Abstract
Degradation of host hemoglobin by the human malaria parasite Plasmodium falciparum is a massive metabolic process. What role this degradation plays and whether it is essential for parasite survival have not been established, nor have the roles of the various degradative enzymes been clearly defined. We report that P. falciparum can grow in medium containing a single amino acid (isoleucine, the only amino acid missing from human hemoglobin). In this medium, growth of hemoglobin-degrading enzyme gene knockout lines (missing falcipain-2 and plasmepsins alone or in combination) is impaired. Blockade of plasmepsins with the potent inhibitor pepstatin A has a minimal effect on WT parasite growth but kills falcipain-2 knockout parasites at low concentrations and is even more potent on falcipain-2, plasmepsin I and IV triple knockout parasites. We conclude that: (i) hemoglobin degradation is necessary for parasite survival; (ii) hemoglobin degradation is sufficient to supply most of the parasite’s amino acid requirements; (iii) external amino acid acquisition and hemoglobin digestion are partially redundant nutrient pathways; (iv) hemoglobin degradation uses dual protease families with overlapping function; and (v) hemoglobin-degrading plasmepsins are not promising drug targets.
Keywords: hemoglobin, malaria, protease
During its intraerythrocytic stage, Plasmodium falciparum ingests and degrades up to 75% of the host cell hemoglobin (1). This is a massive catabolic process that takes place inside an acidic food vacuole and involves a pathway of proteolytic enzymes (2). Despite extensive study, the purpose of hemoglobin degradation and the relative role of different proteases remain unclear. Several functions have been proposed for hemoglobin catabolism. Provision of nutrients for parasite energy and/or protein synthesis needs is an obvious potential role. Some radioactivity from labeled hemoglobin is incorporated into parasite protein (3). However, P. falciparum grows poorly in medium lacking any one of seven amino acids and is sluggish when grown in medium containing only the five amino acids on which it is most reliant (4, 5). These findings suggest a dependence on exogenously supplied amino acids. Also, some strains of P. falciparum digest considerably less hemoglobin, and parasites seem to excrete a substantial portion of the amino acids liberated from hemoglobin (6). Alternatively, it has been suggested that the parasite digests hemoglobin to make room for itself in the erythrocyte (6) or to maintain osmotic balance (7). The assumptions inherent in the osmotic hypothesis are controversial (8), and the association of volume with hemoglobin is only correlative. Thus, it remains unclear why the parasite degrades hemoglobin.
With regard to the enzymology of hemoglobin degradation, two families of proteases play prominent roles. Four aspartic proteases called plasmepsins and three cysteine proteases called falcipains are found in the food vacuole (9–12). All of them are capable of degrading hemoglobin or globin in biochemical assays using recombinant enzyme, although their order of action is unclear (9–14). Inhibitors of aspartic and cysteine proteases kill parasites in culture and animal models, although their specificity is not strict (13–15). Knockouts of plasmepsins individually or in combination (plasmepsin IV/I double knockout) give parasites with only slightly impaired growth (5, 16), and a falcipain-2 knockout shows normal growth (17). The effect of aspartic protease inhibitors is potentiated when combined with cysteine protease inhibitors or used in falcipain-2 knockout parasites (15, 17). Therefore the evidence suggests that there is overlap in and between the food vacuole proteolytic families; their relative roles remain unclear.
We have found that cultured P. falciparum parasites are able to grow well in medium containing isoleucine (the only amino acid that is lacking in human hemoglobin) as the sole exogenously supplied amino acid. We have used this medium to further explore the function of hemoglobin degradation and the roles of food vacuole plasmepsins and falcipains.
Results
Amino Acid Requirements for Parasite Growth.
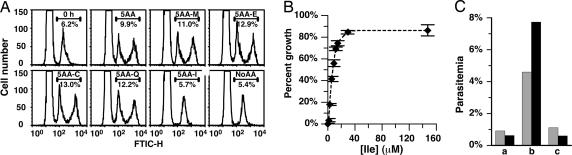
Divo et al. (4) discovered that P. falciparum can grow short term in a culture medium containing only five amino acids (cysteine, glutamate, glutamine, methionine, and isoleucine) (5AA). After 6 days in this medium, parasitemia is 5% of that in rich RPMI medium 1640 containing all 20 amino acids (5). This finding suggested that P. falciparum relies on exogenous amino acid supplementation for most of its growth requirement. The minimal growth that is achieved in 5AA is likely to depend on hemoglobin degradation, because most amino acid biosynthetic pathways are missing from the parasite (3, 18) and the five added amino acids are lacking or limited in hemoglobin. To examine the role of each of the five amino acids, we assessed parasite growth in 5AA lacking each one of the five amino acids separately. Trophozoite-stage parasites were incubated for 25 h and assessed for developmental progression by flow cytometry. Parasites incubated with full 5AA progressed to the schizont stage and many reinvaded fresh RBCs, as evidenced by a rise in parasitemia and emergence of a peak corresponding to ring-stage parasites (Fig. 1A). Parasites incubated in culture medium without any amino acids (no AA) were stalled in their development. When one amino acid at a time was omitted from 5AA, parasites developed well and actually grew slightly better than in full 5AA. In contrast, omission of isoleucine resulted in a stalled culture similar to that grown in complete absence of amino acids.
