest and in the ureteric bud of the developing kidney (9–11), in good agreement with the mutant phenotypes. In addition, ret expression was observed in parts of the central nervous system (CNS) and the peripheral nervous system (PNS), but most of these cells develop normally in the homozygous mutants (7).
ret encodes a transmembrane tyrosine kinase that, in association with another polypeptide, serves as the receptor for glial cell line-derived neurotrophic factor (GDNF) and other members of the GDNF family, which forms a subgroup in the transforming growth factor β superfamily (12–15). GDNF was discovered as a potent survival factor for central dopaminergic and motor neurons in culture (16, 17). However, in GDNF-deficient mice, major morphological defects were found in enteric innervation and kidney formation, whereas the CNS neurons were unaffected (18–20). This result has demonstrated that the obligatory requirement of GDNF/Ret in organogenesis resides outside the CNS. GDNF expression is found adjacent to ret expression throughout the body, supporting the idea that GDNF and Ret mediate histotype-specific communications between juxtaposed tissues (13, 18–21). In the developing kidney, for example, GDNF appears to be an element of the mesenchyme-derived signal that promotes growth and branching of the ureteric bud (7, 18–20).
To gain broader insight into the role of ret, we analyzed a ret homolog in the fruit fly, Drosophila melanogaster. The Drosophila embryo has no neural crest or enteric neurons in the endodermal gut. Nevertheless, D-ret is expressed in cells that are presumed to be the functional equivalents of the ret-expressing cells in vertebrates. Surprisingly, we also found ret expression in the imaginal cells embedded in the larval gut. These nonneuronal cells are highly analogous to the vertebrate enteric neurons in their position. Our finding indicates a previously unrecognized relationship between the enteric neurons in vertebrates and the precursor reserves in the Drosophila gut. This report also serves to illustrate two ret-expressing structures in the embryo that have received little attention in the recent literature, namely midgut imaginal islands and yolk sac.
Materials and Methods
cDNA Cloning.
A PCR fragment carrying the genomic sequence reported in ref. 22 (positions 1724–2545) was used as a probe to screen an oligo(dT)-primed cDNA library (made from wild-type Drosophila embryos by N. Brown, Wellcome/CRC Institute, Cambridge, U.K.), from which clone N3 was isolated. N3 has the 3′ 2.2 kb of the transcript (nucleotides 2663–4813; all cDNA numberings as in the sequence T1 submitted to GenBank). The same PCR probe was used to isolate clone TCC-1 (carrying nucleotides 1546–4446) from a random-primed library. The 5′ portion of the transcript was isolated as follows: Adaptor-ligated cDNAs were synthesized from embryonic mRNAs by using Marathon Amplification Kit (CLONTECH), on which a PCR was performed with the provided “5′ adapter primer” and a gene-specific primer, drL3 (positions 1603–1575). Longest products of this reaction that migrated as 1.6–1.8 kb in an agarose gel were purified and reamplified with a nested primer set, from which clones L12 and L20 were isolated. TaqGold polymerase (Roche) was used in all PCRs.
Genomic Cloning.
A Drosophila genomic library (Lambda FIXII; Stratagene) was screened. Clones TG-1 and TG-2 were isolated with a 900-bp XhoI–XbaI fragment (positions 3116–4022) of N3; clones gD-2 and gD-3 were isolated with a 0.6-kb probe carrying the 5′-most sequence of TCC-1; and clone M2, with a probe that carried the 5′-most 3 kb of gD-2.
Whole-Mount in Situ Hybridization.
The 4.8-kb full-length D-ret cDNA (a mixture of forms T1 and T2) was transcribed in vitro to generate digoxigenin-tagged antisense RNA probes (23). Hybridization to embryos was performed according to a published procedure (23) with the following modifications. All fixatives contained 4.5% formaldehyde; step 18 in ref. 23 was performed for 25 min (five times, 5 min each); step 19 was for 30 min (three times, 10 min each); hybridization was for 9–10 h only; and step 24 was not omitted. Hybridization to ovaries was performed as described in ref. 24.
Results
Isolation of Drosophila ret.
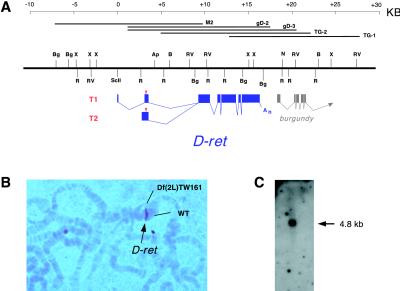
A partial Drosophila genomic clone with an ORF that could encode a ret-like tyrosine kinase was reported by Sugaya et al. (22). However, it remained unknown whether the reported sequence was a part of Drosophila ret, because the genomic clone did not include the sequence for a cadherin motif, a hallmark of Ret (25, 26). Moreover, no cDNA has been identified that corresponds to the reported ORF. To ascertain whether the reading frame encodes a homolog of ret, we isolated cDNAs for two alternatively spliced transcripts, T1 and T2, that correspond to the reported genomic sequence (see Materials and Methods). The cDNAs for T1 and T2 differ only in the 5′ untranslated region, and their lengths, 4.81 and 4.87 kb, respectively, agree well with the observed mRNA size (Fig. 1C). Both clones contain long ORFs preceded by in-frame stop codons, indicating the presence of full coding information. Fig. 1A shows the splicing patterns of T1 and T2, determined with the 35-kb genomic area characterized in this study, which is located within the chromosomal interval of 39B02–39C02 (Fig. 1B).