Fig. 1.
Amino acid requirements for cultured WT parasites. (A) Flow cytometry of the starting culture and cultures incubated in different media for 25 h. FITC-H channel histograms are shown. The truncated, low fluorescence peaks correspond to uninfected RBCs. RBCs infected with mature parasites (schizonts) have high fluorescence intensity and correspond to the far-right peaks. RBCs infected with younger (ring and trophozoite) parasites are closer to the uninfected RBC peak. A single amino acid was omitted from the 5AA as indicated. No AA was used as a control (Lower Right). Parasitemia gates (defined in Materials and Methods) are shown as horizontal bars, and parasitemias are listed underneath the bar. Except for the starting culture in Upper Left, media used are listed above the bar. (B) Isoleucine dose dependence. Parasites were incubated for 51 h in no AA supplemented with varying concentrations of isoleucine. The y axis is percent growth relative to that in full RPMI medium 1640. Error bars indicate the SD of triplicate measurements. (C) Effect of 4-thiaisoleucine. Parasites were cultured in no AA supplemented as follows: lane a, no addition; lane b, 15 μM isoleucine; and lane c, 15 μM isoleucine plus 100 μM 4-thiaisoleucine. Gray bars indicate 51-h incubation; black bars indicate 124-h incubation. Representative experiments that were repeated more than three times are shown.
We next determined whether isoleucine by itself is sufficient to support parasite growth. Remarkably, P. falciparum grown with isoleucine as the sole exogenous amino acid did better than in 5AA, and growth was nearly the same as in rich RPMI medium 1640 containing all 20 amino acids. Initial experiments were done by using 382 μM isoleucine, the same concentration found in RPMI medium 1640 (and in 5AA). A titration of isoleucine addition into no AA showed that parasitemia at 51 h saturated at 86% of that in full RPMI medium 1640, at isoleucine concentrations >20 μM (Fig. 1B). No stimulation of growth was seen upon going to a level even higher than that found in RPMI medium 1640 (1,475 μM; data not shown). We chose 147.5 μM as our standard concentration for no AA supplemented with isoleucine as the sole amino acid (I medium). We have been able to culture P. falciparum clone 3D7 continuously for >2 months in this medium. These data suggest that P. falciparum can use hemoglobin degradation as its main amino acid source. Because isoleucine is absent from human hemoglobin, the parasite is still auxotrophic for this amino acid. In further support of this notion, parasite growth was completely blocked by 100 μM 4-thiaisoleucine (Fig. 1C), and the blockade could be overcome at high exogenous isoleucine concentrations (data not shown).
Growth of Hemoglobin Degradation Enzyme Gene Knockout Lines in I Medium.
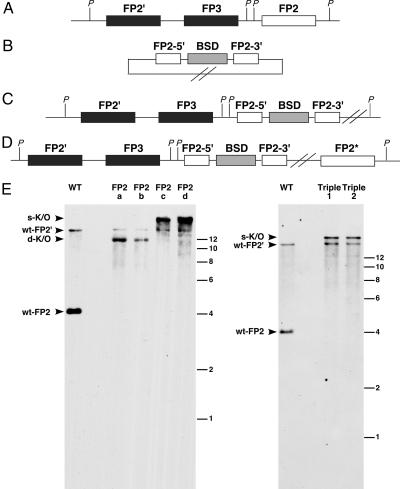
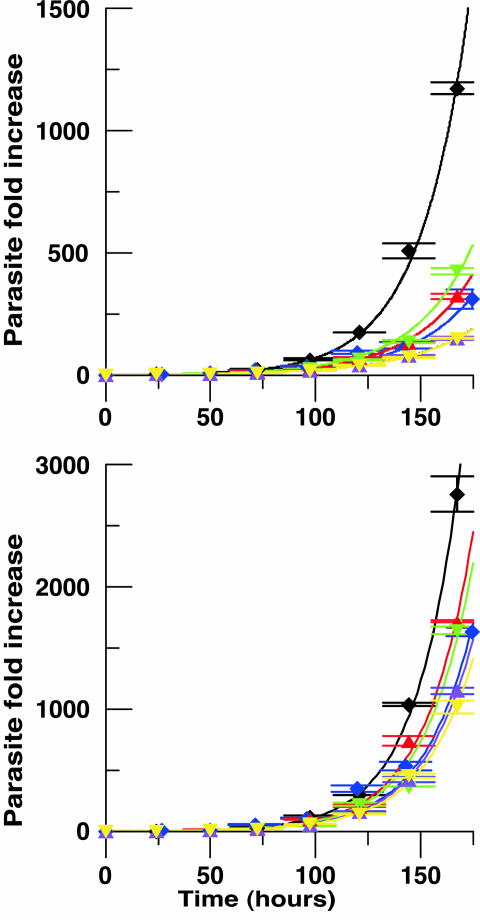
Previous studies have shown that individual knockouts of each of the four food vacuole plasmepsins and falcipain-2 have nearly normal growth phenotypes in RPMI medium 1640 (5, 16, 17). A plasmepsin IV/I double knockout has a doubling time ≈10% longer than the parental strain; this phenotype is accentuated slightly in 5AA (5). We constructed a triple knockout by disrupting falcipain-2 in the plasmepsin IV/I double knockout background. For comparison, we similarly disrupted falcipain-2 in a WT background. Southern blots (Fig. 2) revealed single-crossover recombination