Figure 1.
The D-ret gene and transcripts. (A) Genomic organization of D-ret. The two alternatively spliced transcripts, T1 and T2, are schematically diagramed below the restriction map. Red triangle, translation initiator codon; An, poly(A). A nearby gene, burgundy, is also indicated. Shown above the restriction map are the genomic phage clones analyzed. (B) Heteroduplex loop formed between wild-type and Df(2L)TW161 chromosomes. D-ret (brown signal) is located at 39B02–39C02. (C) Northern blot analysis with 4 μg of poly(A)+ RNA from 0- to 24-h embryos. D-ret mRNA is detected as a 4.8-kb band.
Comparison of Drosophila and Vertebrate Ret Proteins.
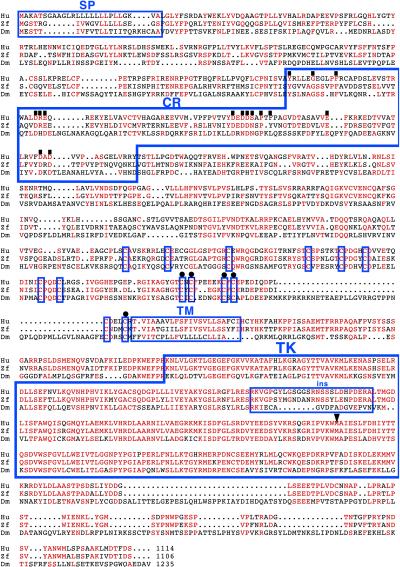
Transcripts T1 and T2 are expected to encode a protein of 1,235 amino acids (Mr = 138,000), which was determined to be Drosophila Ret (D-Ret), on the basis of the following criteria. When searched against the National Center for Biotechnology Information database with the blastp program, the Drosophila Ret sequence showed significantly better score with vertebrate Ret sequences (10−114 with human Ret) than with other receptor tyrosine kinases such as fibroblast growth factor receptor (4 × 10−71 with human fibroblast growth factor receptor). Fig. 2 shows the alignment of Drosophila and vertebrate Ret sequences. The tyrosine kinase domain is highly conserved (52% identity between Drosophila and human; 22). Although long stretches of contiguously conserved residues are not found outside the kinase domain, the domain organization of the proteins is conserved. In addition to a signal peptide and a transmembrane domain, the Drosophila protein has one cadherin repeat (27). It also has a cysteine-rich region immediately N-terminal to the transmembrane domain (22), in which 14 cysteines occur in conserved positions. The five residues mutated in MEN2 type A and the single residue mutated in MEN2 type B (28) are found conserved in the Drosophila protein without exception (Fig. 2). Taken together, these features strongly suggest that this Drosophila gene is a homolog of vertebrate ret.
Figure 2.
Sequence comparison of Drosophila, human, and zebra fish Ret proteins. Identical or chemically similar amino acids are indicated in red. Dm, D. melanogaster; Hu, human; Zf, zebra fish; SP, signal peptide; Cad, cadherin repeat as defined in ref. 27; TM, transmembrane domain; TK, tyrosine kinase domain; ins, insert within the TK domain. Residues conserved between the human Ret and the cadherin superfamily members (26) are indicated with a filled square above them. Immediately N-terminal to TM, conserved cysteine residues in the cysteine-rich area are boxed; the five cysteines mutated in MEN2A are indicated with a dot above them. The methionine residue mutated in MEN2B, at position 918 in the kinase domain, is indicated with an arrowhead above it. Compiled references are in the GenBank entries under the accession numbers P07949 (human Ret) and AAB63283 (zebra fish Ret).
Expression Profile of D-ret.