at the correct locus for both. Additionally, double-crossover recombination clones were found for the single falcipain-2 knockout. Two triple knockout clones [plasmepsins I/IV and falcipain-2 triple knockout clone 1 (Triple1) and plasmepsins I/IV and falcipain-2 triple knockout clone 2 (Triple2)] and one each of the falcipain-2 single and double crossover knockout clones (FP2b and FP2c, respectively) were chosen for further analysis. RT-PCR analysis (as in ref. 17) confirmed the falcipain-2 disruption (data not shown). Each of the falcipain-2 knockout lines (including the triple knockout) displayed a transient swollen food vacuole phenotype, as reported for the single falcipain-2 knockout (17). Knockout clones and their parental lines were grown in RPMI medium 1640 and I medium. In RPMI medium 1640, falcipain-2 knockout clones were only slightly slower-growing than the WT parental line, whereas the double plasmepsin knockout and the triple knockouts had 12–14% slower doubling times than the WT parental line (Fig. 3 and Table 1). In contrast, growth of all knockout lines was substantially impaired in I medium. Falcipain-2 knockouts had 20% longer doubling times than WT and there were 4-fold fewer parasites at the end of the 7-day growth period. The plasmepsin double knockout grew 28% slower than WT (5-fold fewer parasites). The triple knockout grew with 41% slower doubling time than WT (10-fold fewer parasites). Despite this, the culture maintained its slow exponential growth for at least 3 weeks. The data suggest two things. First, parasites grow better when they can degrade hemoglobin well, even in rich medium. Second, when parasites are grown in minimal medium, relying on hemoglobin as an amino acid source, falcipains and plasmepsins are both important to the process and have overlapping functions.
Fig. 2.
Creation of a chromosomal falcipain-2 disruption. (A) Schematic of the falcipain locus on chromosome 11. PacI (P) digestion results in fragments of 4.0 kb (falcipain-2, FP2) and 16.8 kb (falcipain-2′, FP2′). FP3, falcipain-3. (B) Schematic of the falcipain-2 knockout plasmid. FP2–5′ is a 590-bp fragment homologous to the 5′ coding region of the falcipain-2 gene. FP2–3′ is a 605-bp fragment homologous to the 3′ coding region of the falcipain-2 gene. The 7.9-kb plasmid has no PacI site. BSD, blasticidin S-deaminase. (C) Schematic of the falcipain-2 knockout resulting from double crossover. PacI digestion of the integrant with two concatenated plasmids yields a 13.2-kb fragment. (D) Schematic of the falcipain-2 knockout resulting from single crossover. This integration results in a promoter-less copy of falcipain-2, as indicated by FP2*. PacI digestion of the integrant with three concatenated plasmids results in a 26-kb fragment. (E) Southern blot of PacI digested total DNA from untransfected parasites (WT), falcipain-2 single knockout clones (FP2a, FP2b, FP2c, and FP2d), and plasmepsins I and IV, and falcipain-2 triple knockout clones (Triple 1 and Triple 2). Wt-FP2 indicates the fragment corresponding to the WT falcipain-2 locus, d-K/O indicates the falcipain-2 knockout resulting from a double crossover, s-K/O indicates the falcipain-2 knockout resulting from a single crossover. FP2–5′ was used for the Southern probe. Falcipain-2′, an almost identical copy of falcipain-2 that is expressed later in the life cycle (11), cross-hybridizes with this probe. DNA size ladders (in kb) are shown on the right of the blots.
Fig. 3.
Growth of knockout clones in RPMI medium 1640 (Lower) and I medium (Upper). Parasitemias of asynchronous cultures were measured by flow cytometry. Fold increases of parasites were calculated and graphed against time. Black diamonds indicate WT; blue diamonds indicate plasmepsin IV/I double knockout; red triangles indicate falcipain-2 single knockout clone b; green inverse triangles indicate falcipain-2 single knockout clone c; purple triangles indicate Triple 1; yellow inverse triangles indicate Triple 2. Error bars indicate the SD of triplicate measurements. Curves are simulated by an exponential growth model. The same colors for symbols are used for curves. Doubling times were calculated and are listed in Table 1.
Table 1.
Doubling times (T) calculated from the simulation curves in Fig. 3
| Parasite clone | I medium | RPMI medium 1640 | ||
|---|---|---|---|---|
| T, h | % of WT | T, h | % of WT | |
| WT | 16.4 | 100 | 14.6 | 100 |
| PM4/1 | 21.0 | 128 | 16.3 | 112 |
| FP2b | 20.2 | 123 | 15.6 | 107 |
| FP2c | 19.3 | 118 | 15.6 | 108 |
| Triple 1 | 23.2 | 141 | 16.5 | 113 |
| Triple 2 | 23.2 | 141 | 16.7 | 114 |
PM4/1, plasmepsin I/IV double knockout; FP2b, falcipain-2 single knockout clone b; FP2c, falcipain-2 single knockout clone c.
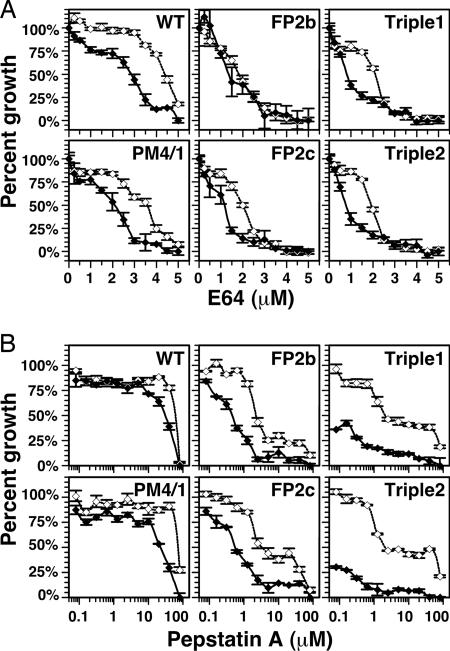
Sensitivity to Protease Inhibitors in I Medium.
To further assess the interrelationship of the two families of hemoglobin-degrading proteases, we treated knockout parasites with the cysteine protease inhibitor trans-epoxysuccinyl-l-leucylamino(4-guanidine)-butane (E64) (Fig. 4A) or the aspartic protease inhibitor pepstatin A (Fig. 4B). Growth of all parasite lines was slightly more sensitive to E64 in I medium than in RPMI medium 1640 (Fig. 4A, linear concentration scale, and Table 2). More dramatic differences were seen with parasites grown in pepstatin A (Fig. 4B, logarithmic concentration scale, and Table 2). In RPMI medium 1640, WT parasites were poorly sensitive to pepstatin A and plasmepsin double knockout parasites were even slightly less sensitive, as has been seen (5). Falcipain-2 knockout clones were ≈30-fold more sensitive, as has also been reported (17). Triple knockout parasites were comparable in sensitivity to the falcipain-2 single knockouts. All parasites were substantially more sensitive to pepstatin A when grown in I medium. WT, falcipain-2, and plasmepsin IV/I knockouts were 3- to 5-fold more sensitive in I medium, suggesting that parasites relying on hemoglobin degradation as their primary amino acid source need full plasmepsin activity for optimal growth. When important components of the plasmepsin and falcipain families were deleted (triple knockouts), growth in I medium was exquisitely sensitive to pepstatin A, with IC50 values in the nanomolar range, hundreds of fold below values determined in full RPMI medium 1640. The heightened sensitivity compared with the falcipain-2 single knockout may be explained by the fact that plasmepsins IV and I are the two food vacuole plasmepsins that are an order of magnitude less sensitive to pepstatin A than the others (9). When they are knocked out, the remaining plasmepsins are more easily inhibited. The data suggest that blocking all plasmepsins and falcipain-2 in the food vacuole forces a reliance on exogenous amino acids for survival. Growth is slowed in parasites with impaired hemoglobin degradation even in RPMI medium 1640 (see Fig. 4) but is halted in I medium. The flip side is that when the falcipain axis is fully operational (WT or plasmepsin double knockout) blockade of all plasmepsins has only a small effect on parasites, even when the organism is relying on hemoglobin degradation for amino acid supply (I medium).