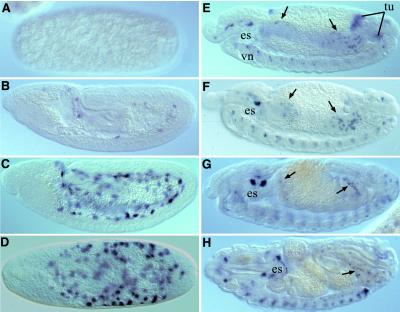
To examine the spatial and temporal pattern of D-ret expression, we performed in situ hybridization on wild-type Drosophila tissues by using a cDNA riboprobe. In the fertilized egg, D-ret expression was first detected in the interior of the egg at the end of gastrulation, as the proctodeum approaches the cephalic furrow (Fig. 3B; stage 8 as defined in ref. 29). During the subsequent stages with momentarily stationary germ-layers (stage 9–11), the D-ret signals develop into intense patches of irregular shape that are located interior to all three germ-layers (Fig. 3C). A dorsal view provides evidence that these signals are scattered throughout the yolk sac, which is an extraembryonic syncytium located at the center of the egg (Fig. 3D; see Discussion); the D-ret signals do not overlap with amnioserosa. D-ret expression in the yolk sac is consistent with the expression of mammalian ret in yolk sac and trophoblasts (9, 30).
Figure 3.
Expression of D-ret in the embryo. Shown are whole-mounts of embryos hybridized with D-ret probe (×80); D-ret mRNA is detected as blue signals. (A) The syncytial blastoderm shows no detectable D-ret signal. (B) D-ret expression begins in the yolk sac at the end of gastrulation. (C) The yolk sac signal becomes stronger at stage 11. Note that the signals are more internal than all embryonic germ-layers. (D) Dorsal view of the stage shown in C. The signal is in the yolk sac, not in the embryo proper (except for the 15–30 cells in the midline neuroectoderm; out of focus). (E) Stage 12. The yolk sac signal clears and de novo expression appears in the SNS anlage [located dorsal to the esophagus (es)], midgut imaginal islands (arrow), malpighian tubule anlage (tu), and in the ventral nerve cord (vn). (F) Stage 13. Malpighian tubule precursors no longer express D-ret. The imaginal island cells migrate, intermingled with the larval midgut cells. (G) The imaginal island cells spread on the developing midgut. (H) A midsagittal optical section of a mature embryo, showing D-ret signals in the head ganglia, in CNS and in some of the midgut island cells that came into focus. For details of organ rudiments, see Fig. 4. Lateral views, anterior left, dorsal up, except in D. Arrows in E–H, midgut imaginal islands.
The yolk sac expression rapidly disappears at stage 12, as the germband retracts and organogenesis begins. At that point, various organ rudiments show de novo expression of D-ret. D-ret is detected dorsal to the esophagus (Fig. 3E) in the anlage for the stomatogastric nervous system (SNS), which consists of foregut-associated ganglia. SNS arises from the epithelial sheets that detach from the mouth rudiment, then migrate posteriorly, settle on the foregut, and differentiate into neurons (31, 32). With no neuron identified on the midgut or hindgut, SNS represents the only gut-associated ganglia in Drosophila. D-ret is expressed in the detaching (Fig. 4A) and migrating (Fig. 4B) SNS cells, as well as in the differentiated SNS neurons (Fig. 4C).
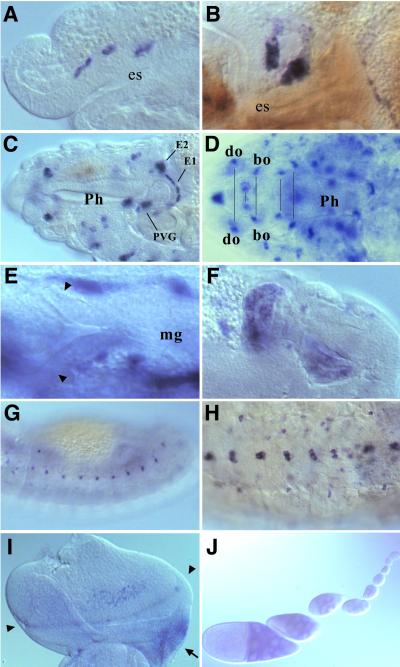
Figure 4.
D-ret expression during organogenesis. In situ hybridization was performed as in Fig. 3. (A) Stage 12. Three invaginations of the stomatogastric nervous system (SNS) anlage express D-ret. (B) Stage 13. SNS cells settling on the foregut. (C) Mature SNS ganglia at stage 16, showing the persistent D-ret expression. EG1 and EG2, first and second esophageal ganglia; PVG, proventricular ganglion (ref. 31). The frontal ganglion of SNS (out of focus) also expresses D-ret. (D) Expression in the cephalopharyngeal ganglia, which appear as paired structures in the head (pairs marked by a line). do, dorsal organ; bo, Bolwig's organ. For the names of the other D-ret-expressing ganglia, see ref. 35. (E) The proctodeum region of a stage 12 embryo. The early malpighian tubule diverticula (arrowheads) do not express D-ret. Pictured signals are the yolk sac expression (see Fig. 3C). (F) Malpighian tubule anlagen transiently show strong D-ret expression at stage 13. Two of the four growing tubules are shown. (G) D-ret expression in the lateral epidermis at stage 13. (H) Ventral neuroectoderm (CNS) expresses D-ret. Stage 16. (I) Eye-antennal imaginal disk of a third-instar larva. D-ret is expressed weakly in the retinal cluster near the morphogenetic furrow (arrowheads). The ocellar region of the presumptive head (arrow) shows strong expression. (J) D-ret expression in germ-line nurse cells in the ovarian follicles, which begins in approximately the fourth follicle from the germarium and gradually increases. However, we were unable to detect D-ret mRNA in the mature oocyte in the egg chamber (not shown) or in the deposited egg at the precellularization stage (Fig. 3A). C and F are composite pictures of two focal planes each. A–C and E–G, lateral views; D, dorsal view; H, ventral view. I, posterior up, medial right.