Fig. 4.
Sensitivities to protease inhibitors. (A) E64 sensitivities. Parasites were incubated with E64 for 62 h. The percent growth of cultures compared with the untreated culture is graphed against E64 concentration in linear scale. Solid diamonds and solid lines indicate I medium; open diamonds and dotted lines indicate RPMI medium 1640. Error bars indicate the SD of triplicate measurements. PM4/1, plasmepsin I/IV double knockout; FP2b, falcipain-2 single knockout clone b; FP2c, falcipain-2 single knockout clone c. (B) Pepstatin A sensitivities. Parasites were incubated with pepstatin A for 63 h. The percent growth of cultures compared with the untreated culture is graphed against pepstatin A concentration in log10 scale. Symbols and lines are the same as in A. E64 and pepstatin A IC50 values are listed in Table 2.
Table 2.
Estimated IC50 values from inhibition curves in Fig. 4
| Parasite clone | Pepstatin A, IC50, μM | E64, IC50, μM | ||
|---|---|---|---|---|
| RPMI medium 1640 | I medium | RPMI medium 1640 | I medium | |
| WT | 60 | 31 | 4.4 | 2.8 |
| PM 4/1 | 70 | 22 | 3.6 | 2.1 |
| FP2b | 2.3 | 0.5 | 1.8 | 1.3 |
| FP2c | 3.3 | 0.6 | 2.0 | 1.2 |
| Triple 1 | 2.1 | <0.2 | 2.1 | 0.8 |
| Triple 2 | 2.1 | <0.1 | 1.9 | 0.8 |
PM4/1, plasmepsin I/IV double knockout; FP2b, falcipain-2 single knockout clone b; FP2c, falcipain-2 single knockout clone c.
Pepstatin A.
We noted a discrepancy between the potency of 5-year-old pepstatin A (from Streptomyces sp., Sigma) and fresh pepstatin A (from Streptomyces sp., Sigma) or synthetic (Calbiochem). The IC50 value for RPMI medium 1640-cultured 3D7 parasites with old pepstatin A (7 μM) was in line with published reports (5, 15, 17), whereas the IC50 of both new preparations was much higher (≈60 μM; Table 2). Inhibition curves of the various knockout clones were quite similar with old pepstatin A (Fig. 5, which is published as supporting information on the PNAS web site) or new, synthetic pepstatin A (Fig. 4B) but shifted an order of magnitude. We chose to present the above data by using new pepstatin A because it is a better-defined reagent (chemically synthesized peptide compared with partially purified microbial extract). Analysis of both preparations by liquid chromatography/MS (Fig. 6, which is published as supporting information on the PNAS web site) revealed the same dominant component, identified as pepstatin A by MS. There were a number of contaminants in the old pepstatin A preparation. A newer Sigma pepstatin A contained fewer contaminants (data not shown) and had potency similar to the synthetic version. Titration of pepstatin A protease inhibitory activity with plasmepsin II enzyme (Fig. 7, which is published as supporting information on the PNAS web site) revealed superimposable quantities of aspartic protease-inhibitory activity in the old and synthetic pepstatin A preparations. We conclude that there is likely to be a contaminant in the old pepstatin A with increased potency.
Discussion
We have found that P. falciparum can grow well in culture with isoleucine as the sole exogenous amino acid. Previous studies, done in rich RPMI medium 1640, suggested that the parasite excretes most of the amino acids from hemoglobin degradation and that therefore hemoglobin degradation is of minimal importance as an amino acid source for the organism (6). In contrast, when we force parasites to rely on hemoglobin degradation, they do so well (except that they need isoleucine supplementation because it is absent from human hemoglobin). The finding may well be physiologically relevant because many of the children in malaria endemic regions are malnourished and contain low, even undetectable, levels of plasma amino acids (19, 20). Indeed, RPMI medium 1640 has higher concentration for most amino acids (up to 20 times) than normal plasma (21, 22).
Our studies were done on P. falciparum strain 3D7, because that is the parental background for our knockouts. We have tested a number of isolates and found that in addition to isoleucine some strains need methionine supplementation for optimal growth (unpublished results). Methionine is present at low (1%) levels in human hemoglobin, and its endogenous supply may be borderline adequate. Seven-day growth of 3D7 is better in I medium than in 5AA (5), even when isoleucine concentration is identical. We do not understand the reason for this difference, although it appears unlikely to be a simple competition for uptake of isoleucine, because increasing the isoleucine in 5AA does not improve growth (data not shown). Perhaps there is a more complex cotransport or countertransport phenomenon that would be interesting to study.