D-ret expression in SNS is consistent with expression of vertebrate ret in the enteric neurons (9–11), because the SNS is a neural network that controls the peristaltic movement of a gut segment (33). However, the position of the SNS (anterior of the animal) does not agree well with that of the vertebrate enteric neurons (located throughout the gastrointestinal track).
The primordia for malpighian tubules, the fly's excretory system, express D-ret. Malpighian tubules arise as epithelial diverticula from the midgut–hindgut junction, which subsequently grow out in four directions (29). D-ret expression in the malpighian tubule anlagen is highly transient. D-ret is not expressed in the initial diverticula (Fig. 4E), but it is strongly expressed in all four branches in the early growing phase, during which the structure is transformed from “bulges” into “ducts” (stage 12; Fig. 4F), and then is rapidly down-regulated in subsequent stages, during which time the “ducts” elongate extensively to form thin tubules (Fig. 3F). The malpighian tubule expression is consistent with the vertebrate ret expression in the pronephric and mesonephric ducts in fish and mammals; in mammals, ret is also expressed in the ureteric bud (see Discussion and refs. 9 and 34).
D-ret is expressed in the numerous peripheral nervous system (PNS) ganglia located in the head (for descriptions, see ref. 35), including the dorsal organ, epiphysis, pharyngeal chordotonal organ (Fig. 4D), and the terminal organ (not shown). D-ret expression in the CNS is found in a small number of cells in the ventral cord (the main nerve trunk in invertebrates; Fig. 3 E–H; Fig. 4H) and in the brain (not shown). The expression of this gene in the CNS and PNS has been recognized by Sugaya et al. (22). These D-ret-expressing organs are analogous to the vertebrate neural structures that express ret, such as the spinal cord, brain, and sensory and autonomic ganglia (9, 11). D-ret expression was also found in the juvenile photoreceptor organ (Bolwig's organ; Fig. 4D) and the imaginal retina (Fig. 4I), in keeping with the vertebrate ret expression in the eye (9, 11, 34).
Thus, most organs that express D-ret are analogous to vertebrate counterparts in their position and function, even though the configuration of organ primordia in Drosophila is very different from that in vertebrates. The expression data strengthen the concept that D-ret is a homolog of vertebrate ret. One notable exception, which is addressed below, is the gut-associated neurons.
D-ret Expression in the Endoderm.
We investigated the curious inconsistency that the ret-expressing enteric neurons in vertebrates occur throughout the length of the gut, whereas in the fly, D-ret-expressing neurons are found only in the foregut (in SNS). In contrast to the vertebrate gut, which is entirely from the endoderm, only the midgut is of endodermal origin in insects (foregut and hindgut are of ectodermal origin; refs. 36 and 37).
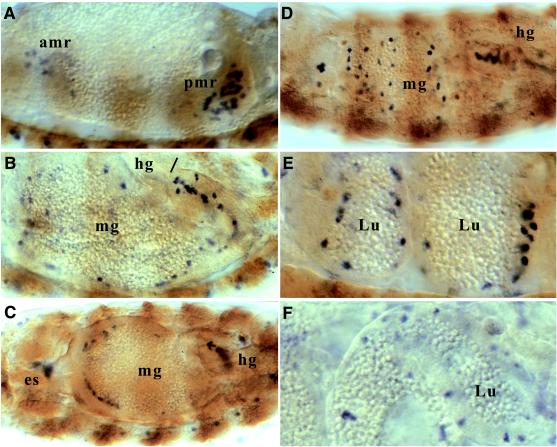
Upon close examination, we found D-ret signals appearing as scattered dots in the endodermal gut, as shown in Fig. 3 and 5. These signals were best visualized by an in situ hybridization protocol that includes a delipidation step (see Materials and Methods). D-ret expression in the endoderm begins at stage 12 in the anterior and posterior midgut rudiments (Fig. 5A). [Each of these two groups, holding about half of the midgut cells, migrates from either end of the body cavity toward the center (29).] There are only three cell types in the midgut rudiments (38): (i) cells that make up the juvenile (larval) midgut; (ii) precursor cells for the adult midgut, which stay dormant in the juvenile midgut as scattered “islands” until metamorphosis; and (iii) interstitial cell precursors (ICPs), which form a narrow band of large cells at the center of the juvenile midgut.
Figure 5.
D-ret expression in the endoderm. In situ hybridization was performed as in Figs. 3 and 4. D-ret mRNA is detected as blue signals, which are difficult to locate in the background of transparent gut. To provide reference points, a shade of brown was added by performing an irrelevant antibody staining (ftz: β-galactosidase). Lateral view, anterior left, except in C. (A) Stage 12. Loose clusters of D-ret-expressing cells appear within the anterior and posterior midgut rudiments. (B) Stage 15. D-ret-expressing cells migrate individually and line the luminal surface of the developing midgut. The junction between the midgut and hindgut is indicated with a bar. (C) Stage 15. Dorsal view. D-ret-expressing cells are spreading from the end of the endoderm toward the center. The posterior end of the midgut, which at this stage is swaying toward the animal's right, contains the highest density of D-ret-expressing cells. (D) Midgut at stage 16. This parasagittal optical section captures the most number of D-ret-expressing cells scattered on the luminal side of the midgut. (E) Stage 16. Close-up view of the stage shown in D. The D-ret-expressing cells are positioned near the luminal side, following the outline of the compartmentalized midgut tissue. (F) Stage 17. The D-ret-expressing cells integrate deeply into the midgut proper. amr, anterior midgut rudiment; pmr, posterior midgut rudiment; mg, midgut; hg, hindgut; es, esophagus; Lu, lumen of the midgut.
The possibility that the cells expressing D-ret might be ICPs can be ruled out, because ICPs occur only in the posterior half of the midgut rudiment (38). During stages 13–16, when the mesenchymal-looking juvenile midgut cells condense into epithelia, the D-ret-positive cells do not participate in epithelium formation. Instead, they migrate individually and spread evenly on the lumen side of the forming midgut (Fig. 5 B and C), identifying themselves as the adult midgut precursor cells (imaginal islands). The fact that they do not converge into a central band further confirms that they are not interstitial cells. At stage 16, when the midgut is compartmentalized, the D-ret signal is found as numerous dots that follow the outline of the juvenile midgut (Fig. 5D), still located near the lumen side (Fig. 5E). These cells then integrate deeply into the juvenile midgut tissue at the last stage of embryogenesis (Fig. 5F), consistent with the previous descriptions of adult midgut precursors (38, 39).
Our finding that the midgut imaginal islands express D-ret was not expected, because they do not give rise to neurons. The striking topographical similarity between the fly midgut imaginal islands and the vertebrate enteric neurons strongly suggests that, despite the disparate cell fates, these two types of ret-expressing cells are evolutionarily related.
Discussion
ret Expression in Endoderm Derivatives.
The structure most sensitive to the dosage level of ret during vertebrate embryogenesis is the enteric nervous system (ENS), a meshwork of neurons in the gut. ENS neurons occur throughout the alimentary tube, at the density of 10–50 embedded neurons per square millimeter of the gut wall (40). ENS neurons are derived from two neural crest populations that originate from an anterior (vagal) and posterior (sacral) location of the body (41). Each of the two groups subsequently migrates toward the opposite end to colonize the endoderm.
Because Drosophila has neither neural crest nor any identified neuron in the endoderm derivatives, we were interested in the expression pattern of D-ret. We have found that, in the embryo where the larval gut is being formed, D-ret is expressed in the midgut imaginal islands, which are the precursors for adult midgut. The island cells arise as two groups at either end of the endoderm, migrate toward the opposite end, and become evenly embedded in the endodermal gut at the end of embryogenesis. Therefore, in their topography, migratory paths, and D-ret expression, the island cells are highly analogous to the vertebrate ENS cells. This compelling parallel suggests evolutionary continuity between the fly imaginal islands and the vertebrate ENS.
In postembryonic development, the imaginal island cells are fated to give rise to the midgut. After a Drosophila embryo hatches, it lives for about 5 days as a larva, during which time the larval gut is functional, until the animal pupates. When the pupa undergoes metamorphosis, the larval gut histolyzes and the gut for the adult (imago) is reconstructed with hidden reserves of precursor cells. The imaginal islands that express D-ret in the embryo are the precursors for the adult midgut (38, 39). At the time of metamorphosis, the island cells form a gut epithelium and engulf the obsolete larval midgut, which is digested and discharged as meconium (42). There is the formal possibility that a small fraction of the island cells differentiate into midgut neurons. However, such have not been reported to date, and all of the island cells are thought to be the precursors for the adult midgut proper, rather than neurons. Our D-ret expression data, together with the known fate of the island cells, suggest that the ret-expressing vertebrate ENS cells are evolutionarily related to the precursor reserves of metamorphosing insects.