Our knockout and inhibitor studies clearly show that parasites with impaired hemoglobin degradation machinery grow better in full RPMI medium 1640 than in I medium. Therefore, provision of exogenous amino acids can overcome slowed hemoglobin degradation. If hemoglobin degradation is drastically attenuated, however, as in falcipain-2 knockout parasites treated with pepstatin A, even full RPMI medium 1640 is not enough to promote growth. Some hemoglobin degradation may be necessary for survival independent of amino acid generation. For example, the two protease classes could have overlapping function in another process such as egress or osmotic regulation. Alternatively, there could be a nutrient that the parasite derives from hemoglobin that cannot be accessed from the medium. Plasmodium iron acquisition is poorly understood, and although much of the heme from hemoglobin is sequestered by the parasite as hemozoin, it is possible that the parasite relies on release of a small amount of iron for its metabolism (23). An amino acid could also conceivably be inaccessible from the external medium.
Amino acid acquisition is clearly an important process for the malaria parasite, which is reflected in multiple layers of redundancy. The ability to use exogenous (RPMI medium 1640) or endogenous (hemoglobin) amino acids as a primary source is one layer, as described above. The presence of multiple plasmepsins with overlapping specificity and function (5, 9) and multiple falcipains that overlap each other (24) is another layer. The final layer, into which we have gained more insight, is the redundancy between the falcipain and plasmepsin families. Knockout of falcipain-2 alone has a minimal effect on parasite growth in rich medium (ref. 17 and this work) and has a significant, but not lethal, effect in I medium (4-fold growth defect over 1 week). Knockout of individual plasmepsins gives similar results (5). Knockout of components from both families in combination was nearly additive, suggesting overlap of function between families. The results suggest that gene duplication of these enzymes may have been selected to provide increased protease dosage rather than as a sophisticated system with complementary cleavage specificities. It should be noted that in the current study, only selected gene knockouts were produced. It is possible that other plasmepsin and falcipain knockouts alone or in combination may be crucial for parasite growth. More study is required.
Pepstatin A treatment had a minimal effect on WT parasites except at very high concentrations but killed falcipain-2 knockout clones potently. The effects were even more dramatic in I medium, and in the triple knockout, low nanomolar concentrations killed parasites. The fact that pepstatin A kills parasites impaired in hemoglobin degradation, especially when they are forced to rely on hemoglobin as an amino acid source, strongly suggests that pepstatin A is able to get into the food vacuole and block aspartic protease function. It had long been suspected that pepstatin A got into cells poorly, accounting for its low potency against cultured parasites. Rather, our data suggest that pepstatin A is not effective on WT parasites because blockade of plasmepsins is not lethal. Dozens of laboratories have made hundreds of compounds targeted to plasmepsins (25). None is more potent than pepstatin A against isolated enzyme, and all show limited efficacy on cultured parasites. Again, bioavailability has been thought to be a limitation, but the current data suggest that bioefficacy may be the problem. Food vacuole plasmepsin inhibition does not appear to be a promising strategy for antimalarial drug development, unless combined with falcipain inhibition.
Materials and Methods
Parasite Culture.
P. falciparum strain 3D7 and derived knockout lines were cultured as described (5). Briefly, asexual-stage parasites were grown in human O+ erythrocytes at 2% hematocrit under 5% CO2, 5% O2, and 90% N2. Full RPMI medium 1640 is the standard RPMI medium 1640 (Invitrogen) containing all 20 amino acids and was supplemented with 27 mM NaHCO3, 22 mM glucose, 0.37 mM hypoxanthine, 10 μg/ml gentamicin, and 5 g/liter Albumax (Invitrogen). RPMI medium 1640 lacking all amino acids (no AA) was custom-made by the tissue culture center at the Washington University School of Medicine. Single amino acids (isoleucine, glutamine, glutamate, methionine, and cysteine; Sigma) were added to the concentrations found in RPMI medium 1640 to make 5AA. Isoleucine alone was added at 147.5 μM to make I medium. Supplements were added as for full RPMI medium 1640 above. I medium supplemented with dialyzed Albumax gave similar results as undialyzed Albumax.
Disruption of the Falcipain-2 Gene.