Given the possible evolutionary continuity, valuable insight into the underlying mechanistic commonality may be obtained through comparative approaches. In this regard, it is noteworthy that the imaginal islands differ in ploidy from the larval midgut cells that surround them. In mid-embryogenesis, the larval midgut cells become polyploid through endoreduplication and never divide again thereafter (43, 44). During the larval stages when the body size increases greatly, the polypoid midgut cells grow only in cell size (42). In contrast, the imaginal islands, kept as a reserve, remain diploid and poised to proliferate. Their proliferation potential is fully realized at the time of metamorphosis (36, 42). We propose that there could be shared components between the mechanism that unleashes the proliferation potential of precursor cells at fly metamorphosis and the mechanism operative in MEN2 cancer cells.
ret Expression in the Excretory System.
D-ret expression was also detected in the malpighian tubules, which are the excretory system in Drosophila, consisting of four relatively simple tubules (29, 36). It appears that ret expression reveals the relatedness between malpighian tubules and the vertebrate nephric duct at the pronephric and mesonephric stages. Pronephros is the vestigial kidney that transiently appears in vertebrate embryos (45); its ductal portion, the nephric duct, expresses ret (9, 34). As the pronephric tubules degenerate, the nephric duct becomes connected to mesonephros, the functional kidney in fish and amphibians. In mammals, the mesonephric tubules degenerate and the ureteric bud grows out from the nephric duct. Subsequently, the ret-expressing ureteric bud induces metanephros, the permanent kidney, through reciprocal interactions with its neighboring mesenchyme (45). In ret-deficient mice, the ureteric bud either fails to be extended into the mesenchyme or fails to be remodeled into fine collecting ducts (7, 46). Our result that the malpighian tubule anlage expresses ret only transiently is consistent with the idea that ret is involved in a specific aspect of tubulogenesis.
ret Expression in Extraembryonic Tissues.
The earliest expression of D-ret was detected in the yolk sac, which finds its counterparts in the yolk sac and trophoblast expression in vertebrates (9, 30). In a Drosophila egg, the yolk sac is a centrally located syncytium, which is formed when the peripherally located blastoderm nuclei become enclosed by infolding membranes. Because D-ret encodes a transmembrane protein, a question arises as to which cytoplasmic membrane would Ret be anchored. At the end of the blastoderm cleavage, the encroaching membrane furrow flattens at the basal side, forming a double sheet of bilayers (47, 48). The double sheet then merges with the same structure of the next furrow to generate two separate bilayers, one belonging to the blastomere and one enclosing the yolk. The latter constitutes the yolk sac membrane and has been identified with scanning electron microscopy (49). The mature Ret protein is expected to be present on the surface of the yolk sac membrane, facing the inner surface of the germband with its extracellular domain. The yolk sac expression peaks shortly before the onset of germband retraction. Because the entire germband glides over the yolk sac during germband retraction, it is possible that D-ret may be involved in germband retraction movement.
Acknowledgments
We thank Drs. T. Kornberg, H. Jäckle, Y. N. Jan, A. Mahowald, and Y. Emori for helpful discussions. M.H. is a Special Fellow of the Leukemia & Lymphoma Society. This work was supported by National Institutes of Health Grant CA 44338 and the G. W. Hooper Research Foundation.
Abbreviations
- MEN2
multiple endocrine neoplasia type 2
- GDNF
glial cell line-derived neurotrophic factor
- CNS
central nervous system
- ENS
enteric nervous system
- SNS
stomatogastric nervous system
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF322652 and AF322653).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.021558598.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.021558598
References
- 1.Edwards P M. Cancer Metastasis Rev. 1999;18:175–180. doi: 10.1023/a:1006304821464. [DOI] [PubMed] [Google Scholar]
- 2.Romeo G, Ronchetto P, Luo Y, Barone V, Seri M, Ceccherini I, Pasini B, Bocciardi R, Lerone M, Kaariainen H, Martucciello G. Nature (London) 1994;367:377–378. doi: 10.1038/367377a0. [DOI] [PubMed] [Google Scholar]
- 3.Ederly P, Lyonnet S, Mulligan L M, Pelet A, Dow E, Abel L, Holder S, Nihoul-Fekete C, Ponder B A J, Munnich A. Nature (London) 1994;367:378–380. doi: 10.1038/367378a0. [DOI] [PubMed] [Google Scholar]
- 4.Mulligan L M, Kwok J B J, Healey C S, Elsdon M J, Eng C, Gardner E, Love D R, Mole S E, Moore J K, Papi L, et al. Nature (London) 1993;363:458–460. doi: 10.1038/363458a0. [DOI] [PubMed] [Google Scholar]
- 5.Hofstra R M W, Landsvater R M, Ceccherini I, Stulp R P, Stelwagen T, Luo Y, Pasini B, Hoppener J W M, van Amstel H K P, Romeo G, et al. Nature (London) 1994;367:375–376. doi: 10.1038/367375a0. [DOI] [PubMed] [Google Scholar]
- 6.Santoro M, Carlomagno F, Romano A, Bottaro D P, Dathan N A, Grieco M, Fusco A, Vecchio G, Matoskova B, Kraus M H, Di Fiore P P. Science. 1995;267:381–383. doi: 10.1126/science.7824936. [DOI] [PubMed] [Google Scholar]
- 7.Schuchardt A, D'Agati V, Larsson-Blomberg L, Costantini F, Pachnis V. Nature (London) 1994;367:380–383. doi: 10.1038/367380a0. [DOI] [PubMed] [Google Scholar]
- 8.Bolk S, Pelet A, Hofstra R M W, Angrist M, Salomon R, Croaker D, Buys C H C M, Lyonnet S, Chakravarti A. Proc Natl Acad Sci USA. 2000;97:268–273. doi: 10.1073/pnas.97.1.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pachnis V, Mankoo B, Costantini F. Development (Cambridge, UK) 1993;119:1005–1017. doi: 10.1242/dev.119.4.1005. [DOI] [PubMed] [Google Scholar]
- 10.Durbec P L, Larsson-Blomberg L B, Schuchardt A, Costantini F, Pachnis V. Development (Cambridge, UK) 1996;122:349–358. doi: 10.1242/dev.122.1.349. [DOI] [PubMed] [Google Scholar]
- 11.Tsuzuki T, Takahashi M, Asiai N, Iwashita T, Matsuyama M, Asai J. Oncogene. 1995;10:191–198. [PubMed] [Google Scholar]
- 12.Trupp M, Arenas E, Fainzilber M, Nilsson A-S, Sieber B-A, Grigoriou M, Kilkenny C, Salazar-Grueso E, Pachnis V, Arumae U, et al. Nature (London) 1996;381:785–789. doi: 10.1038/381785a0. [DOI] [PubMed] [Google Scholar]
- 13.Durbec P, Marcos-Gutierrez C V, Kilkenny C, Grigoriou M, Wartiowaara K, Suvanto P, Smith D, Ponder B, Costantini F, Saarma M, et al. Nature (London) 1996;381:789–792. doi: 10.1038/381789a0. [DOI] [PubMed] [Google Scholar]
- 14.Jing S, Wen D, Yu Y, Hoist P L, Luo Y, Fang M, Tamir R, Antonio L, Hu Z, Cupples R, et al. Neuron. 1996;85:1113–1124. doi: 10.1016/s0092-8674(00)81311-2. [DOI] [PubMed] [Google Scholar]
- 15.Baloh R H, Tansey M G, Golden J P, Creedon D J, Heuckeroth R O, Keck C L, Zimonjic D B, Popescu N C, Johnson E M, Milbrandt J. Neuron. 1997;18:793–802. doi: 10.1016/s0896-6273(00)80318-9. [DOI] [PubMed] [Google Scholar]
- 16.Lin L-F H, Doherty D H, Like J D, Bektesh S, Collins F. Science. 1993;260:1130–1132. doi: 10.1126/science.8493557. [DOI] [PubMed] [Google Scholar]
- 17.Henderson C E, Phillips H S, Pollock R A, Davies A M, Lerneulle C, Armanini M, Simpson L C, Moffet B, Vandlen R A, Koliatsos V E, Rosenthal A. Science. 1994;266:1062–1064. doi: 10.1126/science.7973664. [DOI] [PubMed] [Google Scholar]
- 18.Sanchez M, Silos-Santiago I, Frisen J, He B, Lira S A, Barbacid M. Nature (London) 1996;382:70–73. doi: 10.1038/382070a0. [DOI] [PubMed] [Google Scholar]
- 19.Pichel J G, Shen L, Sheng H Z, Granholm A-C, Drago J, Grinberg A, Lee E J, Huang S P, Saarma M, Hoffer B J, et al. Nature (London) 1996;382:73–76. doi: 10.1038/382073a0. [DOI] [PubMed] [Google Scholar]