Two fragments of the falcipain-2 gene were amplified from genomic DNA by using the primers GGTTAAGCTTATGGATTACAACATGG and GGTTAAGCTTGTTATGCATATTTACTTTG for the 5′ fragment (590 bp, called FP2–5′) or CCTGAATTCGGATCTTGCTGGGCCTTTAG and GGTGAATTCTTAGGCAATTAATGGATG for the 3′ fragment (605 bp, called FP2–3′). FP2–5′ and FP2–3′ were sequenced and cloned into the knockout vector, which is a modified version of pHC1 (26). In the modified vector, the selectable marker BSD (27) is driven by the PcDT promoter element. For the single gene falcipain-2 knockout, parental 3D7 culture (WT) was transfected with plasmid as described (5) and grown in the presence of 3.5 μg/ml blasticidin for 2 weeks. The culture was then subjected to one drug cycle (21-day growth without drug, followed by reselection with blasticidin until parasites reappeared). For the triple knockout of plasmepsins I and IV, and falcipain-2, plasmepsin knockout clone PMIV/I A5 (5) was transfected with the falcipain-2 knockout vector and cycled twice. Parasites were cloned by the method of limiting dilution. Vector integration was analyzed by Southern blot as described (5).
Flow Cytometry.
P. falciparum cultures were stained with 0.5 μg/ml acridine orange (Molecular Probes) in PBS and used within 1 h. A total of 3 × 104 cells per sample were counted on a BD Biosciences Canto flow cytometer. The FITC channel detector records the green fluorescence of acridine orange when it binds to DNA (maximum emission at 525 nm), and the phycoerythrin (PE) channel detector records the red fluorescence of acridine orange when it binds to RNA (maximum emission at 650 nm). Both channels can distinguish parasite-infected RBC from uninfected RBC based on their DNA or RNA content differences. Forward size scattering (FSC) was used to count the total cell number. For all of the three channels, peak height signals (FITC-H, PE-H, and FSC-H) were used for analysis by flowjo software (Treestar, Ashland, OR). Parasitemia gates were defined by the population of cells having intense signals in FITC-H and PE-H channels that are distinct from the main body of cells (uninfected RBCs). Gating of parasite populations was verified by thin blood smears. For the experiment in Fig. 4, culture was fixed in 0.05% glutaraldehyde in PBS and stored at 4°C. The cells were permeabilized with 0.25% Triton X-100 in PBS for 5 min at room temperature and stained with 0.5 μg/ml acridine orange before use. Parasite growth rate was calculated as reported (5).
Inhibition Assay.
Asynchronous cultures at starting parasitemia of 1–1.5% were incubated with varying amounts of either E64 (Sigma) or synthetic pepstatin A (Calbiochem) or without inhibitor (control culture) for 62 or 63 h in the medium indicated. Parasitemias were counted by flow cytometry, and the increase in parasitemia (relative to control culture set to 100%) was graphed against inhibitor concentration. In another series of experiments, culture was incubated with or without 4-thiaisoleucine (Sigma) for the times indicated and analyzed as above.
Supplementary Material
Acknowledgments
This study was supported by National Institutes of Health Grant AI47798.
Abbreviations
- no AA
culture medium without any amino acids
- I medium
no AA supplemented with isoleucine as the sole amino acid
- 5AA
culture medium with only five amino acids (cysteine, glutamic acid, glutamine, isoleucine, and methionine)
- Triple1
plasmepsins I/IV and falcipain-2 triple knockout clone 1
- Triple2
plasmepsins I/IV and falcipain-2 triple knockout clone 2
- E64
trans-epoxysuccinyl-l-leucylamino(4-guanidine)-butane.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Loria P., Miller S., Foley M., Tilley L. Biochem. J. 1999;339:363–370. [PMC free article] [PubMed] [Google Scholar]
- 2.Goldberg D. E. In: Malaria: Drugs, Disease, and Post-Genomic Biology. Sullivan D., Krishna S., editors. Berlin: Springer; 2005. pp. 275–291. [Google Scholar]
- 3.Sherman I. W. Bull. W. H. O. 1977;55:265–276. [PMC free article] [PubMed] [Google Scholar]