- 20.Moore M W, Klein R D, Farinas I, Sauer H, Armanini M, Phillips H, Reichardt L F, Ryan A M, Carver-Moore K, Rosenthal A. Nature (London) 1996;382:76–79. doi: 10.1038/382076a0. [DOI] [PubMed] [Google Scholar]
- 21.Hellmich H, Kos L, Cho E, Mahon K A, Zimmer A. Mech Dev. 1996;54:95–105. doi: 10.1016/0925-4773(95)00464-5. [DOI] [PubMed] [Google Scholar]
- 22.Sugaya R, Ishimaru S, Hosoya T, Saigo K, Emori Y. Mech Dev. 1994;45:139–145. doi: 10.1016/0925-4773(94)90027-2. [DOI] [PubMed] [Google Scholar]
- 23.O'Neill J W, Bier E. BioTechniques. 1994;17:874–875. [PubMed] [Google Scholar]
- 24.Hong C C, Hashimoto C. Cell. 1995;82:785–794. doi: 10.1016/0092-8674(95)90475-1. [DOI] [PubMed] [Google Scholar]
- 25.Schneider R. Trends Biochem Sci. 1992;17:468–469. doi: 10.1016/0968-0004(92)90490-z. [DOI] [PubMed] [Google Scholar]
- 26.Kuma K-I, Iwabe N, Miyata T. Mol Biol Evol. 1993;10:539–551. doi: 10.1093/oxfordjournals.molbev.a040033. [DOI] [PubMed] [Google Scholar]
- 27.Takeichi M. Science. 1991;251:1451–1455. doi: 10.1126/science.2006419. [DOI] [PubMed] [Google Scholar]
- 28.Eng C, Schuffenecker I, Lenoir G, Cote G, Gagel R, van Amstel H K, Lips C J, Nishisho I, Takai S-I, Marsh D J, et al. J Am Med Assoc. 1996;276:1575–1579. [PubMed] [Google Scholar]
- 29.Hartenstein V. Atlas of Drosophila Development. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. [Google Scholar]
- 30.Szentirmay Z, Ishizaka Y, Ohgaki H, Tahira T, Nagao M, Esumi H. Oncogene. 1990;5:701–705. [PubMed] [Google Scholar]
- 31.Hahn M, Jäckle H. EMBO J. 1996;15:3077–3084. [PMC free article] [PubMed] [Google Scholar]
- 32.Hartenstein V, Tepass U, Gruszynski-deFeo E. Dev Biol. 1996;173:213–227. doi: 10.1006/dbio.1996.0018. [DOI] [PubMed] [Google Scholar]
- 33.Penzlin H. In: Comprehensive Insect Physiology, Biochemistry, and Pharmacology. Kerkut G, A, Gilbert L I, editors. Oxford: Pergamon; 1985. pp. 371–406. [Google Scholar]
- 34.Marcos-Gutierrez C V, Wilson S W, Holder N, Pachnis V. Oncogene. 1997;14:879–889. doi: 10.1038/sj.onc.1201048. [DOI] [PubMed] [Google Scholar]
- 35.Schmidt-Ott U, González-Gaitán M, Technau G M. Roux's Arch Dev Biol. 1995;205:31–44. doi: 10.1007/BF00188841. [DOI] [PubMed] [Google Scholar]
- 36.Skaer H. In: The Development of Drosophila melanogaster. Bate M, Martinez Arias A, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 941–1012. [Google Scholar]
- 37.Reuter R, Leptin M. Development (Cambridge, UK) 1994;120:1137–1150. doi: 10.1242/dev.120.5.1137. [DOI] [PubMed] [Google Scholar]
- 38.Tepass U, Hartenstein V. Development (Cambridge, UK) 1995;121:399–405. doi: 10.1242/dev.121.2.393. [DOI] [PubMed] [Google Scholar]
- 39.Hartenstein V, Jan Y N. Roux's Arch Dev Biol. 1992;201:194–220. doi: 10.1007/BF00188752. [DOI] [PubMed] [Google Scholar]
- 40.Furness J B, Costa M. The Enteric Nervous System. New York: Churchill Livingston; 1987. pp. 1–24. [Google Scholar]
- 41.Le Douarin N, Teillet M-A. J Embryol Exp Morphol. 1973;30:31–48. [PubMed] [Google Scholar]
- 42.Bodenstein D. In: Biology of Drosophila. Demerec M, editor. London: Wiley; 1950. pp. 275–367. [Google Scholar]
- 43.Smith A V, Orr-Weaver T L. Development (Cambridge, UK) 1991;112:997–1008. doi: 10.1242/dev.112.4.997. [DOI] [PubMed] [Google Scholar]
- 44.Sonnenblick B P. In: Biology of Drosophila. Demerec M, editor. London: Wiley; 1950. pp. 62–167. [Google Scholar]
- 45.Saxén L. Organogenesis of the Kidney. Cambridge, U.K.: Cambridge Univ. Press; 1987. [Google Scholar]
- 46.Schuchardt A, D'Agati V, Pachnis V, Costantini F. Development (Cambridge, UK) 1996;122:1919–1929. doi: 10.1242/dev.122.6.1919. [DOI] [PubMed] [Google Scholar]
- 47.Mahowald A P. Exp Cell Res. 1963;32:457–468. doi: 10.1016/0014-4827(63)90186-1. [DOI] [PubMed] [Google Scholar]
- 48.Fullilove S, Jacobson A. Dev Biol. 1971;26:560–577. doi: 10.1016/0012-1606(71)90141-2. [DOI] [PubMed] [Google Scholar]
- 49.Turner F R, Mahowald A P. Dev Biol. 1977;57:403–416. doi: 10.1016/0012-1606(77)90225-1. [DOI] [PubMed] [Google Scholar]