- 4.Divo A. A., Geary T. G., Davis N. L., Jensen J. B. J. Protozool. 1985;32:59–64. doi: 10.1111/j.1550-7408.1985.tb03013.x. [DOI] [PubMed] [Google Scholar]
- 5.Liu J., Gluzman I. Y., Drew M. E., Goldberg D. E. J. Biol. Chem. 2005;280:1432–1437. doi: 10.1074/jbc.M409740200. [DOI] [PubMed] [Google Scholar]
- 6.Krugliak M., Zhang J., Ginsburg H. Mol. Biochem. Parasitol. 2002;119:249–256. doi: 10.1016/s0166-6851(01)00427-3. [DOI] [PubMed] [Google Scholar]
- 7.Lew V. L., Tiffert T., Ginsburg H. Blood. 2003;101:4189–4194. doi: 10.1182/blood-2002-08-2654. [DOI] [PubMed] [Google Scholar]
- 8.Allen R. J., Kirk K. Trends Parasitol. 2004;20:7–10. doi: 10.1016/j.pt.2003.10.015. [DOI] [PubMed] [Google Scholar]
- 9.Banerjee R., Liu J., Beatty W., Pelosof L., Klemba M., Goldberg D. E. Proc. Natl. Acad. Sci. USA. 2002;99:990–995. doi: 10.1073/pnas.022630099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shenai B. R., Sijwali P. S., Singh A., Rosenthal P. J. J. Biol. Chem. 2000;275:29000–29010. doi: 10.1074/jbc.M004459200. [DOI] [PubMed] [Google Scholar]
- 11.Singh N., Sijwali P. S., Pandey K. C., Rosenthal P. J. Exp. Parasitol. 2006;112:187–192. doi: 10.1016/j.exppara.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 12.Sijwali P. S., Shenai B. R., Gut J., Singh A., Rosenthal P. J. Biochem. J. 2001;360:481–489. doi: 10.1042/0264-6021:3600481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banerjee R., Goldberg D. E. In: Antimalarial Chemotherapy, Mechanism of Action, Resistance, and New Directions in Drug Discovery. Rosenthal P. J., editor. Totowa, NJ: Humana; 2001. pp. 43–63. [Google Scholar]
- 14.Rosenthal P. J. In: Antimalarial Chemotherapy, Mechanisms of Action, Resistance, and New Directions in Drug Discovery. Rosenthal P. J., editor. Totowa, NJ: Humana; 2001. pp. 325–345. [DOI] [PubMed] [Google Scholar]
- 15.Bailly E., Jambou R., Savel J., Jaureguiberry G. J. Protozool. 1992;39:593–599. doi: 10.1111/j.1550-7408.1992.tb04856.x. [DOI] [PubMed] [Google Scholar]
- 16.Omara-Opyene A. L., Moura P. A., Sulsona C. R., Bonilla J. A., Yowell C. A., Fujioka H., Fidock D. A., Dame J. B. J. Biol. Chem. 2004;279:54088–54096. doi: 10.1074/jbc.M409605200. [DOI] [PubMed] [Google Scholar]
- 17.Sijwali P. S., Rosenthal P. J. Proc. Natl. Acad. Sci. USA. 2004;101:4384–4389. doi: 10.1073/pnas.0307720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Payne S. H., Loomis W. F. Eukaryot. Cell. 2006;5:272–276. doi: 10.1128/EC.5.2.272-276.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whitehead R. G., Dean R. F. Am. J. Clin. Nutr. 1964;14:313–319. doi: 10.1093/ajcn/14.6.313. [DOI] [PubMed] [Google Scholar]
- 20.Baertl J. M., Placko R. P., Graham G. G. Am. J. Clin. Nutr. 1974;27:733–742. doi: 10.1093/ajcn/27.7.733. [DOI] [PubMed] [Google Scholar]
- 21.Aguilo A., Castano E., Tauler P., Guix M. P., Serra N., Pons A. J. Nutr. Biochem. 2000;11:81–86. doi: 10.1016/s0955-2863(99)00073-x. [DOI] [PubMed] [Google Scholar]
- 22.Jones D. P., Carlson J. L., Mody V. C., Cai J., Lynn M. J., Sternberg P. Free Radical Biol. Med. 2000;28:625–635. doi: 10.1016/s0891-5849(99)00275-0. [DOI] [PubMed] [Google Scholar]
- 23.Loyevsky M., Gordeuk V. R. In: Antimalarial Chemotherapy, Mechanism of Action, Resistance, and New Directions in Drug Discovery. Rosenthal P. J., editor. Totowa, NJ: Humana; 2001. pp. 307–324. [Google Scholar]
- 24.Dahl E. L., Rosenthal P. J. Mol. Biochem. Parasitol. 2005;139:205–212. doi: 10.1016/j.molbiopara.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 25.Boss C., Richard-Bildstein S., Weller T., Fischli W., Meyer S., Binkert C. Curr. Med. Chem. 2003;10:883–907. doi: 10.2174/0929867033457674. [DOI] [PubMed] [Google Scholar]
- 26.Crabb B. S., Triglia T., Waterkeyn J. G., Cowman A. F. Mol. Biochem. Parasitol. 1997;90:131–144. doi: 10.1016/s0166-6851(97)00143-6. [DOI] [PubMed] [Google Scholar]
- 27.Mamoun C. B., Gluzman I. Y., Goyard S., Beverley S. M., Goldberg D. E. Proc. Natl. Acad. Sci. USA. 1999;96:8716–8720. doi: 10.1073/pnas.96.15.8716. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.